Empowering Vision Transformer by Network Hyper-Parameter Selection for Whole Pelvis Prostate Planning Target Volume Auto-Segmentation
Abstract
:Simple Summary
Abstract
1. Introduction
- We propose one of the initial auto-segmentation models for the PTV target volume of the prostate, as the previous investigators have predominantly focused on studying organs-at-risk (OARs) and extended it to gross tumor volume/clinical target volume (GTV/CTV) to some extent.
- We demonstrated that the proposed model outperforms the latest state-of-the-art (SOTA) models in the PTV auto-segmentation, including nnU-net and recently proposed hybrid- and fully-vision transformers.
2. Materials and Methods
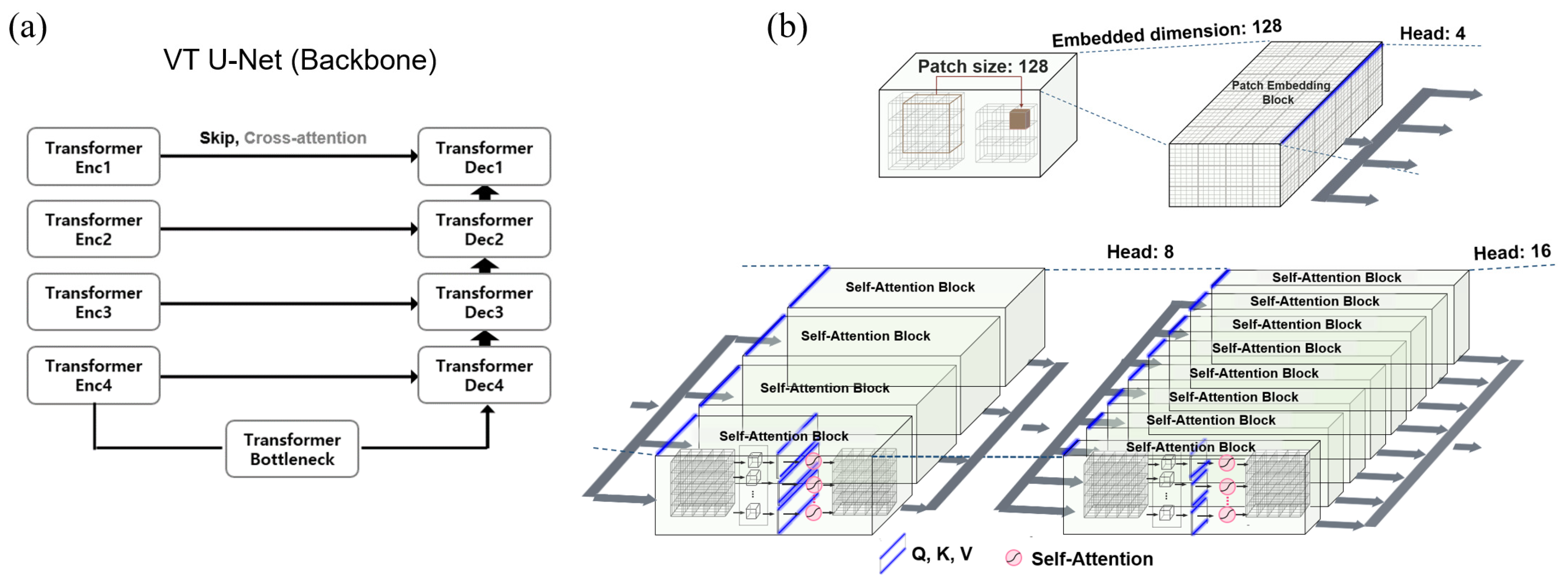
2.1. Transformer VT U-Net
2.2. Hyper-Parameter Selections for Network Architecture of VT U-Net
2.3. Patient Cohorts and Data Pre-Processing
2.4. Implementation and Evaluation
| Network | Learning Rate | Optimizer | Loss Function | Epoch |
|---|---|---|---|---|
| 3D U-Net [42] | 1 × 10−4 | Adam | Dice + BCE | 300 |
| nnU-Net [13,14] | 1 × 10−2 | SGD | Dice + CE + DS | 150 (×250) |
| Swin-Unetr [12] | 1 × 10−4 | AdamW | Dice + CE | 400–1000 |
| nnFormer [20] | 1 × 10−2 | SGD | Dice + CE + DS | 1300 (×250) |
| VT U-Net v.1 [21] | 1 × 10−4 | Adam | Dice + BCE | 400–500 |
| VT U-Net v.2 [22] | 1 × 10−4 | AdamW | Dice + CE | 1300 (×250) |
| Proposed | 1 × 10−4 | AdamW | Dice + CE + DS | 1300 (×250) |
| Network | Embedded Dimension | Patch Size | Number of Blocks | Window Size | Number of Heads | Parameters |
|---|---|---|---|---|---|---|
| Swin-Unetr [12] | 48 | 96 × 96 × 96 | [2, 2, 2, 2] | [7, 7, 7, 7] | [3, 6, 12, 24] | 62.8 M |
| nnFormer [20] | 96 | 128 × 128 × 128 | [2, 2, 2, 2] | [4, 4, 8, 4] | [3, 6, 12, 24] | 37.7 M |
| VT U-Net v.1 [21] | 96 | 128 × 128 × 128 | [2, 2, 2, 1] | [7, 7, 7, 7] | [3, 6, 12, 24] | 20.8 M |
| VT U-Net v.2 [22] | 96 | 128 × 128 × 128 | [2, 2, 2, 1] | [7, 7, 7, 7] | [3, 6, 12, 24] | 30.6 M |
| Proposed | 128 | 128 × 128 × 128 | [2, 2, 2, 1] | [7, 7, 7, 7] | [4, 8, 16, 32] | 36.7 M |
3. Results
3.1. Quantitative Analysis for PTV Auto-Segmentation
3.2. Hyper-Parameter Tuning
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kainmüller, D.; Lange, T.; Lamecker, H. Shape constrained automatic segmentation of the liver based on a heuristic intensity model. In Proceedings of the MICCAI Workshop 3D Segmentation in the Clinic: A Grand Challenge, Brisbane, Australia, 29 October 2007; p. 116. [Google Scholar]
- Park, H.; Bland, P.H.; Meyer, C.R. Construction of an abdominal probabilistic atlas and its application in segmentation. IEEE Trans. Med. Imaging 2003, 22, 483–492. [Google Scholar] [CrossRef]
- Shimizu, A.; Ohno, R.; Ikegami, T.; Kobatake, H.; Nawano, S.; Smutek, D. Segmentation of multiple organs in non-contrast 3D abdominal CT images. Int. J. Comput. Ass. Rad. 2007, 2, 135–142. [Google Scholar] [CrossRef]
- Zhou, Y.; Bai, J. Multiple abdominal organ segmentation: An atlas-based fuzzy connectedness approach. IEEE Trans. Inf. Technol. Biomed. 2007, 11, 348–352. [Google Scholar] [CrossRef]
- Hinton, G.E.; Osindero, S.; Teh, Y.W. A fast learning algorithm for deep belief nets. Neural Comput. 2006, 18, 1527–1554. [Google Scholar] [CrossRef]
- Shrestha, A.; Mahmood, A. Review of deep learning algorithms and architectures. IEEE Access 2019, 7, 53040–53065. [Google Scholar] [CrossRef]
- Aloysius, N.; Geetha, M. A review on deep convolutional neural networks. In Proceedings of the 2017 International Conference on Communication and Signal Processing (ICCSP), Chennai, India, 6–8 April 2017; pp. 0588–0592. [Google Scholar]
- Elizar, E.; Zulkifley, M.A.; Muharar, R.; Zaman, M.H.M.; Mustaza, S.M. A review on multiscale-deep-learning applications. Sensors 2022, 22, 7384. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Lect. Notes Comput. Sci. 2015, 9351, 234–241. [Google Scholar] [CrossRef]
- He, K.; Gan, C.; Li, Z.; Rekik, I.; Yin, Z.; Ji, W.; Gao, Y.; Wang, Q.; Zhang, J.; Shen, D. Transformers in medical image analysis: A review. arXiv 2022, arXiv:2202.12165. [Google Scholar]
- Shamshad, F.; Khan, S.; Zamir, S.W.; Khan, M.H.; Hayat, M.; Khan, F.S.; Fu, H. Transformers in medical imaging: A survey. Med. Image Anal. 2023, 88, 102802. [Google Scholar] [CrossRef]
- Hatamizadeh, A.; Nath, V.; Tang, Y.C.; Yang, D.; Roth, H.R.; Xu, D.G. Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images. In Proceedings of the 7th International Workshop, Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Brainles 2021, Part I, Virtual, 27 September 2022; Volume 12962, pp. 272–284. [Google Scholar] [CrossRef]
- Isensee, F.; Petersen, J.; Klein, A.; Zimmerer, D.; Jaeger, P.F.; Kohl, S.; Wasserthal, J.; Koehler, G.; Norajitra, T.; Wirkert, S. nnu-net: Self-adapting framework for u-net-based medical image segmentation. arXiv 2018, arXiv:1809.10486. [Google Scholar]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Wang, W.X.; Chen, C.; Ding, M.; Yu, H.; Zha, S.; Li, J.Y. TransBTS: Multimodal Brain Tumor Segmentation Using Transformer. In Proceedings of the 24th International Conference, Medical Image Computing and Computer Assisted Intervention—Miccai 2021, Part I, Strasbourg, France, 27 September–1 October 2021; Volume 12901, pp. 109–119. [Google Scholar] [CrossRef]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022; pp. 205–218. [Google Scholar]
- Schlemper, J.; Oktay, O.; Schaap, M.; Heinrich, M.; Kainz, B.; Glocker, B.; Rueckert, D. Attention gated networks: Learning to leverage salient regions in medical images. Med. Image Anal. 2019, 53, 197–207. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R.; Xu, D. Unetr: Transformers for 3D medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 4–8 January 2022; pp. 574–584. [Google Scholar]
- Zhou, H.-Y.; Guo, J.; Zhang, Y.; Yu, L.; Wang, L.; Yu, Y. nnformer: Interleaved transformer for volumetric segmentation. arXiv 2021, arXiv:2109.03201. [Google Scholar]
- Peiris, H.; Hayat, M.; Chen, Z.; Egan, G.; Harandi, M. A robust volumetric transformer for accurate 3D tumor segmentation. In Proceedings of the 25th International Conference on Medical Image Computing and Computer-Assisted Intervention, Sinagpore, 18–22 September 2022; pp. 162–172. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 1–11. [Google Scholar]
- Shaw, P.; Uszkoreit, J.; Vaswani, A. Self-attention with relative position representations. arXiv 2018, arXiv:1803.02155. [Google Scholar]
- Tang, H.; Liu, X.; Han, K.; Xie, X.; Chen, X.; Qian, H.; Liu, Y.; Sun, S.; Bai, N. Spatial context-aware self-attention model for multi-organ segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Virtual, 5–9 January 2021; pp. 939–949. [Google Scholar]
- Ramachandran, P.; Parmar, N.; Vaswani, A.; Bello, I.; Levskaya, A.; Shlens, J. Stand-alone self-attention in vision models. Adv. Neural Inf. Process. Syst. 2019, 32, 1–13. [Google Scholar]
- Laiton-Bonadiez, C.; Sanchez-Torres, G.; Branch-Bedoya, J. Deep 3D neural network for brain structures segmentation using self-attention modules in mri images. Sensors 2022, 22, 2559. [Google Scholar] [CrossRef]
- Liu, Z.; Hu, H.; Lin, Y.; Yao, Z.; Xie, Z.; Wei, Y.; Ning, J.; Cao, Y.; Zhang, Z.; Dong, L. Swin transformer v2: Scaling up capacity and resolution. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 19–24 June 2022; pp. 12009–12019. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Virtual, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Kayalibay, B.; Jensen, G.; van der Smagt, P. CNN-based segmentation of medical imaging data. arXiv 2017, arXiv:1701.03056. [Google Scholar]
- Lee, C.-Y.; Xie, S.; Gallagher, P.; Zhang, Z.; Tu, Z. Deeply-supervised nets. Proc. Artif. Intell. Stat. 2015, 38, 562–570. [Google Scholar]
- Li, S.; Sui, X.; Luo, X.; Xu, X.; Liu, Y.; Goh, R. Medical image segmentation using squeeze-and-expansion transformers. arXiv 2021, arXiv:2105.09511. [Google Scholar]
- Atya, H.B.; Rajchert, O.; Goshen, L.; Freiman, M. Non Parametric Data Augmentations Improve Deep-Learning based Brain Tumor Segmentation. In Proceedings of the 2021 IEEE International Conference on Microwaves, Antennas, Communications and Electronic Systems (COMCAS), Tel Aviv, Israel, 1–3 November 2021; pp. 357–360. [Google Scholar]
- Widesott, L.; Pierelli, A.; Fiorino, C.; Lomax, A.J.; Amichetti, M.; Cozzarini, C.; Soukup, M.; Schneider, R.; Hug, E.; Di Muzio, N. Helical tomotherapy vs. intensity-modulated proton therapy for whole pelvis irradiation in high-risk prostate cancer patients: Dosimetric, normal tissue complication probability, and generalized equivalent uniform dose analysis. Int. J. Radiat. Oncol. Biol. Phys. 2011, 80, 1589–1600. [Google Scholar] [CrossRef]
- Barlow, H.; Mao, S.; Khushi, M. Predicting high-risk prostate cancer using machine learning methods. Data 2019, 4, 129. [Google Scholar] [CrossRef]
- Wang, S.; He, K.; Nie, D.; Zhou, S.; Gao, Y.; Shen, D. CT male pelvic organ segmentation using fully convolutional networks with boundary sensitive representation. Med. Image Anal. 2019, 54, 168–178. [Google Scholar] [CrossRef]
- Miksys, N. Advancements in Monte Carlo Dose Calculations for Prostate and Breast Permanent Implant Brachytherapy. Ph.D. Thesis, Carleton University, Ottawa, ON, Canada, 2016. [Google Scholar]
- Liu, Y.; Khosravan, N.; Liu, Y.; Stember, J.; Shoag, J.; Bagci, U.; Jambawalikar, S. Cross-modality knowledge transfer for prostate segmentation from CT scans. In Proceedings of the Domain Adaptation and Representation Transfer and Medical Image Learning with Less Labels and Imperfect Data: First MICCAI Workshop, DART 2019, and First International Workshop, MIL3ID 2019, Shenzhen, Held in Conjunction with MICCAI 2019, Proceedings 1, Shenzhen, China, 13–17 October 2019; pp. 63–71. [Google Scholar]
- Elmahdy, M.S.; Jagt, T.; Zinkstok, R.T.; Qiao, Y.; Shahzad, R.; Sokooti, H.; Yousefi, S.; Incrocci, L.; Marijnen, C.; Hoogeman, M. Robust contour propagation using deep learning and image registration for online adaptive proton therapy of prostate cancer. Med. Phys. 2019, 46, 3329–3343. [Google Scholar] [CrossRef]
- Shahedi, M.; Ma, L.; Halicek, M.; Guo, R.; Zhang, G.; Schuster, D.M.; Nieh, P.; Master, V.; Fei, B. A semiautomatic algorithm for three-dimensional segmentation of the prostate on CT images using shape and local texture characteristics. In Proceedings of the Medical Imaging 2018: Image-Guided Procedures, Robotic Interventions, and Modeling, Houston, TX, USA, 10–15 February 2018; pp. 280–287. [Google Scholar]
- Shahedi, M.; Dormer, J.D.; Halicek, M.; Fei, B. The effect of image annotation with minimal manual interaction for semiautomatic prostate segmentation in CT images using fully convolutional neural networks. Med. Phys. 2022, 49, 1153–1160. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016: 19th International Conference, Proceedings Part II 19, Athens, Greece, 17–21 October 2016; pp. 424–432. [Google Scholar]
- Ma, J.; Chen, J.; Ng, M.; Huang, R.; Li, Y.; Li, C.; Yang, X.; Martel, A.L. Loss odyssey in medical image segmentation. Med. Image Anal. 2021, 71, 102035. [Google Scholar] [CrossRef]





| nnU-Net | VT U-Net v.1 | VT U-Net v.2 | Modified VT U-Net v.2 (Proposed) | |
|---|---|---|---|---|
| Pre- and post-processing and loss | ||||
| Oversampling 1 | o | × | o | o |
| Post-processing 2 | o | × | o | o |
| Deep supervision | o | × | × | o |
| Hyper-parameter & architecture | ||||
| Batch size | o | × | o | o |
| Patch size | o | × | × | o |
| Embedded dimension | — | × | × | o |
| Head number | — | × | × | △ (adaptive to embedded dimension) |
| Window size | — | × | × | × |
| Architecture | o | × | △ | o (same as nnU-Net) |
| Method | Dice Similarity Coefficient (DSC) ↑ | 95% Hausdorff Distance (HD95) ↓ | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fold1 | Fold2 | Fold3 | Fold4 | Mean | Fold1 | Fold2 | Fold3 | Fold4 | Mean | |
| 3D U-Net | 80.96 | 77.98 | 76.46 | 76.75 | 78.0 *** | 3.23 | 4.19 | 5.55 | 4.45 | 4.4 *** |
| nnU-Net | 83.95 | 82.02 | 79.25 | 82.44 | 81.9 * | 2.70 | 3.60 | 5.14 | 2.94 | 3.6 |
| Swin-Unetr | 82.68 | 81.87 | 79.47 | 83.06 | 81.8 * | 3.00 | 3.65 | 4.97 | 2.75 | 3.6 |
| nnFormer | 83.31 | 81.31 | 79.85 | 82.39 | 81.7 ** | 2.80 | 3.78 | 5.13 | 3.12 | 3.7 * |
| VT U-Net v.1 | 80.21 | 76.65 | 75.08 | 76.25 | 77.0 *** | 3.35 | 4.34 | 5.44 | 3.93 | 4.3 *** |
| VT U-Net v.2 | 84.12 | 82.30 | 79.82 | 82.61 | 82.2 ** | 2.72 | 3.60 | 5.04 | 3.01 | 3.6 |
| Proposed | 84.20 | 82.65 | 80.13 | 82.82 | 82.5 | 2.49 | 3.52 | 4.98 | 3.01 | 3.5 |
| Method | DSC | HD95 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fold1 | Fold2 | Fold3 | Fold4 | All folds | Fold1 | Fold2 | Fold3 | Fold4 | All Folds | |
| 3D U-Net | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.001 | <0.001 | 0.004 | <0.001 | <0.001 |
| nnU-Net | 0.410 | 0.126 | 0.255 | 0.206 | 0.040 | 0.070 | 0.473 | 0.122 | 0.479 | 0.080 |
| Swin-Unetr | 0.022 | 0.020 | 0.030 | 0.339 | 0.020 | 0.007 | 0.123 | 0.091 | 0.392 | 0.210 |
| nnFormer | 0.046 | 0.013 | 0.389 | 0.122 | 0.010 | 0.028 | 0.116 | 0.468 | 0.181 | 0.020 |
| VT U-Net v.1 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.050 | <0.001 | <0.001 |
| VT U-Net v.2 | 0.331 | 0.051 | 0.117 | 0.106 | 0.010 | 0.426 | 0.448 | 0.170 | 0.480 | 0.090 |
| Hyper-Parameter | DSC ↑ | HD95 ↓ | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Patch Size | Embedded Dimension | Fold1 | Fold2 | Fold3 | Fold4 | Fold1 | Fold2 | Fold3 | Fold4 |
| 96 | 96 | 83.57 | 82.09 | 79.57 | 82.33 | 2.72 | 3.78 | 5.02 | 3.12 |
| 96 | 128 | 83.78 | 82.34 | 79.35 | 82.25 | 2.52 | 3.60 | 4.98 | 3.12 |
| 96 | 192 | 84.36 | 82.05 | 79.52 | 81.98 | 2.68 | 4.03 | 5.02 | 3.22 |
| 128 | 96 | 83.72 | 82.32 | 79.77 | 82.39 | 2.73 | 3.70 | 5.05 | 3.07 |
| 128 | 128 | 84.20 | 82.65 | 80.13 | 82.82 | 2.49 | 3.52 | 4.98 | 3.01 |
| 128 | 192 | 83.96 | 81.89 | 80.45 | 82.23 | 2.63 | 3.78 | 4.94 | 3.44 |
| 160 | 96 | 84.16 | 81.87 | 79.37 | 81.87 | 2.78 | 3.59 | 5.07 | 3.26 |
| 160 | 128 | 83.65 | 82.14 | 79.65 | 82.11 | 2.64 | 3.80 | 5.02 | 3.19 |
| 160 | 192 | 84.03 | 81.54 | 79.85 | 81.88 | 2.61 | 3.78 | 4.96 | 3.25 |
| Hyper-Parameter | DSC | HD95 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patch Size | Embedded Dimension | Fold1 | Fold2 | Fold3 | Fold4 | All Folds | Fold1 | Fold2 | Fold3 | Fold4 | All Folds |
| 96 | 96 | 0.111 | 0.018 | 0.061 | 0.043 | 0.001 | 0.021 | 0.157 | 0.149 | 0.190 | 0.006 |
| 96 | 128 | 0.185 | 0.144 | 0.043 | 0.069 | 0.005 | 0.087 | 0.108 | 0.475 | 0.405 | 0.079 |
| 96 | 192 | 0.365 | 0.087 | 0.072 | 0.005 | 0.009 | 0.179 | 0.199 | 0.380 | 0.081 | 0.026 |
| 128 | 96 | 0.101 | 0.124 | 0.021 | 0.046 | 0.004 | 0.026 | 0.236 | 0.222 | 0.166 | 0.019 |
| 128 | 192 | 0.193 | 0.118 | 0.089 | 0.020 | 0.059 | 0.089 | 0.145 | 0.301 | 0.021 | 0.018 |
| 160 | 96 | 0.451 | 0.154 | 0.008 | 0.001 | <0.001 | 0.058 | 0.176 | 0.076 | 0.005 | <0.001 |
| 160 | 128 | 0.137 | 0.066 | 0.127 | 0.007 | 0.002 | 0.228 | 0.080 | 0.389 | 0.007 | 0.006 |
| 160 | 192 | 0.347 | 0.023 | 0.173 | <0.001 | 0.001 | 0.294 | 0.036 | 0.172 | 0.003 | 0.020 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, H.; Lee, J.S.; Kim, J.S.; Koom, W.S.; Kim, H. Empowering Vision Transformer by Network Hyper-Parameter Selection for Whole Pelvis Prostate Planning Target Volume Auto-Segmentation. Cancers 2023, 15, 5507. https://doi.org/10.3390/cancers15235507
Cho H, Lee JS, Kim JS, Koom WS, Kim H. Empowering Vision Transformer by Network Hyper-Parameter Selection for Whole Pelvis Prostate Planning Target Volume Auto-Segmentation. Cancers. 2023; 15(23):5507. https://doi.org/10.3390/cancers15235507
Chicago/Turabian StyleCho, Hyeonjeong, Jae Sung Lee, Jin Sung Kim, Woong Sub Koom, and Hojin Kim. 2023. "Empowering Vision Transformer by Network Hyper-Parameter Selection for Whole Pelvis Prostate Planning Target Volume Auto-Segmentation" Cancers 15, no. 23: 5507. https://doi.org/10.3390/cancers15235507
APA StyleCho, H., Lee, J. S., Kim, J. S., Koom, W. S., & Kim, H. (2023). Empowering Vision Transformer by Network Hyper-Parameter Selection for Whole Pelvis Prostate Planning Target Volume Auto-Segmentation. Cancers, 15(23), 5507. https://doi.org/10.3390/cancers15235507







