PARP1 Characterization as a Potential Biomarker for BCR::ABL1 p190+ Acute Lymphoblastic Leukemia
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Chemical Treatments
2.3. Alamar Blue Cytotoxicity Assay
Alamar Blue Statistical Analysis
2.4. Flow Cytometry Analysis
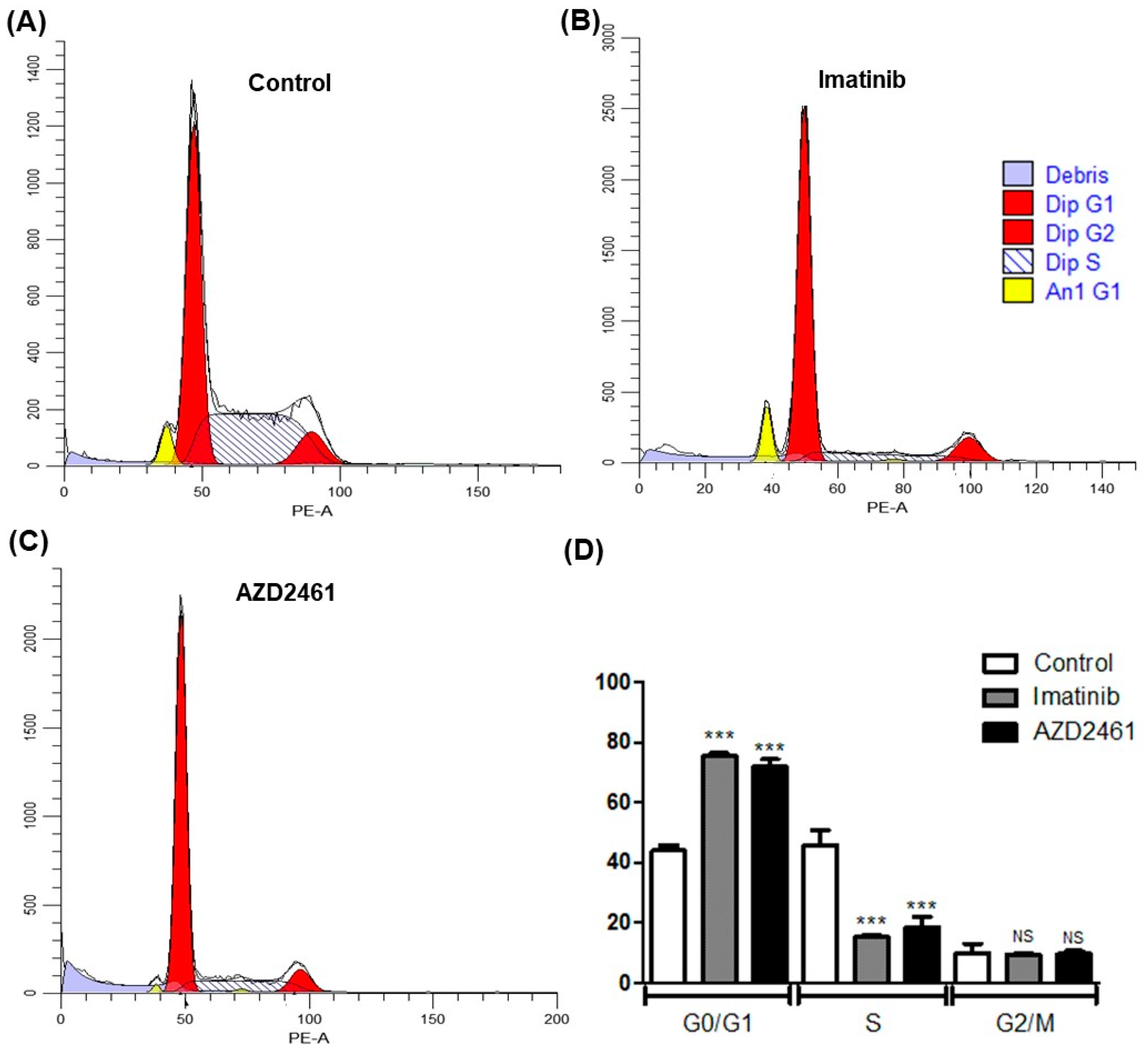
2.4.1. Cell Cycle Arrest
2.4.2. Induction and Quantification of Early Apoptosis
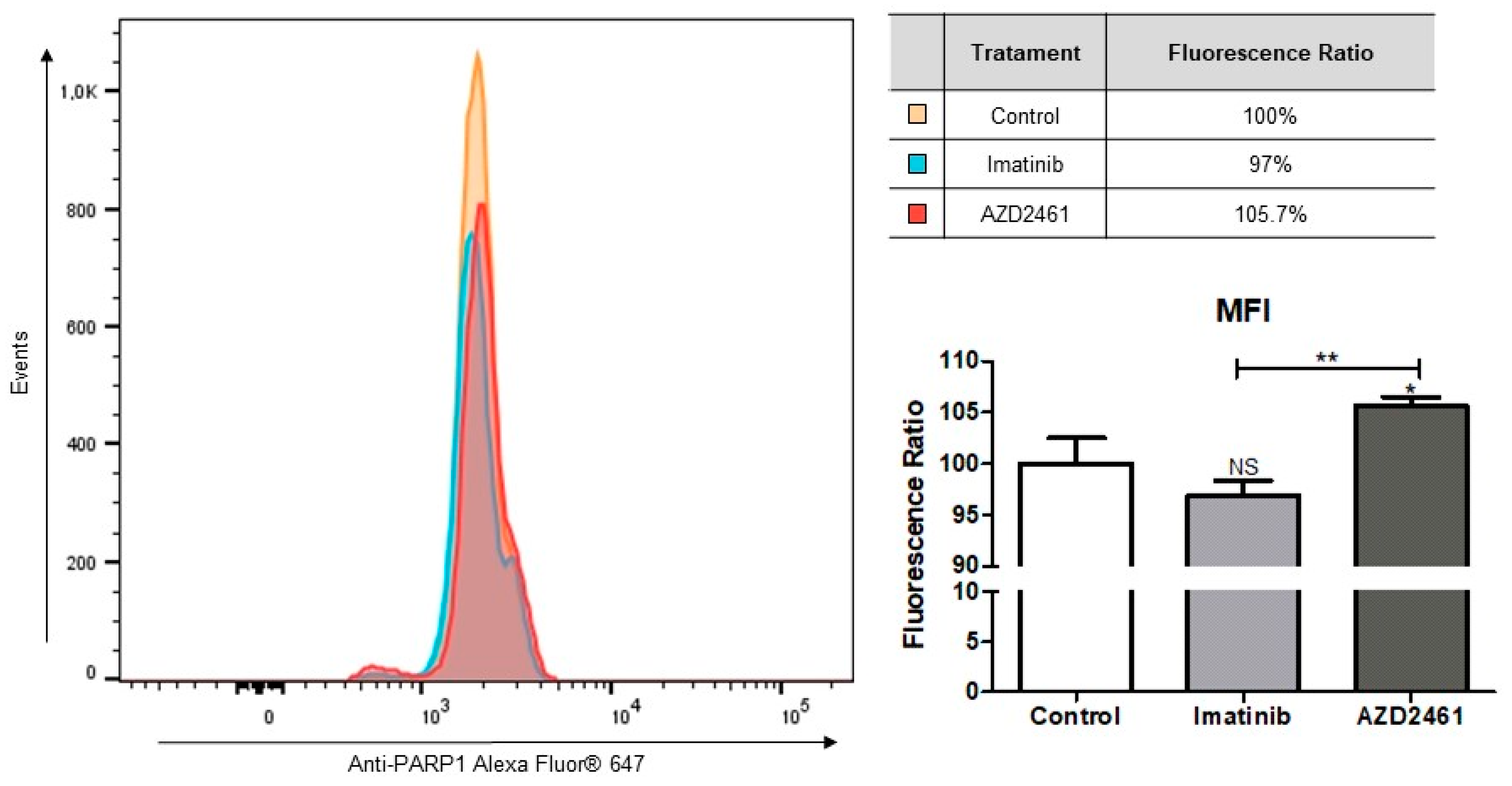
2.4.3. Alterations in PARP1 Intracellular Levels
2.4.4. Flow Cytometry Statistical Analysis
2.5. Expression Analysis
Gene Expression Statistical Analysis
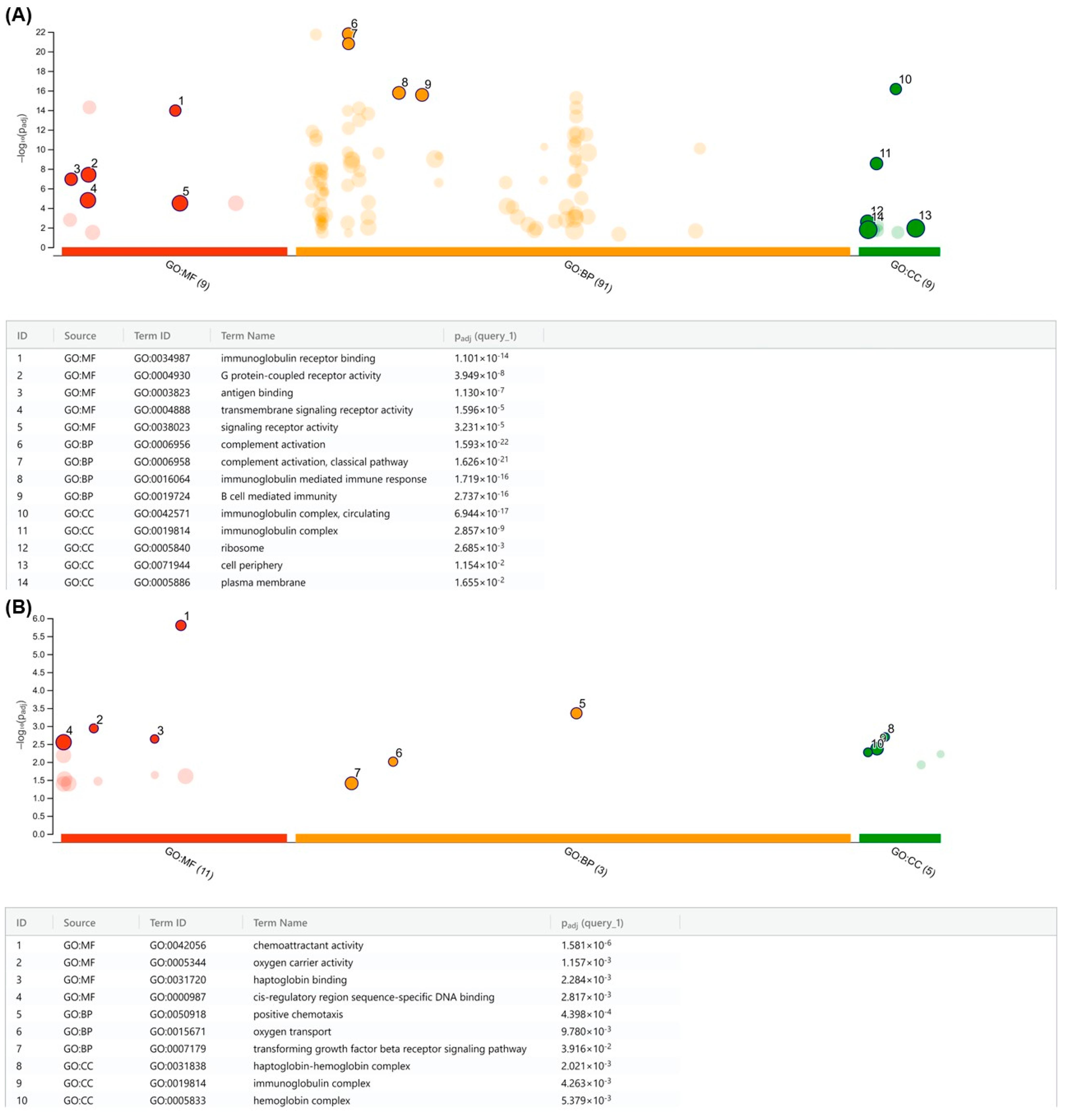
2.6. Chromosomal Analysis and Gene Ontology (GO)
2.6.1. DNA Extraction
2.6.2. Array-Based Comparative Genomic Hybridization (aCGH) Analysis
2.6.3. Biological Interpretation and Statistical Analysis
2.7. Patient Samples
Statistical Analysis of Patient Samples
2.8. Data Analysis Based on the Gene Expression Omnibus (GEO) Database
Statistical Analysis for GEO Expression
2.9. Single-Cell Sequencing Analysis
3. Results
3.1. Significantly Different PARP1 Expression Levels Are Detected among Lymphoblastic Cell Lines
3.2. PARP Inhibitor Is highly Cytotoxic to the BCR::ABL1 p190+ Cell Line
3.3. AZD2461 Promotes Similar Cell Cycle Arrest Profile to That of Imatinib in SUP-B15
3.4. AZD2461 Induces Early Apoptotic Markings in the SUP-B15 Cell Line after Sub-Inhibitory Treatment
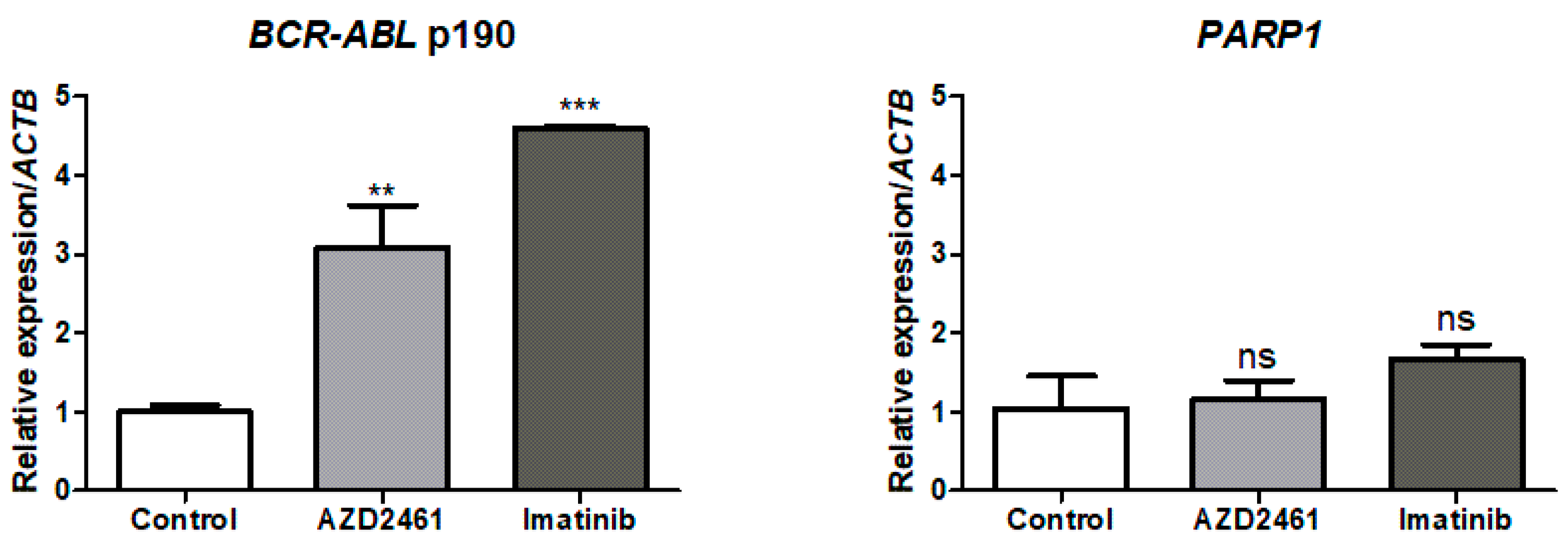
3.5. BCR::ABL1 p190 Expression Is Modulated by Treatment with AZD2461 or Imatinib
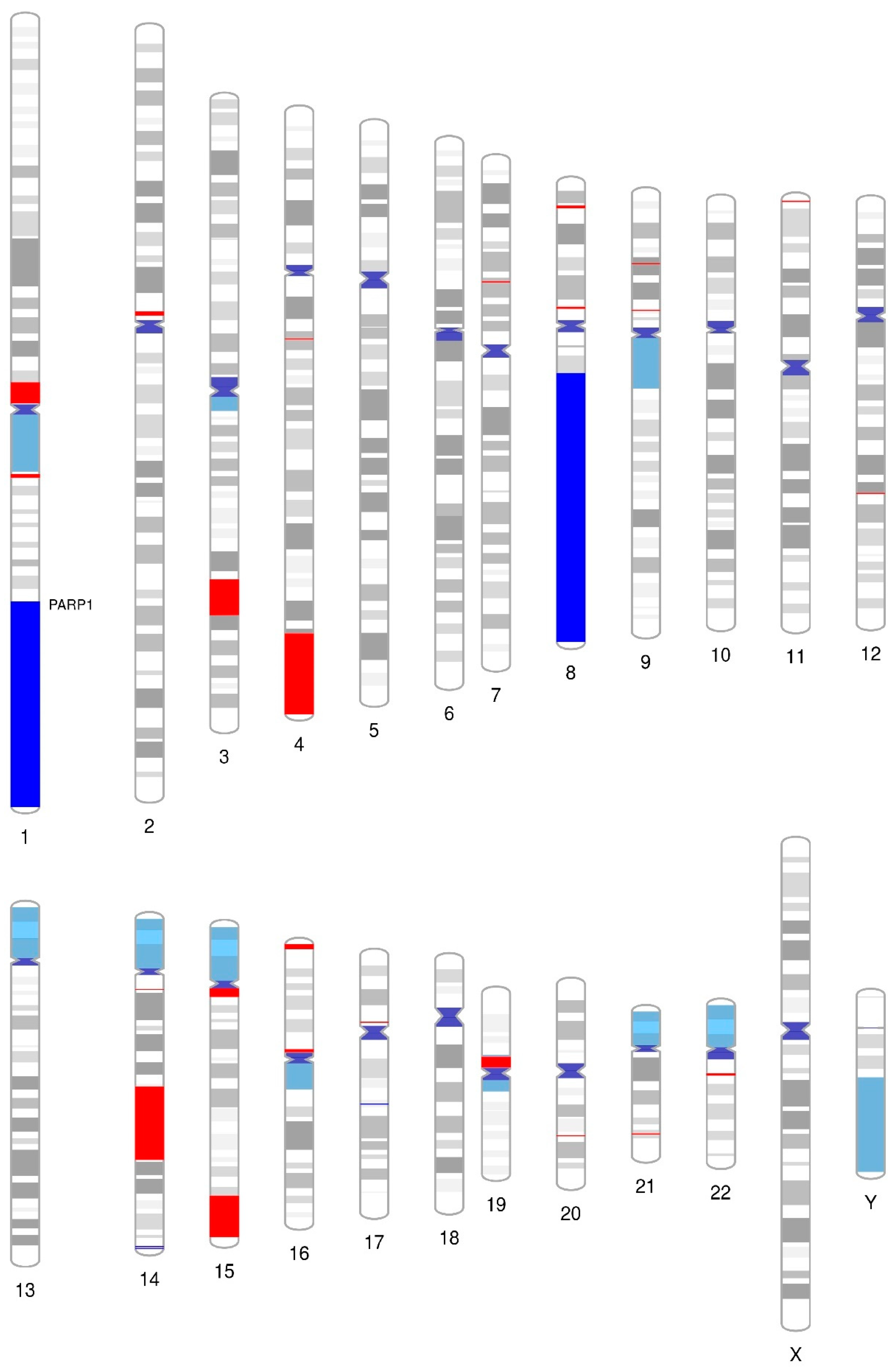
3.6. Array Comparative Genomic Hybridization (aCGH) Revealed That PARP1 Is Amplified in the SUP-B15 Cell Line
3.7. Differential PARP1 Expression Profile among p190+ ALL and CML Patients and Dataset Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luca, D.C. Update on Lymphoblastic Leukemia/Lymphoma. Clin. Lab. Med. 2021, 41, 405–416. [Google Scholar] [CrossRef] [PubMed]
- El Fakih, R.; Jabbour, E.; Ravandi, F.; Hassanein, M.; Anjum, F.; Ahmed, S.; Kantarjian, H. Current Paradigms in the Management of Philadelphia Chromosome Positive Acute Lymphoblastic Leukemia in Adults. Am. J. Hematol. 2018, 93, 286–295. [Google Scholar] [CrossRef]
- Hantschel, O. Structure, Regulation, Signaling, and Targeting of Abl Kinases in Cancer. Genes Cancer 2012, 3, 436. [Google Scholar] [CrossRef] [PubMed]
- Waller, C. Imatinib Mesylate. In Recent Results in Cancer Research; Springer: Cham, Switzerland, 2018; Volume 212, pp. 1–27. [Google Scholar] [CrossRef]
- Adnan-Awad, S.; Kim, D.; Hohtari, H.; Javarappa, K.K.; Brandstoetter, T.; Mayer, I.; Potdar, S.; Heckman, C.A.; Kytölä, S.; Porkka, K.; et al. Characterization of P190-Bcr-Abl Chronic Myeloid Leukemia Reveals Specific Signaling Pathways and Therapeutic Targets. Leukemia 2021, 35, 1964. [Google Scholar] [CrossRef]
- Slupianek, A.; Poplawski, T.; Jozwiakowski, S.K.; Cramer, K.; Pytel, D.; Stoczynska, E.; Nowicki, M.O.; Blasiak, J.; Skorski, T. BCR/ABL Stimulates WRN to Promote Survival and Genomic Instability. Cancer Res. 2011, 71, 842. [Google Scholar] [CrossRef] [PubMed]
- Abdulmawjood, B.; Costa, B.; Roma-rodrigues, C.; Baptista, P.V.; Fernandes, A.R. Genetic Biomarkers in Chronic Myeloid Leukemia: What Have We Learned So Far? Int. J. Mol. Sci. 2021, 22, 12516. [Google Scholar] [CrossRef]
- Nieborowska-Skorska, M.; Kopinski, P.K.; Ray, R.; Hoser, G.; Ngaba, D.; Flis, S.; Cramer, K.; Reddy, M.M.; Koptyra, M.; Penserga, T.; et al. Rac2-MRC-CIII–Generated ROS Cause Genomic Instability in Chronic Myeloid Leukemia Stem Cells and Primitive Progenitors. Blood 2012, 119, 4253. [Google Scholar] [CrossRef]
- Lord, C.J.; Ashworth, A. PARP Inhibitors: Synthetic Lethality in the Clinic. Science 2017, 355, 1152–1158. [Google Scholar] [CrossRef]
- Pascal, J.M. The Comings and Goings of PARP-1 in Response to DNA Damage. DNA Repair 2018, 71, 177. [Google Scholar] [CrossRef]
- Leung, A.K.L. Poly(ADP-Ribose): An Organizer of Cellular Architecture. J. Cell Biol. 2014, 205, 613–619. [Google Scholar] [CrossRef]
- O’Connor, L.O.; Rulten, S.L.; Cranston, A.N.; Odedra, R.; Brown, H.; Jaspers, J.E.; Jones, L.; Knights, C.; Evers, B.; Ting, A.; et al. The PARP Inhibitor AZD2461 Provides Insights into the Role of PARP3 Inhibition for Both Synthetic Lethality and Tolerability with Chemotherapy in Preclinical Models. Cancer Res. 2016, 76, 6084–6094. [Google Scholar] [CrossRef]
- Skelding, K.A.; Lincz, L.F. PARP Inhibitors and Haematological Malignancies—Friend or Foe? Cancers 2021, 13, 5328. [Google Scholar] [CrossRef] [PubMed]
- Fritz, C.; Portwood, S.M.; Przespolewski, A.; Wang, E.S. PARP Goes the Weasel! Emerging Role of PARP Inhibitors in Acute Leukemias. Blood Rev. 2021, 45, 100696. [Google Scholar] [CrossRef]
- Rampersad, S.N. Multiple Applications of Alamar Blue as an Indicator of Metabolic Function and Cellular Health in Cell Viability Bioassays. Sensors 2012, 12, 12347–12360. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Liu, D.; Xu, X.; Wen, J.; Xie, L.; Zhang, J.; Shen, Y.; Jiang, G.; Chen, J.; Fan, M. Integrated Genome-Wide Analysis of Gene Expression and DNA Copy Number Variations Highlights Stem Cell-Related Pathways in Small Cell Esophageal Carcinoma. Stem Cells Int. 2018, 2018, 3481783. [Google Scholar] [CrossRef]
- Wolfe, D.; Dudek, S.; Ritchie, M.D.; Pendergrass, S.A. Visualizing Genomic Information across Chromosomes with PhenoGram. BioData Min. 2013, 6, 18. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Vilo, J.; Peterson, H. Gprofiler2—An R Package for Gene List Functional Enrichment Analysis and Namespace Conversion Toolset g:Profiler. F1000Research 2020, 9, ELIXIR-709. [Google Scholar] [CrossRef]
- Ferreira, W.A.S.; Vitiello, G.A.F.; da Silva Medina, T.; de Oliveira, E.H.C. Comprehensive Analysis of Epigenetics Regulation, Prognostic and the Correlation with Immune Infiltrates of GPX7 in Adult Gliomas. Sci. Rep. 2022, 12, 6442. [Google Scholar] [CrossRef]
- Kohlmann, A.; Kipps, T.J.; Rassenti, L.Z.; Downing, J.R.; Shurtleff, S.A.; Mills, K.I.; Gilkes, A.F.; Hofmann, W.K.; Basso, G.; Dell’Orto, M.C.; et al. An International Standardization Programme towards the Application of Gene Expression Profiling in Routine Leukaemia Diagnostics: The Microarray Innovations in LEukemia Study Prephase. Br. J. Haematol. 2008, 142, 802–807. [Google Scholar] [CrossRef] [PubMed]
- Haferlach, T.; Kohlmann, A.; Wieczorek, L.; Basso, G.; Te Kronnie, G.; Béné, M.C.; De Vos, J.; Hernández, J.M.; Hofmann, W.K.; Mills, K.I.; et al. Clinical Utility of Microarray-Based Gene Expression Profiling in the Diagnosis and Subclassification of Leukemia: Report from the International Microarray Innovations in Leukemia Study Group. J. Clin. Oncol. 2010, 28, 2529–2537. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, Y.; Dong, X.; Sun, D.; Liu, Z.; Yue, J.; Wang, H.; Li, T.; Wang, C. TISCH2: Expanded Datasets and New Tools for Single-Cell Transcriptome Analyses of the Tumor Microenvironment. Nucleic Acids Res. 2023, 51, D1425–D1431. [Google Scholar] [CrossRef] [PubMed]
- Caron, M.; St-Onge, P.; Sontag, T.; Wang, Y.C.; Richer, C.; Ragoussis, I.; Sinnett, D.; Bourque, G. Single-Cell Analysis of Childhood Leukemia Reveals a Link between Developmental States and Ribosomal Protein Expression as a Source of Intra-Individual Heterogeneity. Sci. Rep. 2020, 10, 8079. [Google Scholar] [CrossRef] [PubMed]
- Rabilloud, T.; Potier, D.; Pankaew, S.; Nozais, M.; Loosveld, M.; Payet-Bornet, D. Single-Cell Profiling Identifies Pre-Existing CD19-Negative Subclones in a B-ALL Patient with CD19-Negative Relapse after CAR-T Therapy. Nat. Commun. 2021, 12, 865. [Google Scholar] [CrossRef] [PubMed]
- Bailur, J.K.; McCachren, S.S.; Pendleton, K.; Vasquez, J.C.; Lim, H.S.; Duffy, A.; Doxie, D.B.; Kaushal, A.; Foster, C.; DeRyckere, D.; et al. Risk-Associated Alterations in Marrow T Cells in Pediatric Leukemia. JCI Insight 2020, 5, 16. [Google Scholar] [CrossRef]
- Yu, X.; Xie, L.; Ge, J.; Li, H.; Zhong, S.; Liu, X. Integrating Single-Cell RNA-Seq and Spatial Transcriptomics Reveals MDK-NCL Dependent Immunosuppressive Environment in Endometrial Carcinoma. Front. Immunol. 2023, 14, 1145300. [Google Scholar] [CrossRef]
- Ferreira, W.A.S.; de Oliveira, E.H.C. Expression of GOT2 Is Epigenetically Regulated by DNA Methylation and Correlates with Immune Infiltrates in Clear-Cell Renal Cell Carcinoma. Curr. Issues Mol. Biol. 2022, 44, 2472–2489. [Google Scholar] [CrossRef]
- Dalle, I.A.; Jabbour, E.; Short, N.J.; Ravandi, F. Treatment of Philadelphia Chromosome-Positive Acute Lymphoblastic Leukemia. Curr. Treat. Options Oncol. 2019, 20, 1–13. [Google Scholar] [CrossRef]
- Bukowski, K.; Kciuk, M.; Kontek, R. Mechanisms of Multidrug Resistance in Cancer Chemotherapy. Int. J. Mol. Sci. 2020, 21, 3233. [Google Scholar] [CrossRef]
- Nicoletto, R.E.; Ofner, C.M. Cytotoxic Mechanisms of Doxorubicin at Clinically Relevant Concentrations in Breast Cancer Cells. Cancer Chemother. Pharmacol. 2022, 89, 285–311. [Google Scholar] [CrossRef] [PubMed]
- Xing, C.; Xu, W.; Shi, Y.; Zhou, B.; Wu, D.; Liang, B.; Zhou, Y.; Gao, S.; Feng, J. CD9 Knockdown Suppresses Cell Proliferation, Adhesion, Migration and Invasion, While Promoting Apoptosis and the Efficacy of Chemotherapeutic Drugs and Imatinib in Ph+ ALL SUP-B15 Cells. Mol. Med. Rep. 2020, 22, 2791. [Google Scholar] [CrossRef] [PubMed]
- Stepanenko, A.A.; Dmitrenko, V.V. Pitfalls of the MTT Assay: Direct and off-Target Effects of Inhibitors Can Result in over/Underestimation of Cell Viability. Gene 2015, 574, 193–203. [Google Scholar] [CrossRef]
- Goel, S.; DeCristo, M.J.; McAllister, S.S.; Zhao, J.J. CDK4/6 Inhibition in Cancer: Beyond Cell Cycle Arrest. Trends Cell Biol. 2018, 28, 911. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.H.; Mun, J.G.; Jeon, H.D.; Kee, J.Y. Betulin Inhibits Lung Metastasis by Inducing Cell Cycle Arrest, Autophagy, and Apoptosis of Metastatic Colorectal Cancer Cells. Nutrients 2020, 12, 66. [Google Scholar] [CrossRef]
- Pai, J.T.; Hsu, M.W.; Leu, Y.L.; Chang, K.T.; Weng, M.S. Induction of G2/M Cell Cycle Arrest via P38/P21Waf1/Cip1-Dependent Signaling Pathway Activation by Bavachinin in Non-Small-Cell Lung Cancer Cells. Molecules 2021, 26, 5161. [Google Scholar] [CrossRef]
- Al-Aamri, H.M.; Ku, H.; Irving, H.R.; Tucci, J.; Meehan-Andrews, T.; Bradley, C. Time Dependent Response of Daunorubicin on Cytotoxicity, Cell Cycle and DNA Repair in Acute Lymphoblastic Leukaemia. BMC Cancer 2019, 19, 179. [Google Scholar] [CrossRef]
- Nguyen, A.; Yoshida, M.; Goodarzi, H.; Tavazoie, S.F. Highly Variable Cancer Subpopulations That Exhibit Enhanced Transcriptome Variability and Metastatic Fitness. Nat. Commun. 2016, 7, 11246. [Google Scholar] [CrossRef]
- Van Engeland, M.; Nieland, L.J.W.; Ramaekers, F.C.S.; Schutte, B.; Reutelingsperger, C.P.M. Annexin V-Affinity Assay: A Review on an Apoptosis Detection System Based on Phosphatidylserine Exposure. Cytom. J. Int. Soc. Anal. Cytol. 1998, 31, 1–9. [Google Scholar] [CrossRef]
- Kumar, R.; Saneja, A.; Panda, A.K. An Annexin V-FITC-Propidium Iodide-Based Method for Detecting Apoptosis in a Non-Small Cell Lung Cancer Cell Line. Methods Mol. Biol. 2021, 2279, 213–223. [Google Scholar] [CrossRef]
- Zimmermann, M.; Meyer, N. Annexin V/7-AAD Staining in Keratinocytes. Methods Mol. Biol. 2011, 740, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Zembruski, N.C.L.; Stache, V.; Haefeli, W.E.; Weiss, J. 7-Aminoactinomycin D for Apoptosis Staining in Flow Cytometry. Anal. Biochem. 2012, 429, 79–81. [Google Scholar] [CrossRef] [PubMed]
- Karimiani, E.G.; Marriage, F.; Merritt, A.J.; Burthem, J.; Byers, R.J.; Day, P.J.R. Single-Cell Analysis of K562 Cells: An Imatinib-Resistant Subpopulation Is Adherent and Has Upregulated Expression of BCR-ABL MRNA and Protein. Exp. Hematol. 2014, 42, 183–191.e5. [Google Scholar] [CrossRef] [PubMed]
- Kc, R.; Thapa, B.; Ubeda, A.; Jiang, X.; Uludaǧ, H. BCR-Abl Silencing by SiRNA: A Potent Approach to Sensitize Chronic Myeloid Leukemia Cells to Tyrosine Kinase Inhibitor Therapy. Stem Cells Dev. 2019, 28, 734–744. [Google Scholar] [CrossRef]
- Wang, F.; Gouttia, O.G.; Wang, L.; Peng, A. PARP1 Upregulation in Recurrent Oral Cancer and Treatment Resistance. Front. Cell Dev. Biol. 2022, 9, 3737. [Google Scholar] [CrossRef]
- Ma, X.; Dang, C.; Min, W.; Diao, Y.; Hui, W.; Wang, X.; Dai, Z.; Wang, X.; Kang, H. Downregulation of APE1 Potentiates Breast Cancer Cells to Olaparib by Inhibiting PARP-1 Expression. Breast Cancer Res. Treat. 2019, 176, 109–117. [Google Scholar] [CrossRef]
- Gu, Z.; Churchman, M.L.; Roberts, K.G.; Moore, I.; Zhou, X.; Nakitandwe, J.; Hagiwara, K.; Pelletier, S.; Gingras, S.; Berns, H.; et al. PAX5-Driven Subtypes of B-Progenitor Acute Lymphoblastic Leukemia. Nat. Genet. 2019, 51, 296–307. [Google Scholar] [CrossRef]
- Conceição Barbosa, T.; Terra-Granado, E.; Quezado Magalhães, I.M.; Neves, G.R.; Gadelha, A.; Guedes Filho, G.E.; Souza, M.S.; Melaragno, R.; Emerenciano, M.; Pombo-de-Oliveira, M.S. Frequency of Copy Number Abnormalities in Common Genes Associated with B-Cell Precursor Acute Lymphoblastic Leukemia Cytogenetic Subtypes in Brazilian Children. Cancer Genet. 2015, 208, 492–501. [Google Scholar] [CrossRef]
- Pour Feizi, A.H.; Zeinali, S.; Toporski, J.; Sheervalilou, R.; Mehranfar, S. Frequency and Correlation of Common Genes Copy Number Alterations in Childhood Acute Lymphoblastic Leukemia with Prognosis. Asian Pac. J. Cancer Prev. 2020, 21, 3493–3500. [Google Scholar] [CrossRef]
- Ribera, J.; Zamora, L.; García, O.; Hernández-Rivas, J.M.; Genescà, E.; Ribera, J.M. [Copy Number Alterations in Adult Patients with Mature B Acute Lymphoblastic Leukemia Treated with Specific Immunochemotherapy]. Med. Clin. 2016, 147, 488–491. [Google Scholar] [CrossRef]
- Galindo-Campos, M.A.; Bedora-Faure, M.; Farrés, J.; Lescale, C.; Moreno-Lama, L.; Martínez, C.; Martín-Caballero, J.; Ampurdanés, C.; Aparicio, P.; Dantzer, F.; et al. Coordinated Signals from the DNA Repair Enzymes PARP-1 and PARP-2 Promotes B-Cell Development and Function. Cell Death Differ. 2019, 26, 2667–2681. [Google Scholar] [CrossRef] [PubMed]
- Slade, D. Mitotic Functions of Poly(ADP-Ribose) Polymerases. Biochem. Pharmacol. 2019, 167, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, Z.Y.; Wu, N.; Chen, Y.C.; Cheng, Q.; Wang, J. PARP Inhibitor Resistance: The Underlying Mechanisms and Clinical Implications. Mol. Cancer 2020, 19, 107. [Google Scholar] [CrossRef]
- Schmitz, R.; Young, R.M.; Ceribelli, M.; Jhavar, S.; Xiao, W.; Zhang, M.; Wright, G.; Shaffer, A.L.; Hodson, D.J.; Buras, E.; et al. Burkitt Lymphoma Pathogenesis and Therapeutic Targets from Structural and Functional Genomics. Nature 2012, 490, 116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Y.; Zhang, Y.J.; Zhao, H.Y.; Zhai, Q.L.; Shen, Y.F. The Impact of R213 Mutation on P53-Mediated P21 Activity. Biochimie 2014, 99, 215–218. [Google Scholar] [CrossRef]
- Engeland, K. Cell Cycle Regulation: P53-P21-RB Signaling. Cell Death Differ. 2022, 29, 946–960. [Google Scholar] [CrossRef]
- Balasundaram, A.; Doss, C.G.P. Unraveling the Structural Changes in the DNA-Binding Region of Tumor Protein P53 (TP53) upon Hotspot Mutation P53 Arg248 by Comparative Computational Approach. Int. J. Mol. Sci. 2022, 23, 15499. [Google Scholar] [CrossRef]
- Ireno, I.C.; Wiehe, R.S.; Stahl, A.I.; Hampp, S.; Aydin, S.; Troester, M.A.; Selivanova, G.; Wiesmüller, L. Modulation of the Poly (ADP-Ribose) Polymerase Inhibitor Response and DNA Recombination in Breast Cancer Cells by Drugs Affecting Endogenous Wild-Type P53. Carcinogenesis 2014, 35, 2273–2282. [Google Scholar] [CrossRef]
- Xiao, G.; Lundine, D.; Annor, G.K.; Canar, J.; Ellison, V.; Polotskaia, A.; Donabedian, P.L.; Reiner, T.; Khramtsova, G.F.; Olopade, O.I.; et al. Gain-of-Function Mutant P53 R273H Interacts with Replicating DNA and PARP1 in Breast Cancer. Cancer Res. 2020, 80, 394. [Google Scholar] [CrossRef]
- Guan, H.; Miao, H.; Ma, N.; Lu, W.; Luo, B. Correlations between Epstein-Barr Virus and Acute Leukemia. J. Med. Virol. 2017, 89, 1453–1460. [Google Scholar] [CrossRef]
- Shannon-Lowe, C.; Rickinson, A.B.; Bell, A.I. Epstein–Barr Virus-Associated Lymphomas. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160271. [Google Scholar] [CrossRef] [PubMed]
- Morgan, S.M.; Tanizawa, H.; Caruso, L.B.; Hulse, M.; Kossenkov, A.; Madzo, J.; Keith, K.; Tan, Y.; Boyle, S.; Lieberman, P.M.; et al. The Three-Dimensional Structure of Epstein-Barr Virus Genome Varies by Latency Type and Is Regulated by PARP1 Enzymatic Activity. Nat. Commun. 2022, 13, 187. [Google Scholar] [CrossRef]
- Lupey-Green, L.N.; Caruso, L.B.; Madzo, J.; Martin, K.A.; Tan, Y.; Hulse, M.; Tempera, I. PARP1 Stabilizes CTCF Binding and Chromatin Structure to Maintain Epstein-Barr Virus Latency Type. J. Virol. 2018, 92, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Bavaro, L.; Martelli, M.; Cavo, M.; Soverini, S. Mechanisms of Disease Progression and Resistance to Tyrosine Kinase Inhibitor Therapy in Chronic Myeloid Leukemia: An Update. Int. J. Mol. Sci. 2019, 20, 6141. [Google Scholar] [CrossRef] [PubMed]
- Arena, A.; Gilardini Montani, M.S.; Romeo, M.A.; Benedetti, R.; Gaeta, A.; Cirone, M. DNA Damage Triggers an Interplay between Wtp53 and C-Myc Affecting Lymphoma Cell Proliferation and Kaposi Sarcoma Herpesvirus Replication. Biochim. Biophys. Acta Mol. Cell Res. 2022, 1869, 119168. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Li, W.; Li, X.; Bai, H.; Zhang, Z. Current Status and Future Prospects of PARP Inhibitor Clinical Trials in Ovarian Cancer. Cancer Manag. Res. 2019, 11, 4371–4390. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.R.; Nussenzweig, A. The Multifaceted Roles of PARP1 in DNA Repair and Chromatin Remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, N.; Wang, X.; Chen, J.L.; Ma, Y. Mechanism Underlying Tumorigenesis Induced by Bcr-Abl Oncogene and A-MuLV Virus. Chin. J. Biotechnol. 2018, 34, 1943–1952. [Google Scholar] [CrossRef]
- Cavone, L.; Aldinucci, A.; Ballerini, C.; Biagioli, T.; Moroni, F.; Chiarugi, A. PARP-1 Inhibition Prevents CNS Migration of Dendritic Cells during EAE, Suppressing the Encephalitogenic Response and Relapse Severity. Mult. Scler. 2011, 17, 794–807. [Google Scholar] [CrossRef]
- Zhang, P.; Nakatsukasa, H.; Tu, E.; Kasagi, S.; Cui, K.; Ishikawa, M.; Konkel, J.E.; Maruyama, T.; Wei, G.; Abbatiello, B.; et al. PARP-1 Regulates Expression of TGF-β Receptors in T Cells. Blood 2013, 122, 2224. [Google Scholar] [CrossRef][Green Version]
- Martínez-Bosch, N.; Iglesias, M.; Munné-Collado, J.; Martínez-Cáceres, C.; Moreno, M.; Guerra, C.; Yélamos, J.; Navarro, P. Parp-1 Genetic Ablation in Ela-Myc Mice Unveils Novel Roles for Parp-1 in Pancreatic Cancer. J. Pathol. 2014, 234, 214–227. [Google Scholar] [CrossRef] [PubMed]
- Navarro, J.; Gozalbo-López, B.; Méndez, A.C.; Dantzer, F.; Schreiber, V.; Martínez, C.; Arana, D.M.; Farrés, J.; Revilla-Nuin, B.; Bueno, M.F.; et al. PARP-1/PARP-2 Double Deficiency in Mouse T Cells Results in Faulty Immune Responses and T Lymphomas. Sci. Rep. 2017, 7, 41962. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Kim, H.J.; Wang, Q.; Kearns, M.; Jiang, T.; Ohlson, C.E.; Li, B.B.; Xie, S.; Liu, J.F.; Stover, E.H.; et al. PARP Inhibition Elicits STING-Dependent Antitumor Immunity in Brca1-Deficient Ovarian Cancer. Cell Rep. 2018, 25, 2972. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.K.; Konstantinopoulos, P.A. PARP Inhibition and Immune Modulation: Scientific Rationale and Perspectives for the Treatment of Gynecologic Cancers. Ther. Adv. Med. Oncol. 2020, 12, 1758835920944116. [Google Scholar] [CrossRef]
- Jiao, S.; Xia, W.; Yamaguchi, H.; Wei, Y.; Chen, M.K.; Hsu, J.M.; Hsu, J.L.; Yu, W.H.; Du, Y.; Lee, H.H.; et al. PARP Inhibitor Upregulates PD-L1 Expression and Enhances Cancer-Associated Immunosuppression. Clin. Cancer Res. 2017, 23, 3711–3720. [Google Scholar] [CrossRef]
- Sen, T.; Rodriguez, B.L.; Chen, L.; Della Corte, C.M.; Morikawa, N.; Fujimoto, J.; Cristea, S.; Nguyen, T.; Diao, L.; Li, L.; et al. Targeting DNA Damage Response Promotes Antitumor Immunity through STING-Mediated T-Cell Activation in Small Cell Lung Cancer. Cancer Discov. 2019, 9, 646–661. [Google Scholar] [CrossRef]
- De Jonge, M.M.; Auguste, A.; Van Wijk, L.M.; Schouten, P.C.; Meijers, M.; Ter Haar, N.T.; Smit, V.T.H.B.M.; Nout, R.A.; Glaire, M.A.; Church, D.N.; et al. Frequent Homologous Recombination Deficiency in High-Grade Endometrial Carcinomas. Clin. Cancer Res. 2019, 25, 1087–1097. [Google Scholar] [CrossRef]
- Muvarak, N.; Kelley, S.; Robert, C.; Baer, M.R.; Perrotti, D.; Gambacorti-Passerini, C.; Civin, C.; Scheibner, K.; Rassool, F.V. C-MYC Generates Repair Errors via Increased Transcription of Alternative-NHEJ Factors, LIG3 and PARP1, in Tyrosine Kinase-Activated Leukemias. Mol. Cancer Res. 2015, 13, 699–712. [Google Scholar] [CrossRef]
- Bian, X.; Wang, X.; Zhang, Q.; Ma, L.; Cao, G.; Xu, A.; Han, J.; Huang, J.; Lin, W. The MYC Paralog-PARP1 Axis as a Potential Therapeutic Target in MYC Paralog-Activated Small Cell Lung Cancer. Front. Oncol. 2020, 10, 565820. [Google Scholar] [CrossRef]
- Caracciolo, D.; Scionti, F.; Juli, G.; Altomare, E.; Golino, G.; Todoerti, K.; Grillone, K.; Riillo, C.; Arbitrio, M.; Iannone, M.; et al. Exploiting MYC-Induced PARPness to Target Genomic Instability in Multiple Myeloma. Haematologica 2021, 106, 185. [Google Scholar] [CrossRef]
- Galindo-Campos, M.A.; Lutfi, N.; Bonnin, S.; Martínez, C.; Velasco-Hernandez, T.; García-Hernández, V.; Martín-Caballero, J.; Ampurdanés, C.; Gimeno, R.; Colomo, L.; et al. Distinct Roles for PARP-1 and PARP-2 in c-Myc–Driven B-Cell Lymphoma in Mice. Blood 2022, 139, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Nieborowska-Skorska, M.; Sullivan, K.; Dasgupta, Y.; Podszywalow-Bartnicka, P.; Hoser, G.; Maifrede, S.; Martinez, E.; Marcantonio, D.D.; Bolton-Gillespie, E.; Cramer-Morales, K.; et al. Gene Expression and Mutation-Guided Synthetic Lethality Eradicates Proliferating and Quiescent Leukemia Cells. J. Clin. Investig. 2017, 127, 2392. [Google Scholar] [CrossRef] [PubMed]




| Cell Line | SUP-B15 | Raji | Namalwa | |
|---|---|---|---|---|
| Drug | ||||
| AZD2461 | 344.3 nM (240.1–493.7) (R2: 0.9358) | NR | 20,938 nM (16,152–27,143) (R2: 0.9806) | |
| Imatinib | 329.2 nM (215.7–502.2) (R2: 0.9385) | 2509 nM (1622–3882) (R2: 0.9431) | 1296 nM (803.8–2089) (R2: 0.9284) | |
| Doxorubicin | 20.32 nM (11.41–36.19) (R2: 0.9233) | 85 nM (65.13–110.9) (R2: 0.9694) | 75.93 nM (49.63–116.2) (R2: 0.9622) | |
| Period | 24 h | 48 h | |
|---|---|---|---|
| Drug | |||
| AZD2461 | 3925 nM (2502–6157) (R2: 0.8763) | 1421 nM (766.9–2632) (R2: 0.8087) | |
| Imatinib | 3966 nM (2398–6558) (R2: 0.8612) | 1230 nM (747.5–2024) (R2: 0.8905) | |
| Doxorubicin | 141.6 nM (93.92–213.6) (R2: 0.9305) | 57.33 nM (36.53–89.97) (R2: 0.9342) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Machado, C.B.; da Silva, E.L.; Ferreira, W.A.S.; Pessoa, F.M.C.d.P.; de Quadros, A.U.; Fantacini, D.M.C.; Furtado, I.P.; Rossetti, R.; Silveira, R.M.; de Lima, S.C.G.; et al. PARP1 Characterization as a Potential Biomarker for BCR::ABL1 p190+ Acute Lymphoblastic Leukemia. Cancers 2023, 15, 5510. https://doi.org/10.3390/cancers15235510
Machado CB, da Silva EL, Ferreira WAS, Pessoa FMCdP, de Quadros AU, Fantacini DMC, Furtado IP, Rossetti R, Silveira RM, de Lima SCG, et al. PARP1 Characterization as a Potential Biomarker for BCR::ABL1 p190+ Acute Lymphoblastic Leukemia. Cancers. 2023; 15(23):5510. https://doi.org/10.3390/cancers15235510
Chicago/Turabian StyleMachado, Caio Bezerra, Emerson Lucena da Silva, Wallax Augusto Silva Ferreira, Flávia Melo Cunha de Pinho Pessoa, Andreza Urba de Quadros, Daianne Maciely Carvalho Fantacini, Izadora Peter Furtado, Rafaela Rossetti, Roberta Maraninchi Silveira, Sarah Caroline Gomes de Lima, and et al. 2023. "PARP1 Characterization as a Potential Biomarker for BCR::ABL1 p190+ Acute Lymphoblastic Leukemia" Cancers 15, no. 23: 5510. https://doi.org/10.3390/cancers15235510
APA StyleMachado, C. B., da Silva, E. L., Ferreira, W. A. S., Pessoa, F. M. C. d. P., de Quadros, A. U., Fantacini, D. M. C., Furtado, I. P., Rossetti, R., Silveira, R. M., de Lima, S. C. G., Mello Júnior, F. A. R., Seabra, A. D., Moreira, E. C. d. O., de Moraes Filho, M. O., de Moraes, M. E. A., Montenegro, R. C., Ribeiro, R. M., Khayat, A. S., Burbano, R. M. R., ... Moreira-Nunes, C. d. F. A. (2023). PARP1 Characterization as a Potential Biomarker for BCR::ABL1 p190+ Acute Lymphoblastic Leukemia. Cancers, 15(23), 5510. https://doi.org/10.3390/cancers15235510










