Diffuse Gliomas with FGFR3-TACC3 Fusions: Oncogenic Mechanisms, Hallmarks, and Therapeutic Perspectives
Abstract
:Simple Summary
Abstract
1. Introduction
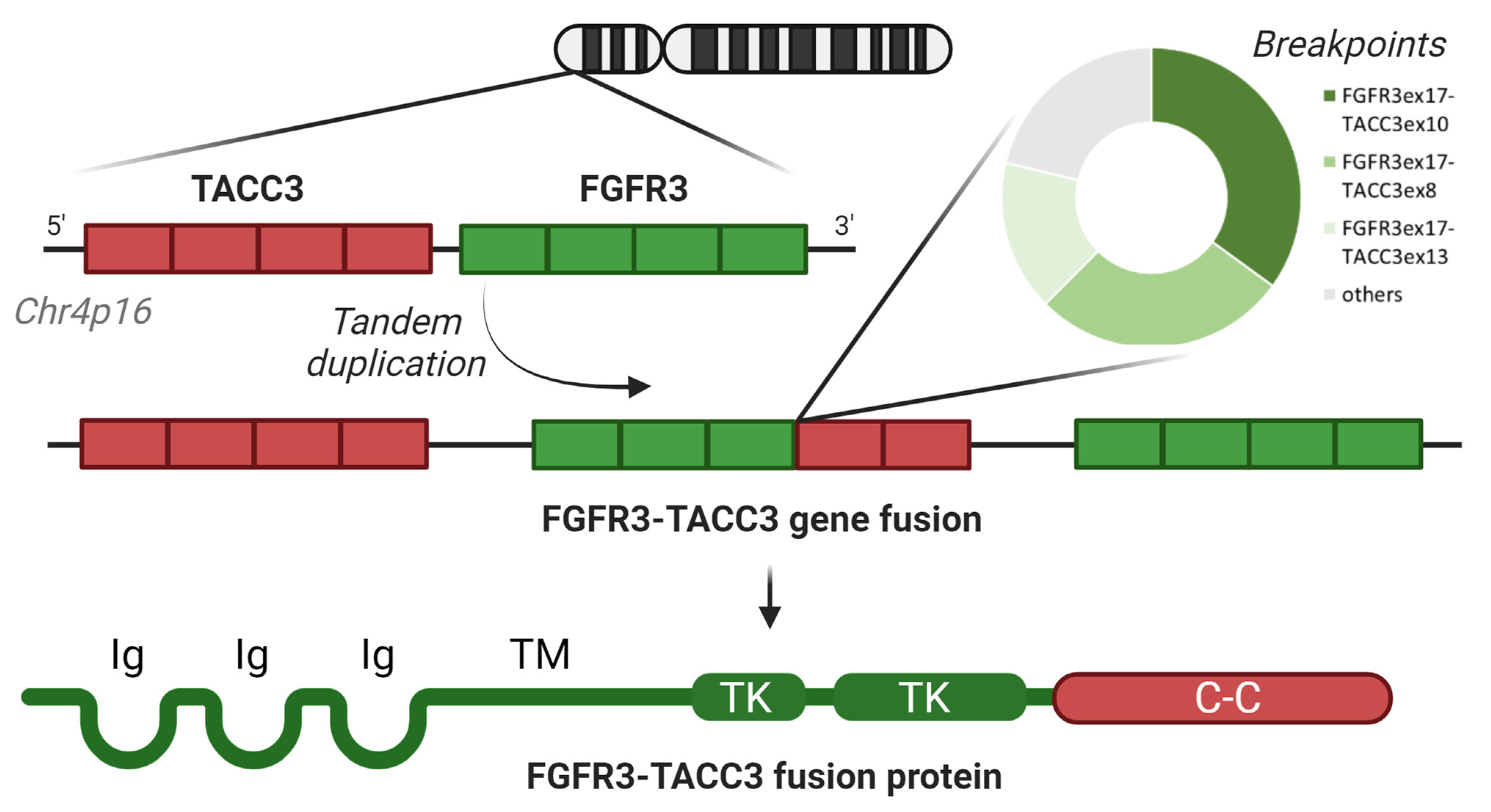
1.1. Screening and Identification of F3T3 in Gliomas
1.2. Prevalence of F3T3 and Its Structural Variants in Gliomas
1.3. The Conundrum of Oncogenic Mechanisms of F3T3
2. The Hallmarks of F3T3 Glioblastomas
2.1. Pathology
2.2. Clinical Features
2.3. Radiological Features
2.4. Genetic Landscape
2.5. Methylation Profiling
2.6. Metabolism
2.7. F3T3 as a Theranostic Marker?
3. FGFR Inhibition in F3T3 Glioblastomas
3.1. Erdafitinib
3.2. Infigratinib
3.3. Fexagratinib
3.4. Pemigatinib
3.5. Zoligratinib
3.6. Futibatinib
4. New Approaches and Perspectives in the Treatment of F3T3 Glioblastomas
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Wen, P.Y.; Weller, M.; Lee, E.Q.; Alexander, B.M.; Barnholtz-Sloan, J.S.; Barthel, F.P.; Batchelor, T.T.; Bindra, R.S.; Chang, S.M.; Chiocca, E.A.; et al. Glioblastoma in Adults: A Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) Consensus Review on Current Management and Future Directions. Neuro-Oncology 2020, 22, 1073–1113. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Chan, J.M.; Zoppoli, P.; Niola, F.; Sullivan, R.; Castano, A.; Liu, E.M.; Reichel, J.; Porrati, P.; Pellegatta, S.; et al. Transforming Fusions of FGFR and TACC Genes in Human Glioblastoma. Science 2012, 337, 1231–1235. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, A.L.; Picca, A.; Saragoussi, E.; Bielle, F.; Ducray, F.; Villa, C.; Eoli, M.; Paterra, R.; Bellu, L.; Mathon, B.; et al. Clinical, Molecular, and Radiomic Profile of Gliomas with FGFR3-TACC3 Fusions. Neuro-Oncology 2020, 22, 1614–1624. [Google Scholar] [CrossRef]
- Mata, D.A.; Benhamida, J.K.; Lin, A.L.; Vanderbilt, C.M.; Yang, S.-R.; Villafania, L.B.; Ferguson, D.C.; Jonsson, P.; Miller, A.M.; Tabar, V.; et al. Genetic and Epigenetic Landscape of IDH-Wildtype Glioblastomas with FGFR3-TACC3 Fusions. Acta Neuropathol. Commun. 2020, 8, 186. [Google Scholar] [CrossRef]
- Touat, M.; Ileana, E.; Postel-Vinay, S.; André, F.; Soria, J.-C. Targeting FGFR Signaling in Cancer. Clin. Cancer Res. 2015, 21, 2684–2694. [Google Scholar] [CrossRef] [PubMed]
- Greenman, C.; Stephens, P.; Smith, R.; Dalgliesh, G.L.; Hunter, C.; Bignell, G.; Davies, H.; Teague, J.; Butler, A.; Stevens, C.; et al. Patterns of Somatic Mutation in Human Cancer Genomes. Nature 2007, 446, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Picca, A.; Berzero, G.; Bielle, F.; Touat, M.; Savatovsky, J.; Polivka, M.; Trisolini, E.; Meunier, S.; Schmitt, Y.; Idbaih, A.; et al. FGFR1 Actionable Mutations, Molecular Specificities, and Outcome of Adult Midline Gliomas. Neurology 2018, 90, e2086–e2094. [Google Scholar] [CrossRef]
- Hood, F.E.; Royle, S.J. Pulling It Together. Bioarchitecture 2011, 1, 105–109. [Google Scholar] [CrossRef]
- Ding, Z.-M.; Huang, C.-J.; Jiao, X.-F.; Wu, D.; Huo, L.-J. The Role of TACC3 in Mitotic Spindle Organization. Cytoskeleton 2017, 74, 369–378. [Google Scholar] [CrossRef]
- Lasorella, A.; Sanson, M.; Iavarone, A. FGFR-TACC Gene Fusions in Human Glioma. Neuro-Oncology 2017, 19, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Parker, B.C.; Annala, M.J.; Cogdell, D.E.; Granberg, K.J.; Sun, Y.; Ji, P.; Li, X.; Gumin, J.; Zheng, H.; Hu, L.; et al. The Tumorigenic FGFR3-TACC3 Gene Fusion Escapes miR-99a Regulation in Glioblastoma. J. Clin. Investig. 2013, 123, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.V.; Hurst, C.D.; Knowles, M.A. Oncogenic FGFR3 Gene Fusions in Bladder Cancer. Hum. Mol. Genet. 2013, 22, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-M.; Su, F.; Kalyana-Sundaram, S.; Khazanov, N.; Ateeq, B.; Cao, X.; Lonigro, R.J.; Vats, P.; Wang, R.; Lin, S.-F.; et al. Identification of Targetable FGFR Gene Fusions in Diverse Cancers. Cancer Discov. 2013, 3, 636–647. [Google Scholar] [CrossRef]
- Capelletti, M.; Dodge, M.E.; Ercan, D.; Hammerman, P.S.; Park, S.-I.; Kim, J.; Sasaki, H.; Jablons, D.M.; Lipson, D.; Young, L.; et al. Identification of Recurrent FGFR3–TACC3 Fusion Oncogenes from Lung Adenocarcinoma. Clin. Cancer Res. 2014, 20, 6551–6558. [Google Scholar] [CrossRef]
- Carneiro, B.A.; Elvin, J.A.; Kamath, S.D.; Ali, S.M.; Paintal, A.S.; Restrepo, A.; Berry, E.; Giles, F.J.; Johnson, M.L. FGFR3–TACC3: A Novel Gene Fusion in Cervical Cancer. Gynecol. Oncol. Rep. 2015, 13, 53–56. [Google Scholar] [CrossRef]
- Yuan, L.; Liu, Z.-H.; Lin, Z.-R.; Xu, L.-H.; Zhong, Q.; Zeng, M.-S. Recurrent FGFR3-TACC3 Fusion Gene in Nasopharyngeal Carcinoma. Cancer Biol. Ther. 2014, 15, 1613–1621. [Google Scholar] [CrossRef]
- Stransky, N.; Cerami, E.; Schalm, S.; Kim, J.L.; Lengauer, C. The Landscape of Kinase Fusions in Cancer. Nat. Commun. 2014, 5, 4846. [Google Scholar] [CrossRef]
- Shaver, T.M.; Lehmann, B.D.; Beeler, J.S.; Li, C.-I.; Li, Z.; Jin, H.; Stricker, T.P.; Shyr, Y.; Pietenpol, J.A. Diverse, Biologically Relevant, and Targetable Gene Rearrangements in Triple-Negative Breast Cancer and Other Malignancies. Cancer Res. 2016, 76, 4850–4860. [Google Scholar] [CrossRef]
- TACC2-FGFR2 Fusion—My Cancer Genome. Available online: https://www.mycancergenome.org/content/alteration/tacc2-fgfr2-fusion/ (accessed on 28 October 2023).
- Neumann, O.; Burn, T.C.; Allgäuer, M.; Ball, M.; Kirchner, M.; Albrecht, T.; Volckmar, A.-L.; Beck, S.; Endris, V.; Goldschmid, H.; et al. Genomic Architecture of FGFR2 Fusions in Cholangiocarcinoma and Its Implication for Molecular Testing. Br. J. Cancer 2022, 127, 1540–1549. [Google Scholar] [CrossRef]
- Arai, Y.; Totoki, Y.; Hosoda, F.; Shirota, T.; Hama, N.; Nakamura, H.; Ojima, H.; Furuta, K.; Shimada, K.; Okusaka, T.; et al. Fibroblast Growth Factor Receptor 2 Tyrosine Kinase Fusions Define a Unique Molecular Subtype of Cholangiocarcinoma. Hepatology 2014, 59, 1427–1434. [Google Scholar] [CrossRef] [PubMed]
- FGFR1-TACC1 Fusion—My Cancer Genome. Available online: https://www.mycancergenome.org/content/alteration/fgfr1-tacc1-fusion/ (accessed on 28 October 2023).
- Zhang, J.; Wu, G.; Miller, C.P.; Tatevossian, R.G.; Dalton, J.D.; Tang, B.; Orisme, W.; Punchihewa, C.; Parker, M.; Qaddoumi, I.; et al. Whole-Genome Sequencing Identifies Genetic Alterations in Pediatric Low-Grade Gliomas. Nat. Genet. 2013, 45, 602–612. [Google Scholar] [CrossRef] [PubMed]
- Ryall, S.; Zapotocky, M.; Fukuoka, K.; Nobre, L.; Guerreiro Stucklin, A.; Bennett, J.; Siddaway, R.; Li, C.; Pajovic, S.; Arnoldo, A.; et al. Integrated Molecular and Clinical Analysis of 1000 Pediatric Low-Grade Gliomas. Cancer Cell 2020, 37, 569–583.e5. [Google Scholar] [CrossRef] [PubMed]
- Sievers, P.; Stichel, D.; Schrimpf, D.; Sahm, F.; Koelsche, C.; Reuss, D.E.; Wefers, A.K.; Reinhardt, A.; Huang, K.; Ebrahimi, A.; et al. FGFR1:TACC1 Fusion Is a Frequent Event in Molecularly Defined Extraventricular Neurocytoma. Acta Neuropathol. 2018, 136, 293–302. [Google Scholar] [CrossRef]
- Di Stefano, A.L.; Fucci, A.; Frattini, V.; Labussiere, M.; Mokhtari, K.; Zoppoli, P.; Marie, Y.; Bruno, A.; Boisselier, B.; Giry, M.; et al. Detection, Characterization, and Inhibition of FGFR-TACC Fusions in IDH Wild-Type Glioma. Clin. Cancer Res. 2015, 21, 3307–3317. [Google Scholar] [CrossRef]
- Heydt, C.; Wölwer, C.B.; Velazquez Camacho, O.; Wagener-Ryczek, S.; Pappesch, R.; Siemanowski, J.; Rehker, J.; Haller, F.; Agaimy, A.; Worm, K.; et al. Detection of Gene Fusions Using Targeted Next-Generation Sequencing: A Comparative Evaluation. BMC Med. Genom. 2021, 14, 62. [Google Scholar] [CrossRef]
- Bielle, F.; Di Stefano, A.-L.; Meyronet, D.; Picca, A.; Villa, C.; Bernier, M.; Schmitt, Y.; Giry, M.; Rousseau, A.; Figarella-Branger, D.; et al. Diffuse Gliomas with FGFR3-TACC3 Fusion Have Characteristic Histopathological and Molecular Features. Brain Pathol. 2018, 28, 674–683. [Google Scholar] [CrossRef]
- Granberg, K.J.; Annala, M.; Lehtinen, B.; Kesseli, J.; Haapasalo, J.; Ruusuvuori, P.; Yli-Harja, O.; Visakorpi, T.; Haapasalo, H.; Nykter, M.; et al. Strong FGFR3 Staining Is a Marker for FGFR3 Fusions in Diffuse Gliomas. Neuro-Oncology 2017, 19, 1206–1216. [Google Scholar] [CrossRef]
- Métais, A.; Tauziède-Espariat, A.; Garcia, J.; Appay, R.; Uro-Coste, E.; Meyronet, D.; Maurage, C.-A.; Vandenbos, F.; Rigau, V.; Chiforeanu, D.C.; et al. Clinico-Pathological and Epigenetic Heterogeneity of Diffuse Gliomas with FGFR3::TACC3 Fusion. Acta Neuropathol. Commun. 2023, 11, 14. [Google Scholar] [CrossRef]
- Schittenhelm, J.; Ziegler, L.; Sperveslage, J.; Mittelbronn, M.; Capper, D.; Burghardt, I.; Poso, A.; Biskup, S.; Skardelly, M.; Tabatabai, G. FGFR3 Overexpression Is a Useful Detection Tool for FGFR3 Fusions and Sequence Variations in Glioma. Neuro-Oncol. Pract. 2021, 8, 209–221. [Google Scholar] [CrossRef]
- Capper, D.; Reifenberger, G.; French, P.J.; Schweizer, L.; Weller, M.; Touat, M.; Niclou, S.P.; Euskirchen, P.; Haberler, C.; Hegi, M.E.; et al. EANO Guideline on Rational Molecular Testing of Gliomas, Glioneuronal, and Neuronal Tumors in Adults for Targeted Therapy Selection. Neuro-Oncology 2023, 25, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Nelson, K.N.; Meyer, A.N.; Siari, A.; Campos, A.R.; Motamedchaboki, K.; Donoghue, D.J. Oncogenic Gene Fusion FGFR3-TACC3 Is Regulated by Tyrosine Phosphorylation. Mol. Cancer Res. 2016, 14, 458–469. [Google Scholar] [CrossRef] [PubMed]
- Frattini, V.; Pagnotta, S.M.; Tala; Fan, J.J.; Russo, M.V.; Lee, S.B.; Garofano, L.; Zhang, J.; Shi, P.; Lewis, G.; et al. A Metabolic Function of FGFR3-TACC3 Gene Fusions in Cancer. Nature 2018, 553, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Nelson, K.N.; Meyer, A.N.; Wang, C.G.; Donoghue, D.J. Oncogenic Driver FGFR3-TACC3 Is Dependent on Membrane Trafficking and ERK Signaling. Oncotarget 2018, 9, 34306–34319. [Google Scholar] [CrossRef]
- Lombardi, B.; Ashford, P.; Moya-Garcia, A.A.; Rust, A.; Crawford, M.; Williams, S.V.; Knowles, M.A.; Katan, M.; Orengo, C.; Godovac-Zimmermann, J. Unique Signalling Connectivity of FGFR3-TACC3 Oncoprotein Revealed by Quantitative Phosphoproteomics and Differential Network Analysis. Oncotarget 2017, 8, 102898–102911. [Google Scholar] [CrossRef]
- Murugesan, K.; Necchi, A.; Burn, T.C.; Gjoerup, O.; Greenstein, R.; Krook, M.; López, J.A.; Montesion, M.; Nimeiri, H.; Parikh, A.R.; et al. Pan-Tumor Landscape of Fibroblast Growth Factor Receptor 1-4 Genomic Alterations. ESMO Open 2022, 7, 100641. [Google Scholar] [CrossRef] [PubMed]
- Yemelyanenko, J.; Zingg, D.; Bhin, J.; Lee, J.K.; Klarenbeek, S.; Song, J.-Y.; Lutz, C.; Annunziato, S.; Proost, N.; Siteur, B.; et al. Deciphering FGFR3-TACC3 Oncogenic Fusions. In Proceedings of the Abstract P-0232, EACR 2023: Innovative Cancer Science, Torino, Italy, 12–15 June 2023. [Google Scholar]
- Stephens, P.J.; McBride, D.J.; Lin, M.-L.; Varela, I.; Pleasance, E.D.; Simpson, J.T.; Stebbings, L.A.; Leroy, C.; Edkins, S.; Mudie, L.J.; et al. Complex Landscapes of Somatic Rearrangement in Human Breast Cancer Genomes. Nature 2009, 462, 1005–1010. [Google Scholar] [CrossRef]
- Sarkar, S.; Ryan, E.L.; Royle, S.J. FGFR3–TACC3 Cancer Gene Fusions Cause Mitotic Defects by Removal of Endogenous TACC3 from the Mitotic Spindle. Open Biol. 2017, 7, 170080. [Google Scholar] [CrossRef]
- Zingg, D.; Bhin, J.; Yemelyanenko, J.; Kas, S.M.; Rolfs, F.; Lutz, C.; Lee, J.K.; Klarenbeek, S.; Silverman, I.M.; Annunziato, S.; et al. Truncated FGFR2 Is a Clinically Actionable Oncogene in Multiple Cancers. Nature 2022, 608, 609–617. [Google Scholar] [CrossRef]
- Huse, J.T.; Snuderl, M.; Jones, D.T.W.; Brathwaite, C.D.; Altman, N.; Lavi, E.; Saffery, R.; Sexton-Oates, A.; Blumcke, I.; Capper, D.; et al. Polymorphous Low-Grade Neuroepithelial Tumor of the Young (PLNTY): An Epileptogenic Neoplasm with Oligodendroglioma-like Components, Aberrant CD34 Expression, and Genetic Alterations Involving the MAP Kinase Pathway. Acta Neuropathol. 2017, 133, 417–429. [Google Scholar] [CrossRef]
- Kleinschmidt-DeMasters, B.K.; Gilani, A. Extra-CNS and Dural Metastases in FGFR3::TACC3 Fusion+ Adult Glioblastoma, IDH-Wildtype. Neuro-Oncol. Pract. 2022, 9, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Ballester, L.Y.; Moghadamtousi, S.Z.; Leeds, N.E.; Huse, J.T.; Fuller, G.N. Coexisting FGFR3 p.K650T Mutation in Two FGFR3-TACC3 Fusion Glioma Cases. Acta Neuropathol. Commun. 2019, 7, 63. [Google Scholar] [CrossRef] [PubMed]
- Gilani, A.; Davies, K.D.; Kleinschmidt-DeMasters, B.K. Can Adult IDH-Wildtype Glioblastomas with FGFR3:TACC3 Fusions Be Reliably Predicted by Histological Features? Clin. Neuropathol. 2021, 40, 165–167. [Google Scholar] [CrossRef]
- McDonald, M.F.; Athukuri, P.; Anand, A.; Gopakumar, S.; Jalali, A.; Patel, A.J.; Rao, G.; Goodman, J.C.; Lu, H.-C.; Mandel, J.J. Varied Histomorphology and Clinical Outcomes of FGFR3-TACC3 Fusion Gliomas. Neurosurg. Focus 2022, 53, E16. [Google Scholar] [CrossRef] [PubMed]
- Broggi, G.; Piombino, E.; Altieri, R.; Romano, C.; Certo, F.; Barbagallo, G.M.V.; Vigneri, P.; Condorelli, D.; Colarossi, L.; Colarossi, C.; et al. Glioblastoma, IDH-Wild Type With FGFR3-TACC3 Fusion: When Morphology May Reliably Predict the Molecular Profile of a Tumor. A Case Report and Literature Review. Front. Neurol. 2022, 13, 823015. [Google Scholar] [CrossRef]
- Garofano, L.; Migliozzi, S.; Oh, Y.T.; D’Angelo, F.; Najac, R.D.; Ko, A.; Frangaj, B.; Caruso, F.P.; Yu, K.; Yuan, J.; et al. Pathway-Based Classification of Glioblastoma Uncovers a Mitochondrial Subtype with Therapeutic Vulnerabilities. Nat. Cancer 2021, 2, 141–156. [Google Scholar] [CrossRef]
- Wu, Z.; Lopes Abath Neto, O.; Bale, T.A.; Benhamida, J.; Mata, D.; Turakulov, R.; Abdullaev, Z.; Marker, D.; Ketchum, C.; Chung, H.-J.; et al. DNA Methylation Analysis of Glioblastomas Harboring FGFR3-TACC3 Fusions Identifies a Methylation Subclass with Better Patient Survival. Acta Neuropathol. 2022, 144, 155–157. [Google Scholar] [CrossRef]
- Takahashi, H.; Natsumeda, M.; Tsukamoto, Y.; Mizu, H.; Okamoto, K.; Mineharu, Y.; Arakawa, Y.; Oishi, M.; Kakita, A.; Fujii, Y. Macrocalcification on CT Imaging Is Milestone in Detecting Diffuse Glioma with FGFR3-TACC3 Fusion. Brain Tumor Pathol. 2023, 40, 099. [Google Scholar]
- Picca, A.; Gareau, T.; Mohand Oumoussa, B.; Carpentier, C.; Dridi-Aloulou, A.; Di Stefano, A.; Mokhtari, K.; Bielle, F.; Sanson, M. P05.03.B Methylome Profiling of Glioblastomas with FGFR3/TACC3 Fusion. Neuro-Oncology 2023, 25, ii44. [Google Scholar] [CrossRef]
- Sturm, D.; Orr, B.A.; Toprak, U.H.; Hovestadt, V.; Jones, D.T.W.; Capper, D.; Sill, M.; Buchhalter, I.; Northcott, P.A.; Leis, I.; et al. New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs. Cell 2016, 164, 1060–1072. [Google Scholar] [CrossRef]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA Methylation-Based Classification of Central Nervous System Tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Frederick, M.; Skinner, H.D.; Kazi, S.A.; Sikora, A.G.; Sandulache, V.C. High Expression of Oxidative Phosphorylation Genes Predicts Improved Survival in Squamous Cell Carcinomas of the Head and Neck and Lung. Sci. Rep. 2020, 10, 6380. [Google Scholar] [CrossRef] [PubMed]
- Migliozzi, S.; Oh, Y.T.; Hasanain, M.; Garofano, L.; D’Angelo, F.; Najac, R.D.; Picca, A.; Bielle, F.; Di Stefano, A.L.; Lerond, J.; et al. Integrative Multi-Omics Networks Identify PKCδ and DNA-PK as Master Kinases of Glioblastoma Subtypes and Guide Targeted Cancer Therapy. Nat. Cancer 2023, 4, 181–202. [Google Scholar] [CrossRef]
- Sansone, G.; Vivori, N.; Vivori, C.; Di Stefano, A.L.; Picca, A. Basic Premises: Searching for New Targets and Strategies in Diffuse Gliomas. Clin. Transl. Imaging 2022, 10, 517–534. [Google Scholar] [CrossRef]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-Cell RNA-Seq Highlights Intratumoral Heterogeneity in Primary Glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef]
- Snuderl, M.; Fazlollahi, L.; Le, L.P.; Nitta, M.; Zhelyazkova, B.H.; Davidson, C.J.; Akhavanfard, S.; Cahill, D.P.; Aldape, K.D.; Betensky, R.A.; et al. Mosaic Amplification of Multiple Receptor Tyrosine Kinase Genes in Glioblastoma. Cancer Cell 2011, 20, 810–817. [Google Scholar] [CrossRef]
- van den Bent, M.J.; Gao, Y.; Kerkhof, M.; Kros, J.M.; Gorlia, T.; van Zwieten, K.; Prince, J.; van Duinen, S.; Sillevis Smitt, P.A.; Taphoorn, M.; et al. Changes in the EGFR Amplification and EGFRvIII Expression between Paired Primary and Recurrent Glioblastomas. Neuro-Oncology 2015, 17, 935–941. [Google Scholar] [CrossRef]
- Wang, J.; Cazzato, E.; Ladewig, E.; Frattini, V.; Rosenbloom, D.I.S.; Zairis, S.; Abate, F.; Liu, Z.; Elliott, O.; Shin, Y.-J.; et al. Clonal Evolution of Glioblastoma under Therapy. Nat. Genet. 2016, 48, 768–776. [Google Scholar] [CrossRef]
- Mateo, J.; Chakravarty, D.; Dienstmann, R.; Jezdic, S.; Gonzalez-Perez, A.; Lopez-Bigas, N.; Ng, C.K.Y.; Bedard, P.L.; Tortora, G.; Douillard, J.-Y.; et al. A Framework to Rank Genomic Alterations as Targets for Cancer Precision Medicine: The ESMO Scale for Clinical Actionability of Molecular Targets (ESCAT). Ann. Oncol. 2018, 29, 1895–1902. [Google Scholar] [CrossRef]
- Tabernero, J.; Bahleda, R.; Dienstmann, R.; Infante, J.R.; Mita, A.; Italiano, A.; Calvo, E.; Moreno, V.; Adamo, B.; Gazzah, A.; et al. Phase I Dose-Escalation Study of JNJ-42756493, an Oral Pan-Fibroblast Growth Factor Receptor Inhibitor, in Patients with Advanced Solid Tumors. J. Clin. Oncol. 2015, 33, 3401–3408. [Google Scholar] [CrossRef]
- Pant, S.; Schuler, M.; Iyer, G.; Witt, O.; Doi, T.; Qin, S.; Tabernero, J.; Reardon, D.A.; Massard, C.; Minchom, A.; et al. Erdafitinib in Patients with Advanced Solid Tumours with FGFR Alterations (RAGNAR): An International, Single-Arm, Phase 2 Study. Lancet Oncol. 2023, 24, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Lassman, A.B.; Sepúlveda-Sánchez, J.M.; Cloughesy, T.F.; Gil-Gil, M.J.; Puduvalli, V.K.; Raizer, J.J.; De Vos, F.Y.F.; Wen, P.Y.; Butowski, N.A.; Clement, P.M.J.; et al. Infigratinib in Patients with Recurrent Gliomas and FGFR Alterations: A Multicenter Phase II Study. Clin. Cancer Res. 2022, 28, 2270–2277. [Google Scholar] [CrossRef] [PubMed]
- Spanggaard, I.; Matrana, M.; Rocha-Lima, C.; Mahipal, A.; Vieito, M.; Hervieu, A.; Ahn, M.-J.; Goyal, L.; Ahnert, J.R.; Veronese, L.; et al. Pemigatinib For Previously Treated Central Nervous System Tumors With Activating FGFR Mutations or Translocations: Results From FIGHT-207 (S17.004). Neurology 2023, 100, 4218. [Google Scholar] [CrossRef]
- Cleary, J.M.; Iyer, G.; Oh, D.-Y.; Mellinghoff, I.K.; Goyal, L.; Ng, M.C.H.; Meric-Bernstam, F.; Matos, I.; Chao, T.-Y.; Ait Sarkouh, R.; et al. Final Results from the Phase I Study Expansion Cohort of the Selective FGFR Inhibitor Debio 1347 in Patients with Solid Tumors Harboring an FGFR Gene Fusion. JCO 2020, 38, 3603. [Google Scholar] [CrossRef]
- Farouk Sait, S.; Gilheeney, S.W.; Bale, T.A.; Haque, S.; Dinkin, M.J.; Vitolano, S.; Rosenblum, M.K.; Ibanez, K.; Prince, D.E.; Spatz, K.H.; et al. Debio1347, an Oral FGFR Inhibitor: Results From a Single-Center Study in Pediatric Patients With Recurrent or Refractory FGFR-Altered Gliomas. JCO Precis Oncol. 2021, 5, 876–883. [Google Scholar] [CrossRef] [PubMed]
- Meric-Bernstam, F.; Bahleda, R.; Hierro, C.; Sanson, M.; Bridgewater, J.; Arkenau, H.-T.; Tran, B.; Kelley, R.K.; Park, J.O.; Javle, M.; et al. Futibatinib, an Irreversible FGFR1–4 Inhibitor, in Patients with Advanced Solid Tumors Harboring FGF/FGFR Aberrations: A Phase I Dose-Expansion Study. Cancer Discov. 2022, 12, 402–415. [Google Scholar] [CrossRef]
- Loriot, Y.; Necchi, A.; Park, S.H.; Garcia-Donas, J.; Huddart, R.; Burgess, E.; Fleming, M.; Rezazadeh, A.; Mellado, B.; Varlamov, S.; et al. Erdafitinib in Locally Advanced or Metastatic Urothelial Carcinoma. N. Engl. J. Med. 2019, 381, 338–348. [Google Scholar] [CrossRef]
- Javle, M.; Roychowdhury, S.; Kelley, R.K.; Sadeghi, S.; Macarulla, T.; Weiss, K.H.; Waldschmidt, D.-T.; Goyal, L.; Borbath, I.; El-Khoueiry, A.; et al. Infigratinib (BGJ398) in Previously Treated Patients with Advanced or Metastatic Cholangiocarcinoma with FGFR2 Fusions or Rearrangements: Mature Results from a Multicentre, Open-Label, Single-Arm, Phase 2 Study. Lancet Gastroenterol. Hepatol. 2021, 6, 803–815. [Google Scholar] [CrossRef]
- Picca, A.; Di Stefano, A.L.; Savatovsky, J.; Ducray, F.; Chinot, O.; Cohen-Moyal, E.; Augereau, P.; Schmidt, Y.; Lerond, J.; Rousseaux, N.; et al. TARGET Trial: A Phase I/II Open-Label Multicentre Study to Assess Safety, Tolerability, and Clinical Efficacy of AZD4547 in Patients with Relapsed/Refractory FGFR Fusion Positive Glioma. In Proceedings of the Abstract CTNI-33, 28th Annual Meeting of the Society for Neuro-Oncology, Vancouver, BC, Canada, 15–19 November 2023. [Google Scholar]
- Abou-Alfa, G.K.; Sahai, V.; Hollebecque, A.; Vaccaro, G.; Melisi, D.; Al-Rajabi, R.; Paulson, A.S.; Borad, M.J.; Gallinson, D.; Murphy, A.G.; et al. Pemigatinib for Previously Treated, Locally Advanced or Metastatic Cholangiocarcinoma: A Multicentre, Open-Label, Phase 2 Study. Lancet Oncol. 2020, 21, 671–684. [Google Scholar] [CrossRef]
- Subbiah, V.; Iannotti, N.O.; Gutierrez, M.; Smith, D.C.; Féliz, L.; Lihou, C.F.; Tian, C.; Silverman, I.M.; Ji, T.; Saleh, M. FIGHT-101, a First-in-Human Study of Potent and Selective FGFR 1-3 Inhibitor Pemigatinib in Pan-Cancer Patients with FGF/FGFR Alterations and Advanced Malignancies. Ann. Oncol. 2022, 33, 522–533. [Google Scholar] [CrossRef]
- Rodon, J.; Damian, S.; Furqan, M.; Garcia-Donas, J.; Imai, H.; Italiano, A.; Spanggaard, I.; Ueno, M.; Yokota, T.; Veronese, L.; et al. Abstract CT016: Clinical and Translational Findings of Pemigatinib in Previously Treated Solid Tumors with Activating FGFR1-3 Alterations in the FIGHT-207 Study. Cancer Res. 2023, 83, CT016. [Google Scholar] [CrossRef]
- Ahluwalia, M.; Franceschi, E.; Veronese, L.; Oliveira, N.; Li, X.; van den Bent, M. CTNI-39. FIGHT-209: A phase 2, open-label, multicenter study of pemigatinib in patients with previously treated glioblastoma or other primary central nervous system tumors with activating FGFR1-3 alterations. Neuro-Oncology 2022, 24, vii80. [Google Scholar] [CrossRef]
- Hyman, D.M.; Goyal, L.; Grivas, P.; Meric-Bernstam, F.; Tabernero, J.; Hu, Y.; Kirpicheva, Y.; Nicolas-Metral, V.; Pokorska-Bocci, A.; Vaslin, A.; et al. FUZE Clinical Trial: A Phase 2 Study of Debio 1347 in FGFR Fusion-Positive Advanced Solid Tumors Irrespectively of Tumor Histology. JCO 2019, 37, TPS3157. [Google Scholar] [CrossRef]
- Goyal, L.; Meric-Bernstam, F.; Hollebecque, A.; Valle, J.W.; Morizane, C.; Karasic, T.B.; Abrams, T.A.; Furuse, J.; Kelley, R.K.; Cassier, P.A.; et al. Futibatinib for FGFR2-Rearranged Intrahepatic Cholangiocarcinoma. N. Engl. J. Med. 2023, 388, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Sanai, N.; Margaryan, T.; Molloy, J.; DeSantis, A.; Harmon, J.; Hong, A.; Braun, K.; Wanebo, J.; Yoo, W.; Tien, A.-C.; et al. A Phase 0 Pharmacokinetic Trigger Trial of Infigratinib in Patients with Recurrent High-Grade Glioma. JCO 2023, 41, 2051. [Google Scholar] [CrossRef]
- Yap, T.A.; Daver, N.; Mahendra, M.; Zhang, J.; Kamiya-Matsuoka, C.; Meric-Bernstam, F.; Kantarjian, H.M.; Ravandi, F.; Collins, M.E.; Francesco, M.E.D.; et al. Complex I Inhibitor of Oxidative Phosphorylation in Advanced Solid Tumors and Acute Myeloid Leukemia: Phase I Trials. Nat. Med. 2023, 29, 115–126. [Google Scholar] [CrossRef]
- Shi, Y.; Lim, S.K.; Liang, Q.; Iyer, S.V.; Wang, H.-Y.; Wang, Z.; Xie, X.; Sun, D.; Chen, Y.-J.; Tabar, V.; et al. Gboxin Is an Oxidative Phosphorylation Inhibitor That Targets Glioblastoma. Nature 2019, 567, 341–346. [Google Scholar] [CrossRef]
- Zou, Y.; Sun, Y.; Wang, Y.; Zhang, D.; Yang, H.; Wang, X.; Zheng, M.; Shi, B. Cancer Cell-Mitochondria Hybrid Membrane Coated Gboxin Loaded Nanomedicines for Glioblastoma Treatment. Nat. Commun. 2023, 14, 4557. [Google Scholar] [CrossRef]
- Lazow, M.; Thomas, D.; Cottrell, C.; Kobolt, D.; Ramadesikan, S.; Salloum, R.; Shaikhouni, A.; Mardis, E.; Jones, J.; Leonard, J.; et al. LGG-14. Treatment of two pediatric fgfr-altered low-grade glioneuronal tumors with mek inhibition. Neuro-Oncology 2023, 25, i58. [Google Scholar] [CrossRef]
- Parker Kerrigan, B.C.; Ledbetter, D.; Kronowitz, M.; Phillips, L.; Gumin, J.; Hossain, A.; Yang, J.; Mendt, M.; Singh, S.; Cogdell, D.; et al. RNAi Technology Targeting the FGFR3-TACC3 Fusion Breakpoint: An Opportunity for Precision Medicine. Neurooncol. Adv. 2020, 2, vdaa132. [Google Scholar] [CrossRef]
- Wu, Y.; Jin, W.; Wang, Q.; Zhou, J.; Wang, Y.; Tan, Y.; Cui, X.; Tong, F.; Yang, E.; Wang, J.; et al. Precise Editing of FGFR3-TACC3 Fusion Genes with CRISPR-Cas13a in Glioblastoma. Mol. Ther. 2021, 29, 3305–3318. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Ghosh, B.; Biswas, S. Nanocarriers for Cancer-Targeted Drug Delivery. J. Drug Target. 2016, 24, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Herrero, E.; Fernández-Medarde, A. Advanced Targeted Therapies in Cancer: Drug Nanocarriers, the Future of Chemotherapy. Eur. J. Pharm. Biopharm. 2015, 93, 52–79. [Google Scholar] [CrossRef] [PubMed]

| Study | Study Design | Target Population | Selected Results |
|---|---|---|---|
| Erdafitinib (JNJ-42756493) | |||
| NCT01703481 [63] | Phase I | Advanced solid tumors | 3 brain tumor patients included. 1 PR in F3T3 GBM. |
| NCT04083976 (RAGNAR) [64] | Phase II | Advanced solid tumors with FGFR1-4 fusions or mutations | 10 HGGs included. 3 PR (3 F3T3, 1 F1T1). |
| NCT05859334 | Phase II | Recurrent gliomas with FGFR-TACC fusions | Ongoing, not yet recruiting. |
| Infigratinib (BGJ398) | |||
| NCT01975701 [65] | Phase II | Recurrent gliomas with FGFR1-3 amplifications, fusions, or activating mutations | 26 patients included, of which 10 with F3T3. ORR 5%, 6-mo PFS 16%. One prolonged response (32+ mo) in an F3T3 GBM |
| Fexagratinib (AZD4547) | |||
| TARGET | Phase I/II | Recurrent gliomas with FGFR gene fusions | 12 F3T3 patients included. ORR 8% (1 delayed PR after 13 mo of treatment), 6-mo PFS 25% |
| Pemigatinib (INCB054828) | |||
| NCT03822117 (FIGHT-207) [66] | Phase II | Advanced solid tumors with FGFR1-4 fusions or mutations | 13 patients with FGFR-altered gliomas included (9 with F3T3). 1 CR, 1 PR, 2 SD in the 9 F3T3 |
| FIGHT-209 NCT05267106 | Phase II | Recurrent gliomas with FGFR1-3 alterations | Ongoing, recruiting |
| Zoligratinib (Debio-1347) | |||
| NCT01948297 [67] | Phase II | Advanced solid tumors with FGFR1-3 fusions | Five brain tumor patients included, 0% disease control |
| [68] | Single-patient use protocols | Pediatric FGFR-altered gliomas | Five patients, including 1 F3T3 and 1 F1T1. In FGFR-TACC fusions, 1 PR and 1 sustained SD |
| Futibatinib (TAS120) | |||
| NCT02052778 [69] | Phase I | Advanced solid tumors | 36 CNS tumors (23 F3T3 fus, 2 F1T1 fus). 1 PR in F1T1, 8 tumor volume reductions in F3T3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Picca, A.; Sansone, G.; Santonocito, O.S.; Mazzanti, C.M.; Sanson, M.; Di Stefano, A.L. Diffuse Gliomas with FGFR3-TACC3 Fusions: Oncogenic Mechanisms, Hallmarks, and Therapeutic Perspectives. Cancers 2023, 15, 5555. https://doi.org/10.3390/cancers15235555
Picca A, Sansone G, Santonocito OS, Mazzanti CM, Sanson M, Di Stefano AL. Diffuse Gliomas with FGFR3-TACC3 Fusions: Oncogenic Mechanisms, Hallmarks, and Therapeutic Perspectives. Cancers. 2023; 15(23):5555. https://doi.org/10.3390/cancers15235555
Chicago/Turabian StylePicca, Alberto, Giulio Sansone, Orazio Santo Santonocito, Chiara Maria Mazzanti, Marc Sanson, and Anna Luisa Di Stefano. 2023. "Diffuse Gliomas with FGFR3-TACC3 Fusions: Oncogenic Mechanisms, Hallmarks, and Therapeutic Perspectives" Cancers 15, no. 23: 5555. https://doi.org/10.3390/cancers15235555
APA StylePicca, A., Sansone, G., Santonocito, O. S., Mazzanti, C. M., Sanson, M., & Di Stefano, A. L. (2023). Diffuse Gliomas with FGFR3-TACC3 Fusions: Oncogenic Mechanisms, Hallmarks, and Therapeutic Perspectives. Cancers, 15(23), 5555. https://doi.org/10.3390/cancers15235555







