Breaking Barriers: AI’s Influence on Pathology and Oncology in Resource-Scarce Medical Systems
Abstract
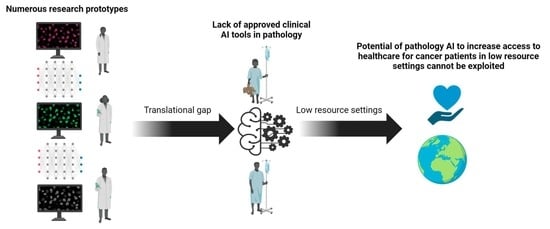
:Simple Summary
Abstract
1. Introduction
2. Artificial Intelligence in Pathology: Research
2.1. AI Tools for Tumor Classification
2.2. AI Tools for Biomarker Quantification
2.3. AI Tools for Survival Prediction
2.4. AI Tools for Predicting Molecular Alterations
2.5. Generative AI and Synthetic Data
3. Artificial Intelligence in Pathology: Clinical-Grade Tools
3.1. Implementation of AI in Routine Practice
3.2. Examples of AI Tools Approved for Clinical Use in the USA and/or EU
3.3. The Problem of Cost-Efficiency
4. Digitalization and AI in Countries with Scarce Resources
4.1. The Challenge of Digitalization
4.2. The Challenges of Pathology and Digitalization in Developing Countries
4.3. Telepathology as a Stepping Stone for Digitalization in Developing Countries
4.4. The Potential Uses of Social Media
5. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shreve, J.T.; Khanani, S.A.; Haddad, T.C. Artificial Intelligence in Oncology: Current Capabilities, Future Opportunities, and Ethical Considerations. Am. Soc. Clin. Oncol. Educ. Book 2022, 42, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Pedro, A.R.; Dias, M.B.; Laranjo, L.; Cunha, A.S.; Cordeiro, J.V. Artificial intelligence in medicine: A comprehensive survey of medical doctor’s perspectives in Portugal. PLoS ONE 2023, 18, e0290613. [Google Scholar] [CrossRef]
- on behalf of the European Society of Pathology (ESP); Matias-Guiu, X.; Stanta, G.; Carneiro, F.; Ryska, A.; Hoefler, G.; Moch, H. The leading role of pathology in assessing the somatic molecular alterations of cancer: Position Paper of the European Society of Pathology. Virchows Arch. 2020, 476, 491–497. [Google Scholar] [CrossRef]
- Jahn, S.W.; Plass, M.; Moinfar, F. Digital Pathology: Advantages, Limitations and Emerging Perspectives. J. Clin. Med. 2020, 9, 3697. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Cheng, M.; Huang, S.; Pei, Z.; Zuo, Y.; Liu, J.; Yang, K.; Zhu, Q.; Zhang, J.; Hong, H.; et al. Recent Advances of Deep Learning for Computational Histopathology: Principles and Applications. Cancers 2022, 14, 1199. [Google Scholar] [CrossRef]
- Steiner, D.F.; Chen, P.-H.C.; Mermel, C.H. Closing the translation gap: AI applications in digital pathology. Biochim. Biophys. Acta (BBA) Rev. Cancer 2021, 1875, 188452. [Google Scholar] [CrossRef]
- Kearney, S.J.; Lowe, A.; Lennerz, J.K.; Parwani, A.; Bui, M.M.; Wack, K.; Giannini, G.; Abels, E. Bridging the Gap: The Critical Role of Regulatory Affairs and Clinical Affairs in the Total Product Life Cycle of Pathology Imaging Devices and Software. Front. Med. 2021, 8, 765385. [Google Scholar] [CrossRef]
- Bulten, W.; Pinckaers, H.; van Boven, H.; Vink, R.; de Bel, T.; van Ginneken, B.; van der Laak, J.; Hulsbergen-van de Kaa, C.; Litjens, G. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: A diagnostic study. Lancet Oncol. 2020, 21, 233–241. [Google Scholar] [CrossRef]
- Abele, N.; Tiemann, K.; Krech, T.; Wellmann, A.; Schaaf, C.; Länger, F.; Peters, A.; Donner, A.; Keil, F.; Daifalla, K.; et al. Noninferiority of Artificial Intelligence–Assisted Analysis of Ki-67 and Estrogen/Progesterone Receptor in Breast Cancer Routine Diagnostics. Mod. Pathol. 2023, 36, 100033. [Google Scholar] [CrossRef]
- Echle, A.; Laleh, N.G.; Quirke, P.; Grabsch, H.; Muti, H.; Saldanha, O.; Brockmoeller, S.; Brandt, P.v.D.; Hutchins, G.; Richman, S.; et al. Artificial intelligence for detection of microsatellite instability in colorectal cancer—A multicentric analysis of a pre-screening tool for clinical application. ESMO Open 2022, 7, 100400. [Google Scholar] [CrossRef]
- Echle, A.; Rindtorff, N.T.; Brinker, T.J.; Luedde, T.; Pearson, A.T.; Kather, J.N. Deep learning in cancer pathology: A new generation of clinical biomarkers. Br. J. Cancer 2021, 124, 686–696. [Google Scholar] [CrossRef]
- Kanavati, F.; Toyokawa, G.; Momosaki, S.; Takeoka, H.; Okamoto, M.; Yamazaki, K.; Takeo, S.; Iizuka, O.; Tsuneki, M. A deep learning model for the classification of indeterminate lung carcinoma in biopsy whole slide images. Sci. Rep. 2021, 11, 8110. [Google Scholar] [CrossRef]
- Yang, H.; Chen, L.; Cheng, Z.; Yang, M.; Wang, J.; Lin, C.; Wang, Y.; Huang, L.; Chen, Y.; Peng, S.; et al. Deep learning-based six-type classifier for lung cancer and mimics from histopathological whole slide images: A retrospective study. BMC Med. 2021, 19, 80. [Google Scholar] [CrossRef]
- Iizuka, O.; Kanavati, F.; Kato, K.; Rambeau, M.; Arihiro, K.; Tsuneki, M. Deep Learning Models for Histopathological Classification of Gastric and Colonic Epithelial Tumours. Sci. Rep. 2020, 10, 1504. [Google Scholar] [CrossRef]
- Byeon, S.-J.; Park, J.; Cho, Y.A.; Cho, B.-J. Automated histological classification for digital pathology images of colonoscopy specimen via deep learning. Sci. Rep. 2022, 12, 12804. [Google Scholar] [CrossRef] [PubMed]
- Bulten, W.; Balkenhol, M.; Belinga, J.-J.A.; Brilhante, A.; Çakır, A.; Egevad, L.; Eklund, M.; Farré, X.; Geronatsiou, K.; Molinié, V.; et al. Artificial intelligence assistance significantly improves Gleason grading of prostate biopsies by pathologists. Mod. Pathol. 2021, 34, 660–671. [Google Scholar] [CrossRef]
- Lu, M.Y.; Chen, T.Y.; Williamson, D.F.K.; Zhao, M.; Shady, M.; Lipkova, J.; Mahmood, F. AI-based pathology predicts origins for cancers of unknown primary. Nature 2021, 594, 106–110. [Google Scholar] [CrossRef]
- Baez-Navarro, X.; van Bockstal, M.R.; Nawawi, D.; Broeckx, G.; Colpaert, C.; Doebar, S.C.; Hogenes, M.C.; Koop, E.; Lambein, K.; Peeters, D.J.; et al. Interobserver Variation in the Assessment of Immunohistochemistry Expression Levels in HER2-Negative Breast Cancer: Can We Improve the Identification of Low Levels of HER2 Expression by Adjusting the Criteria? An International Interobserver Study. Mod. Pathol. 2023, 36, 100009. [Google Scholar] [CrossRef] [PubMed]
- Robert, M.E.; Rüschoff, J.; Jasani, B.; Graham, R.P.; Badve, S.S.; Rodriguez-Justo, M.; Kodach, L.L.; Srivastava, A.; Wang, H.L.; Tang, L.H.; et al. High Interobserver Variability Among Pathologists Using Combined Positive Score to Evaluate PD-L1 Expression in Gastric, Gastroesophageal Junction, and Esophageal Adenocarcinoma. Mod. Pathol. 2023, 36, 100154. [Google Scholar] [CrossRef] [PubMed]
- Butter, R.; Hondelink, L.M.; van Elswijk, L.; Blaauwgeers, J.L.; Bloemena, E.; Britstra, R.; Bulkmans, N.; van Gullik, A.L.; Monkhorst, K.; de Rooij, M.J.; et al. The impact of a pathologist’s personality on the interobserver variability and diagnostic accuracy of predictive PD-L1 immunohistochemistry in lung cancer. Lung Cancer 2022, 166, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Yue, M.; Zhang, J.; Li, X.; Li, Z.; Zhang, H.; Wang, X.; Han, X.; Cai, L.; Shang, J.; et al. The Role of Artificial Intelligence in Accurate Interpretation of HER2 Immunohistochemical Scores 0 and 1+ in Breast Cancer. Mod. Pathol. 2023, 36, 100054. [Google Scholar] [CrossRef]
- Qaiser, T.; Mukherjee, A.; Pb, C.R.; Munugoti, S.D.; Tallam, V.; Pitkäaho, T.; Lehtimäki, T.; Naughton, T.; Berseth, M.; Pedraza, A.; et al. HER2 challenge contest: A detailed assessment of automated HER2 scoring algorithms in whole slide images of breast cancer tissues. Histopathology 2018, 72, 227–238. [Google Scholar] [CrossRef]
- Wang, X.; Wang, L.; Bu, H.; Zhang, N.; Yue, M.; Jia, Z.; Cai, L.; He, J.; Wang, Y.; Xu, X.; et al. How can artificial intelligence models assist PD-L1 expression scoring in breast cancer: Results of multi-institutional ring studies. NPJ Breast Cancer 2021, 7, 61. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [PubMed]
- Naso, J.R.; Povshedna, T.; Wang, G.; Banyi, N.; MacAulay, C.; Ionescu, D.N.; Zhou, C. Automated PD-L1 Scoring for Non-Small Cell Lung Carcinoma Using Open-Source Software. Pathol. Oncol. Res. 2021, 27, 609717. [Google Scholar] [CrossRef]
- Liu, J.; Zheng, Q.; Mu, X.; Zuo, Y.; Xu, B.; Jin, Y.; Wang, Y.; Tian, H.; Yang, Y.; Xue, Q.; et al. Automated tumor proportion score analysis for PD-L1 (22C3) expression in lung squamous cell carcinoma. Sci. Rep. 2021, 11, 15907. [Google Scholar] [CrossRef]
- Li, L.; Han, D.; Yu, Y.; Li, J.; Liu, Y. Artificial intelligence-assisted interpretation of Ki-67 expression and repeatability in breast cancer. Diagn. Pathol. 2022, 17, 20. [Google Scholar] [CrossRef]
- Fulawka, L.; Blaszczyk, J.; Tabakov, M.; Halon, A. Assessment of Ki-67 proliferation index with deep learning in DCIS (ductal carcinoma in situ). Sci. Rep. 2022, 12, 3166. [Google Scholar] [CrossRef]
- Zhu, W.; Xie, L.; Han, J.; Guo, X. The Application of Deep Learning in Cancer Prognosis Prediction. Cancers 2020, 12, 603. [Google Scholar] [CrossRef] [PubMed]
- Katzman, J.L.; Shaham, U.; Cloninger, A.; Bates, J.; Jiang, T.; Kluger, Y. DeepSurv: Personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med. Res. Methodol. 2018, 18, 24. [Google Scholar] [CrossRef]
- Xu, Z.; Lim, S.; Shin, H.-K.; Uhm, K.-H.; Lu, Y.; Jung, S.-W.; Ko, S.-J. Risk-aware survival time prediction from whole slide pathological images. Sci. Rep. 2022, 12, 21948. [Google Scholar] [CrossRef]
- Zhu, X.; Yao, J.; Zhu, F.; Huang, J. WSISA: Making Survival Prediction from Whole Slide Histopathological Images. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 6855–6863. [Google Scholar]
- Vale-Silva, L.A.; Rohr, K. Long-term cancer survival prediction using multimodal deep learning. Sci. Rep. 2021, 11, 13505. [Google Scholar] [CrossRef]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyö, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef]
- Wagner, S.J.; Reisenbüchler, D.; West, N.P.; Niehues, J.M.; Zhu, J.; Foersch, S.; Veldhuizen, G.P.; Quirke, P.; Grabsch, H.I.; Brandt, P.A.v.D.; et al. Transformer-based biomarker prediction from colorectal cancer histology: A large-scale multicentric study. Cancer Cell 2023, 41, 1650–1661.e4. [Google Scholar] [CrossRef] [PubMed]
- Fremond, S.; Andani, S.; Wolf, J.B.; Dijkstra, J.; Melsbach, S.; Jobsen, J.J.; Brinkhuis, M.; Roothaan, S.; Jurgenliemk-Schulz, I.; Lutgens, L.C.H.W.; et al. Interpretable deep learning model to predict the molecular classification of endometrial cancer from haematoxylin and eosin-stained whole-slide images: A combined analysis of the PORTEC randomised trials and clinical cohorts. Lancet Digit. Health 2023, 5, e71–e82. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, B.; Topatana, W.; Cao, J.; Zhu, H.; Juengpanich, S.; Mao, Q.; Yu, H.; Cai, X. Classification and mutation prediction based on histopathology H&E images in liver cancer using deep learning. npj Precis. Oncol. 2020, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zanazzi, G.J.; Hassanpour, S. Predicting prognosis and IDH mutation status for patients with lower-grade gliomas using whole slide images. Sci. Rep. 2021, 11, 16849. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Shen, Y.; Zhang, X.; Xu, P.; Wang, J.; Li, J.; Ren, F.; Ye, D.; Zhou, S.K. Histopathological bladder cancer gene mutation prediction with hierarchical deep multiple-instance learning. Med. Image Anal. 2023, 87, 102824. [Google Scholar] [CrossRef] [PubMed]
- Qu, H.; Zhou, M.; Yan, Z.; Wang, H.; Rustgi, V.K.; Zhang, S.; Gevaert, O.; Metaxas, D.N. Genetic mutation and biological pathway prediction based on whole slide images in breast carcinoma using deep learning. npj Precis. Oncol. 2021, 5, 87. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zou, C.; Zhang, Y.; Li, X.; Wang, C.; Ke, F.; Chen, J.; Wang, W.; Wang, D.; Xu, X.; et al. Prediction of BRCA Gene Mutation in Breast Cancer Based on Deep Learning and Histopathology Images. Front. Genet. 2021, 12, 661109. [Google Scholar] [CrossRef] [PubMed]
- Bourgade, R.; Rabilloud, N.; Perennec, T.; Pécot, T.; Garrec, C.; Guédon, A.F.; Delnatte, C.; Bézieau, S.; Lespagnol, A.; de Tayrac, M.; et al. Deep Learning for Detecting BRCA Mutations in High-Grade Ovarian Cancer Based on an Innovative Tumor Segmentation Method From Whole Slide Images. Mod. Pathol. 2023, 36, 100304. [Google Scholar] [CrossRef] [PubMed]
- Dolezal, J.M.; Trzcinska, A.; Liao, C.-Y.; Kochanny, S.; Blair, E.; Agrawal, N.; Keutgen, X.M.; Angelos, P.; Cipriani, N.A.; Pearson, A.T. Deep learning prediction of BRAF-RAS gene expression signature identifies noninvasive follicular thyroid neoplasms with papillary-like nuclear features. Mod. Pathol. 2021, 34, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.H.; Nomikou, S.; Coudray, N.; Jour, G.; Dawood, Z.; Hong, R.; Esteva, E.; Sakellaropoulos, T.; Donnelly, D.; Moran, U.; et al. Deep Learning and Pathomics Analyses Reveal Cell Nuclei as Important Features for Mutation Prediction of BRAF-Mutated Melanomas. J. Investig. Dermatol. 2022, 142, 1650–1658.e6. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.S.; Massoud, T.F. Predicting tumour mutational burden from histopathological images using multiscale deep learning. Nat. Mach. Intell. 2020, 2, 356–362. [Google Scholar] [CrossRef]
- Morrison, D.; Harris-Birtill, D.; Caie, P.D. Generative Deep Learning in Digital Pathology Workflows. Am. J. Pathol. 2021, 191, 1717–1723. [Google Scholar] [CrossRef]
- Quiros, A.C.; Murray-Smith, R.; Yuan, K. PathologyGAN: Learning deep representations of cancer tissue. arXiv 2021, arXiv:1907.02644. [Google Scholar] [CrossRef]
- Wei, J.; Suriawinata, A.; Vaickus, L.; Ren, B.; Liu, X.; Wei, J.; Hassanpour, S. Generative Image Translation for Data Augmentation in Colorectal Histopathology Images. Proc. Mach. Learn. Res. 2019, 116, 10–24. [Google Scholar]
- Jose, L.; Liu, S.; Russo, C.; Nadort, A.; Di Ieva, A. Generative Adversarial Networks in Digital Pathology and Histopathological Image Processing: A Review. J. Pathol. Inform. 2021, 12, 43. [Google Scholar] [CrossRef]
- Acs, B.; Rantalainen, M.; Hartman, J. Artificial intelligence as the next step towards precision pathology. J. Intern. Med. 2020, 288, 62–81. [Google Scholar] [CrossRef]
- Kaur, S.; Singh, P. How does object-oriented code refactoring influence software quality? Research landscape and challenges. J. Syst. Softw. 2019, 157, 110394. [Google Scholar] [CrossRef]
- Eljasik-Swoboda, T.; Rathgeber, C.; Hasenauer, R. Assessing Technology Readiness for Artificial Intelligence and Machine Learning Based Innovations. In Proceedings of the 8th International Conference on Data Science, Technology and Applications, Prague, Czech Republic, 26–28 July 2019; pp. 281–288. [Google Scholar]
- Homeyer, A.; Lotz, J.; Schwen, L.O.; Weiss, N.; Romberg, D.; Höfener, H.; Zerbe, N.; Hufnagl, P. Artificial Intelligence in Pathology: From Prototype to Product. J. Pathol. Inform. 2021, 12, 13. [Google Scholar] [CrossRef]
- Abels, E.; Pantanowitz, L.; Aeffner, F.; Zarella, M.D.; van der Laak, J.; Bui, M.M.; Vemuri, V.N.; Parwani, A.V.; Gibbs, J.; Agosto-Arroyo, E.; et al. Computational pathology definitions, best practices, and recommendations for regulatory guidance: A white paper from the Digital Pathology Association. J. Pathol. 2019, 249, 286–294. [Google Scholar] [CrossRef]
- Sandbank, J.; Bataillon, G.; Nudelman, A.; Krasnitsky, I.; Mikulinsky, R.; Bien, L.; Thibault, L.; Shach, A.A.; Sebag, G.; Clark, D.P.; et al. Validation and real-world clinical application of an artificial intelligence algorithm for breast cancer detection in biopsies. NPJ Breast Cancer 2022, 8, 129. [Google Scholar] [CrossRef] [PubMed]
- Pantanowitz, L.; Quiroga-Garza, G.M.; Bien, L.; Heled, R.; Laifenfeld, D.; Linhart, C.; Sandbank, J.; Shach, A.A.; Shalev, V.; Vecsler, M.; et al. An artificial intelligence algorithm for prostate cancer diagnosis in whole slide images of core needle biopsies: A blinded clinical validation and deployment study. Lancet Digit. Health 2020, 2, e407–e416. [Google Scholar] [CrossRef] [PubMed]
- Sandbank, J.; Sebag, G.; Linhart, C. Validation and Clinical Deployment of an AI-Based Solution for Detection of Gastric Adenocarcinoma and Helicobacter Pylori in Gastric Biopsies. Lab. Investig. 2022, 102 (Suppl. S1), 419–565. [Google Scholar] [CrossRef]
- Tolkach, Y.; Ovtcharov, V.; Pryalukhin, A.; Eich, M.-L.; Gaisa, N.T.; Braun, M.; Radzhabov, A.; Quaas, A.; Hammerer, P.; Dellmann, A.; et al. An international multi-institutional validation study of the algorithm for prostate cancer detection and Gleason grading. NPJ Precis. Oncol. 2023, 7, 77. [Google Scholar] [CrossRef]
- Kleppe, A.; Skrede, O.-J.; De Raedt, S.; Hveem, T.S.; Askautrud, H.A.; Jacobsen, J.E.; Church, D.N.; Nesbakken, A.; Shepherd, N.A.; Novelli, M.; et al. A clinical decision support system optimising adjuvant chemotherapy for colorectal cancers by integrating deep learning and pathological staging markers: A development and validation study. Lancet Oncol. 2022, 23, 1221–1232. [Google Scholar] [CrossRef]
- Saillard, C.; Dubois, R.; Tchita, O.; Loiseau, N.; Garcia, T.; Adriansen, A.; Carpentier, S.; Reyre, J.; Enea, D.; von Loga, K.; et al. Validation of MSIntuit as an AI-based pre-screening tool for MSI detection from colorectal cancer histology slides. Nat. Commun. 2023, 14, 6695. [Google Scholar] [CrossRef]
- Raciti, P.; Sue, J.; Retamero, J.A.; Ceballos, R.; Godrich, R.; Kunz, J.D.; Casson, A.; Thiagarajan, D.; Ebrahimzadeh, Z.; Viret, J.; et al. Clinical Validation of Artificial Intelligence–Augmented Pathology Diagnosis Demonstrates Significant Gains in Diagnostic Accuracy in Prostate Cancer Detection. Arch. Pathol. Lab. Med. 2023, 147, 1178–1185. [Google Scholar] [CrossRef]
- Eloy, C.; Marques, A.; Pinto, J.; Pinheiro, J.; Campelos, S.; Curado, M.; Vale, J.; Polónia, A. Artificial intelligence–assisted cancer diagnosis improves the efficiency of pathologists in prostatic biopsies. Virchows Arch. 2023, 482, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Shafi, S.; Kellough, D.A.; Lujan, G.; Satturwar, S.; Parwani, A.V.; Li, Z. Integrating and validating automated digital imaging analysis of estrogen receptor immunohistochemistry in a fully digital workflow for clinical use. J. Pathol. Inform. 2022, 13, 100122. [Google Scholar] [CrossRef] [PubMed]
- Anand, D.; Kurian, N.C.; Dhage, S.; Kumar, N.; Rane, S.; Gann, P.H.; Sethi, A. Deep Learning to Estimate Human Epidermal Growth Factor Receptor 2 Status from Hematoxylin and Eosin-Stained Breast Tissue Images. J. Pathol. Inform. 2020, 11, 19. [Google Scholar] [CrossRef]
- Hondelink, L.M.; Hüyük, M.; Postmus, P.E.; Smit, V.T.H.B.M.; Blom, S.; von der Thüsen, J.H.; Cohen, D. Development and validation of a supervised deep learning algorithm for automated whole-slide programmed death-ligand 1 tumour proportion score assessment in non-small cell lung cancer. Histopathology 2022, 80, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.S.; Jin, M.-S.; Park, J.H.; Lee, S.; Cho, J.; Oh, S.; Kwak, T.-Y.; Woo, J.I.; Mun, Y.; Kim, S.W.; et al. Automated Gleason Scoring and Tumor Quantification in Prostate Core Needle Biopsy Images Using Deep Neural Networks and Its Comparison with Pathologist-Based Assessment. Cancers 2019, 11, 1860. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.; Ahlers, S.M.; Stratman, C.; Aridor, O.; Pantanowitz, L.; Fine, J.L.; Kuzmishin, J.A.; Montalto, M.C.; Parwani, A.V. Can digital pathology result in cost savings? A financial projection for digital pathology implementation at a large integrated health care organization. J. Pathol. Inform. 2014, 5, 33. [Google Scholar] [CrossRef]
- Griffin, J.; Treanor, D. Digital pathology in clinical use: Where are we now and what is holding us back? Histopathology 2017, 70, 134–145. [Google Scholar] [CrossRef]
- Hanna, M.G.; Reuter, V.E.; Samboy, J.; England, C.; Corsale, L.; Fine, S.W.; Agaram, N.P.; Stamelos, E.; Yagi, Y.; Hameed, M.; et al. Implementation of Digital Pathology Offers Clinical and Operational Increase in Efficiency and Cost Savings. Arch. Pathol. Lab. Med. 2019, 143, 1545–1555. [Google Scholar] [CrossRef]
- Berbís, M.A.; McClintock, D.S.; Bychkov, A.; Van der Laak, J.; Pantanowitz, L.; Lennerz, J.K.; Cheng, J.Y.; Delahunt, B.; Egevad, L.; Eloy, C.; et al. Computational pathology in 2030: A Delphi study forecasting the role of AI in pathology within the next decade. EBioMedicine 2023, 88, 104427. [Google Scholar] [CrossRef]
- Kacew, A.J.; Strohbehn, G.W.; Saulsberry, L.; Laiteerapong, N.; Cipriani, N.A.; Kather, J.N.; Pearson, A.T. Artificial Intelligence Can Cut Costs While Maintaining Accuracy in Colorectal Cancer Genotyping. Front. Oncol. 2021, 11, 630953. [Google Scholar] [CrossRef]
- Fraggetta, F.; Caputo, A.; Guglielmino, R.; Pellegrino, M.G.; Runza, G.; L’Imperio, V. A Survival Guide for the Rapid Transition to a Fully Digital Workflow: The “Caltagirone Example”. Diagnostics 2021, 11, 1916. [Google Scholar] [CrossRef]
- Fraggetta, F.; Garozzo, S.; Zannoni, G.F.; Pantanowitz, L.; Rossi, E.D. Routine Digital Pathology Workflow: The Catania Experience. J. Pathol. Inform. 2017, 8, 51. [Google Scholar] [CrossRef] [PubMed]
- Schüffler, P.J.; Geneslaw, L.; Yarlagadda, D.V.K.; Hanna, M.G.; Samboy, J.; Stamelos, E.; Vanderbilt, C.; Philip, J.; Jean, M.-H.; Corsale, L.; et al. Integrated digital pathology at scale: A solution for clinical diagnostics and cancer research at a large academic medical center. J. Am. Med. Inform. Assoc. 2021, 28, 1874–1884. [Google Scholar] [CrossRef] [PubMed]
- Montezuma, D.; Monteiro, A.; Fraga, J.; Ribeiro, L.; Gonçalves, S.; Tavares, A.; Monteiro, J.; Macedo-Pinto, I. Digital Pathology Implementation in Private Practice: Specific Challenges and Opportunities. Diagnostics 2022, 12, 529. [Google Scholar] [CrossRef] [PubMed]
- Retamero, J.A.; Aneiros-Fernandez, J.; del Moral, R.G. Complete Digital Pathology for Routine Histopathology Diagnosis in a Multicenter Hospital Network. Arch. Pathol. Lab. Med. 2020, 144, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Hartman, D.; Pantanowitz, L.; McHugh, J.; Piccoli, A.; Oleary, M.; Lauro, G. Enterprise Implementation of Digital Pathology: Feasibility, Challenges, and Opportunities. J. Digit. Imaging 2017, 30, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.C.; Kayamba, V.; Peek, M.R., Jr.; Heimburger, D. Cancer Control in Low- and Middle-Income Countries: Is It Time to Consider Screening? J. Glob. Oncol. 2019, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Nanda, M.; Fronterre, C.; Sewagudde, P.; Ssentongo, A.E.; Yenney, K.; Arhin, N.D.; Oh, J.; Amponsah-Manu, F.; Ssentongo, P. Mapping Cancer in Africa: A Comprehensive and Comparable Characterization of 34 Cancer Types Using Estimates From GLOBOCAN 2020. Front. Public Health 2022, 10, 839835. [Google Scholar] [CrossRef]
- Sayed, S.; Lukande, R.; Fleming, K.A. Providing Pathology Support in Low-Income Countries. J. Glob. Oncol. 2015, 1, 3–6. [Google Scholar] [CrossRef]
- Bychkov, A.; Schubert, M. Constant Demand, Patchy Supply. Pathologist 2023, 88, 18–27. [Google Scholar]
- Wamala, D.; Katamba, A.; Dworak, O. Feasibility and diagnostic accuracy of Internet-based dynamic telepathology between Uganda and Germany. J. Telemed. Telecare 2011, 17, 222–225. [Google Scholar] [CrossRef]
- Pagni, F.; Bono, F.; Di Bella, C.; Faravelli, A.; Cappellini, A. Virtual Surgical Pathology in Underdeveloped Countries: The Zambia Project. Arch. Pathol. Lab. Med. 2011, 135, 215–219. [Google Scholar] [CrossRef]
- Fischer, M.K.; Kayembe, M.K.; Scheer, A.J.; Introcaso, C.E.; Binder, S.W.; Kovarik, C.L. Establishing telepathology in Africa: Lessons from Botswana. J. Am. Acad. Dermatol. 2011, 64, 986–987. [Google Scholar] [CrossRef]
- Montgomery, N.D.; Tomoka, T.; Krysiak, R.; Powers, E.; Mulenga, M.; Kampani, C.; Chimzimu, F.; Owino, M.K.; Dhungel, B.M.; Gopal, S.; et al. Practical Successes in Telepathology Experiences in Africa. Clin. Lab. Med. 2018, 38, 141–150. [Google Scholar] [CrossRef]
- Hamnvåg, H.M.; McHenry, A.; Ahmed, A.; Trabzonlu, L.; Arnold, C.A.; Mirza, K.M. #TwitterHomework During Pathology Electives: Transforming Pathology Pedagogy. Arch. Pathol. Lab. Med. 2021, 145, 1438–1447. [Google Scholar] [CrossRef] [PubMed]
- Folaranmi, O.O.; Ibiyeye, K.M.; Odetunde, O.A.; Kerr, D.A. The Influence of Social Media in Promoting Knowledge Acquisition and Pathology Excellence in Nigeria. Front. Med. 2022, 9, 906950. [Google Scholar] [CrossRef]
- Crane, G.M.; Gardner, J.M. Pathology Image-Sharing on Social Media: Recommendations for Protecting Privacy While Motivating Education. AMA J. Ethics 2016, 18, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Schaumberg, A.J.; Juarez-Nicanor, W.C.; Choudhury, S.J.; Pastrián, L.G.; Pritt, B.S.; Pozuelo, M.P.; Sánchez, R.S.; Ho, K.; Zahra, N.; Sener, B.D.; et al. Interpretable multimodal deep learning for real-time pan-tissue pan-disease pathology search on social media. Mod. Pathol. 2020, 33, 2169–2185. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Bianchi, F.; Yuksekgonul, M.; Montine, T.J.; Zou, J. A visual–language foundation model for pathology image analysis using medical Twitter. Nat. Med. 2023, 29, 2307–2316. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vigdorovits, A.; Köteles, M.M.; Olteanu, G.-E.; Pop, O. Breaking Barriers: AI’s Influence on Pathology and Oncology in Resource-Scarce Medical Systems. Cancers 2023, 15, 5692. https://doi.org/10.3390/cancers15235692
Vigdorovits A, Köteles MM, Olteanu G-E, Pop O. Breaking Barriers: AI’s Influence on Pathology and Oncology in Resource-Scarce Medical Systems. Cancers. 2023; 15(23):5692. https://doi.org/10.3390/cancers15235692
Chicago/Turabian StyleVigdorovits, Alon, Maria Magdalena Köteles, Gheorghe-Emilian Olteanu, and Ovidiu Pop. 2023. "Breaking Barriers: AI’s Influence on Pathology and Oncology in Resource-Scarce Medical Systems" Cancers 15, no. 23: 5692. https://doi.org/10.3390/cancers15235692
APA StyleVigdorovits, A., Köteles, M. M., Olteanu, G.-E., & Pop, O. (2023). Breaking Barriers: AI’s Influence on Pathology and Oncology in Resource-Scarce Medical Systems. Cancers, 15(23), 5692. https://doi.org/10.3390/cancers15235692