Determinants Affecting the Clinical Implementation of a Molecularly Informed Molecular Tumor Board Recommendation: Experience from a Tertiary Cancer Center
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. The Molecular Tumor Board
2.3. Next-Generation Sequencing
2.4. Variant Classification
2.5. Evidence Levels for Biomarker Stratification
2.6. Data Visualization
3. Results
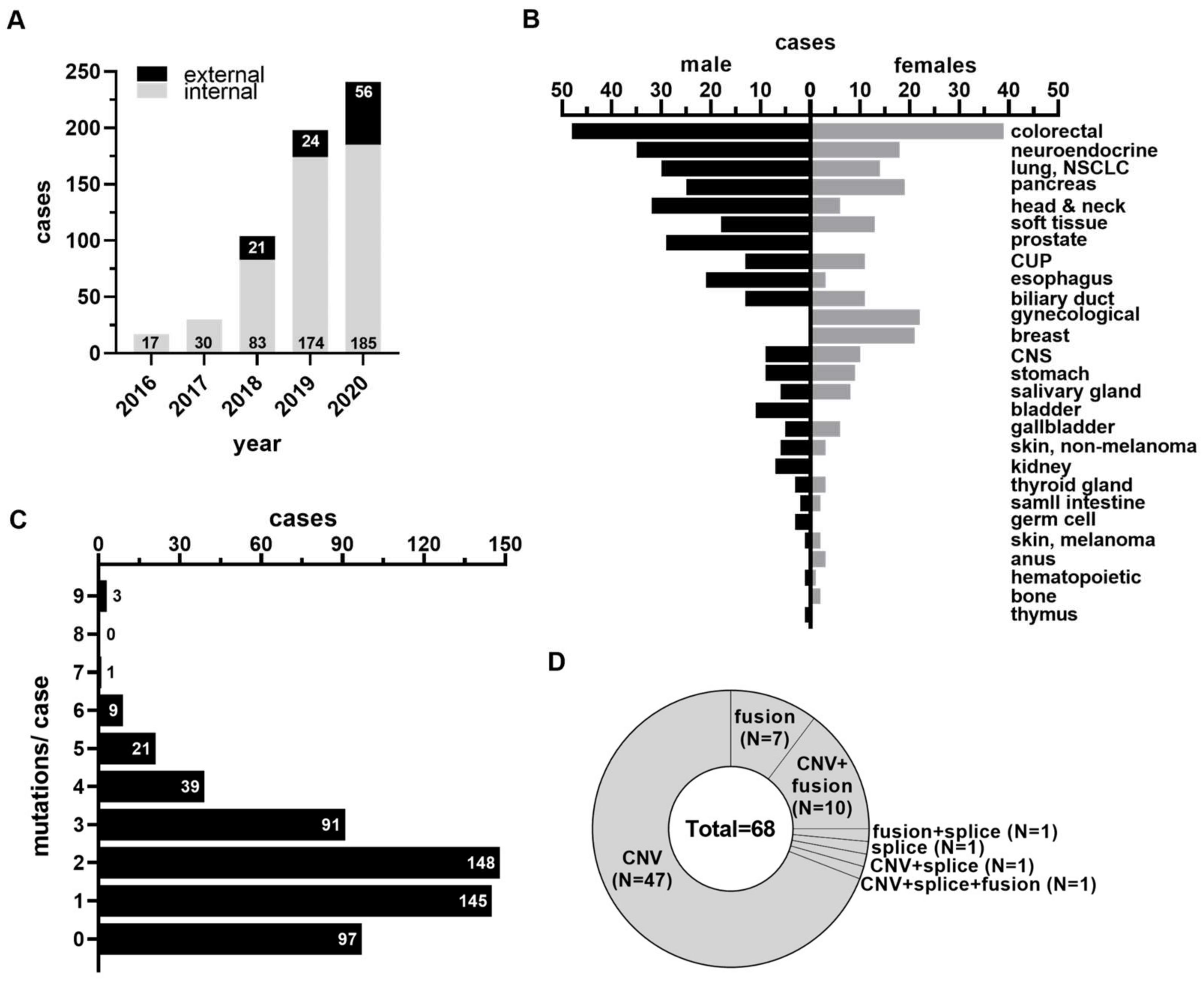
3.1. Study Cohort, Patient Demographics, and Molecular Findings
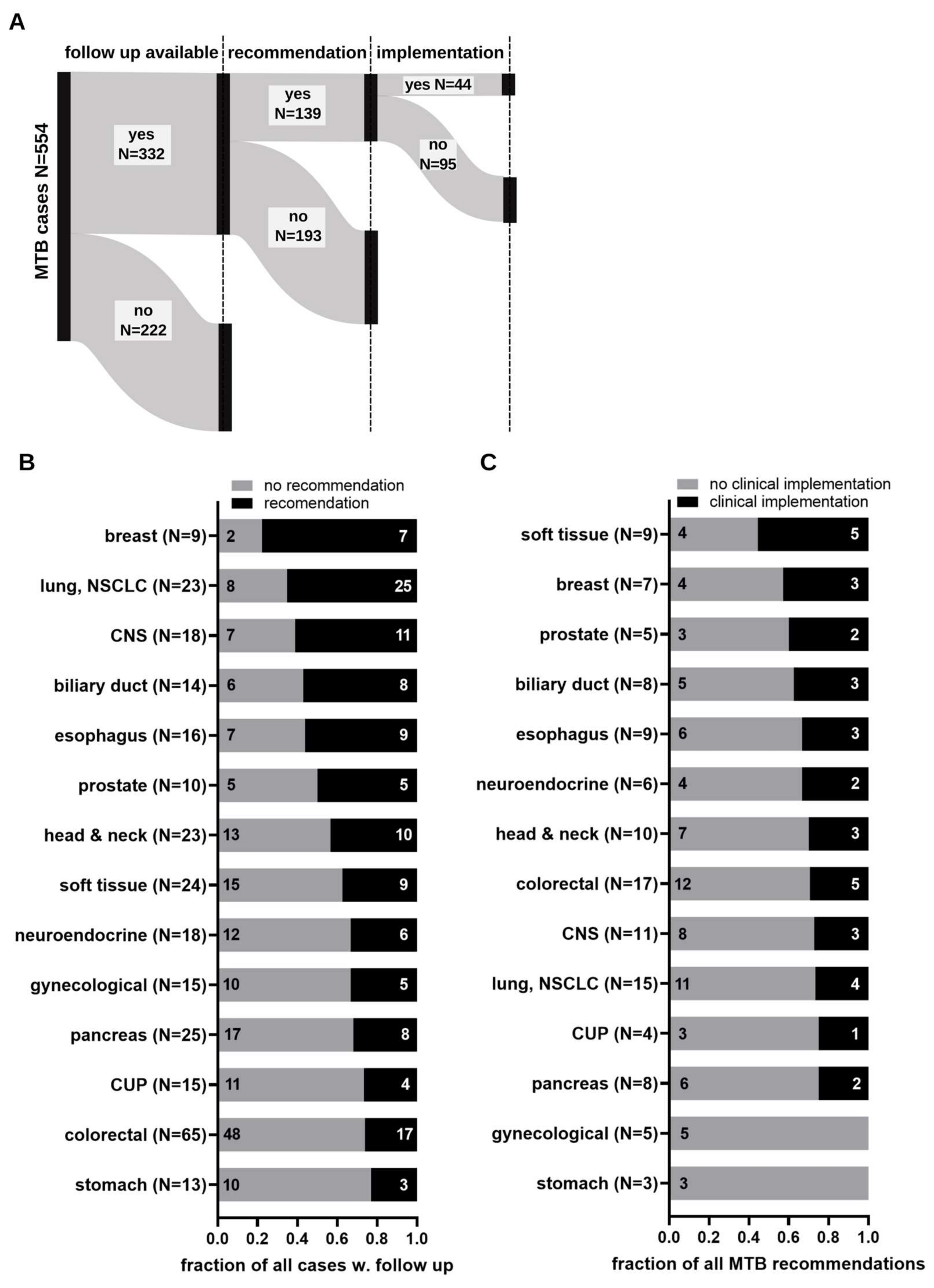
3.2. Implementation of Molecular Tumor Board Recommendations
3.3. Determinants Impacting on Clinical Implementation of Molecular Tumor Board Recommendations
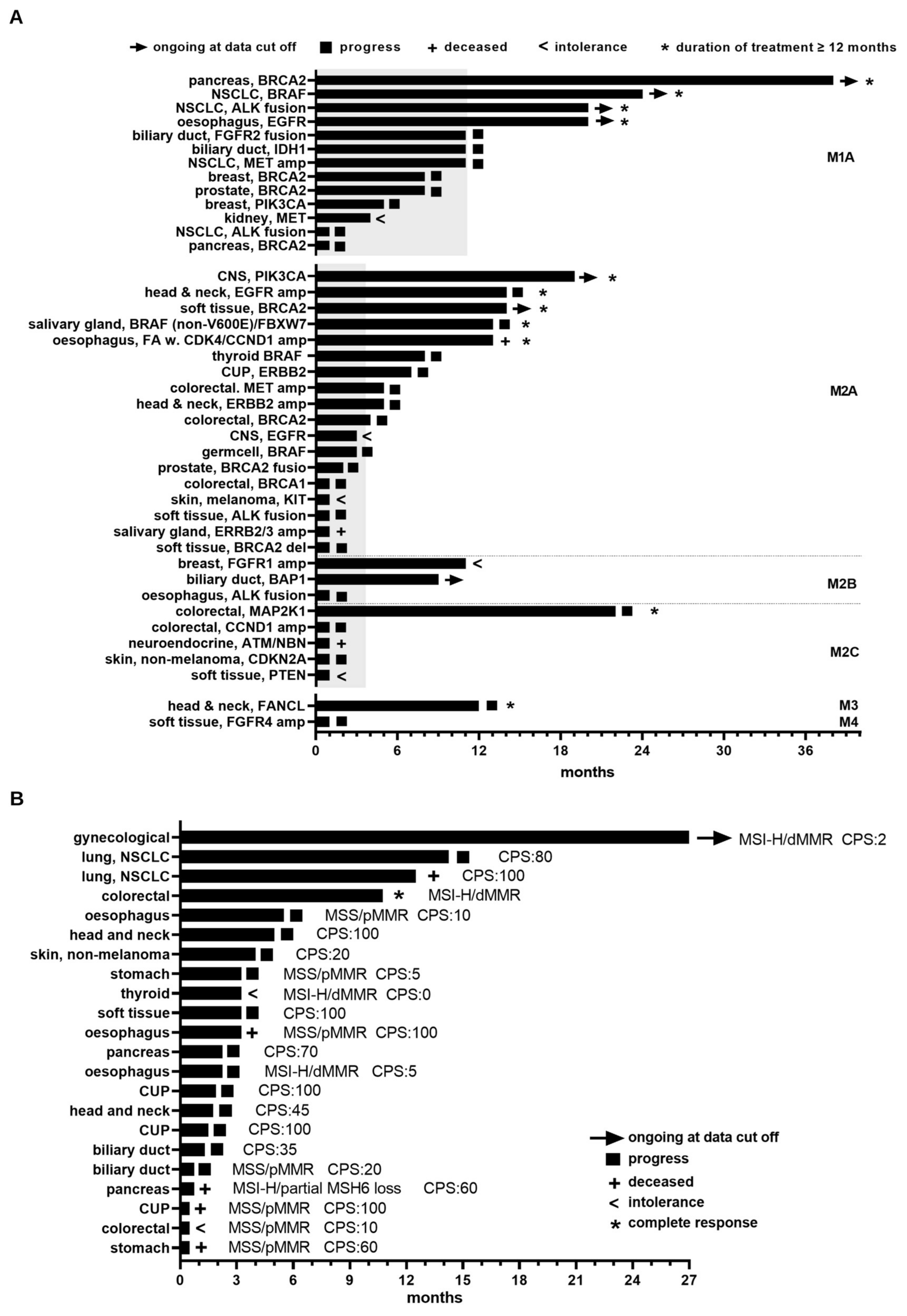
3.4. Clinical Benefit Arising from MTB Recommendations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337.e10. [Google Scholar] [CrossRef] [PubMed]
- Sholl, L.M.; Do, K.; Shivdasani, P.; Cerami, E.; Dubuc, A.M.; Kuo, F.C.; Garcia, E.P.; Jia, Y.; Davineni, P.; Abo, R.P.; et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight 2016, 1, e87062. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, D.; Gao, J.; Phillips, S.M.; Kundra, R.; Zhang, H.; Wang, J.; Rudolph, J.E.; Yaeger, R.; Soumerai, T.; Nissan, M.H.; et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017, 2017, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Walters, M.K.; Ackerman, A.T.; Weese, J.L.; Ruggeri, A.; Mullane, M.P.; Hunt, A.; Wilson, A.; Ramczyk, B.L.; Thompson, M.A. Quantifying the Value of the Molecular Tumor Board: Discordance Recommendation Rate and Drug Cost Avoidance. JCO Precis. Oncol. 2022, 6, e2200132. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Kim, K.H.; Lim, H.J.; Boichard, A.; Nikanjam, M.; Weihe, E.; Kuo, D.J.; Eskander, R.N.; Goodman, A.; Galanina, N.; et al. Real-world data from a molecular tumor board demonstrates improved outcomes with a precision N-of-One strategy. Nat. Commun. 2020, 11, 4965. [Google Scholar] [CrossRef] [PubMed]
- Rodon, J.; Soria, J.C.; Berger, R.; Miller, W.H.; Rubin, E.; Kugel, A.; Tsimberidou, A.; Saintigny, P.; Ackerstein, A.; Brana, I.; et al. Genomic and transcriptomic profiling expands precision cancer medicine: The WINTHER trial. Nat. Med. 2019, 25, 751–758. [Google Scholar] [CrossRef]
- Schwaederle, M.; Parker, B.A.; Schwab, R.B.; Daniels, G.A.; Piccioni, D.E.; Kesari, S.; Helsten, T.L.; Bazhenova, L.A.; Romero, J.; Fanta, P.T.; et al. Precision Oncology: The UC San Diego Moores Cancer Center PREDICT Experience. Mol. Cancer Ther. 2016, 15, 743–752. [Google Scholar] [CrossRef]
- Heinrich, K.; Miller-Phillips, L.; Ziemann, F.; Hasselmann, K.; Ruhlmann, K.; Flach, M.; Biro, D.; von Bergwelt-Baildon, M.; Holch, J.; Herold, T.; et al. Lessons learned: The first consecutive 1000 patients of the CCCMunich(LMU) Molecular Tumor Board. J. Cancer Res. Clin. Oncol. 2023, 149, 1905–1915. [Google Scholar] [CrossRef]
- Hoefflin, R.; Lazarou, A.; Hess, M.E.; Reiser, M.; Wehrle, J.; Metzger, P.; Frey, A.V.; Becker, H.; Aumann, K.; Berner, K.; et al. Transitioning the Molecular Tumor Board from Proof of Concept to Clinical Routine: A German Single-Center Analysis. Cancers 2021, 13, 1151. [Google Scholar] [CrossRef]
- Rieke, D.T.; de Bortoli, T.; Horak, P.; Lamping, M.; Benary, M.; Jelas, I.; Ruter, G.; Berger, J.; Zettwitz, M.; Kagelmann, N.; et al. Feasibility and outcome of reproducible clinical interpretation of high-dimensional molecular data: A comparison of two molecular tumor boards. BMC Med. 2022, 20, 367. [Google Scholar] [CrossRef] [PubMed]
- Scheiter, A.; Hierl, F.; Luke, F.; Keil, F.; Heudobler, D.; Einhell, S.; Klier-Richter, M.; Konstandin, N.P.; Weber, F.; Scheiter, A.; et al. Critical evaluation of molecular tumour board outcomes following 2 years of clinical practice in a Comprehensive Cancer Centre. Br. J. Cancer 2023, 128, 1134–1147. [Google Scholar] [CrossRef] [PubMed]
- Luke, F.; Haller, F.; Utpatel, K.; Krebs, M.; Meidenbauer, N.; Scheiter, A.; Spoerl, S.; Heudobler, D.; Sparrer, D.; Kaiser, U.; et al. Identification of Disparities in Personalized Cancer Care-A Joint Approach of the German WERA Consortium. Cancers 2022, 14, 5040. [Google Scholar] [CrossRef] [PubMed]
- Horak, P.; Heining, C.; Kreutzfeldt, S.; Hutter, B.; Mock, A.; Hullein, J.; Frohlich, M.; Uhrig, S.; Jahn, A.; Rump, A.; et al. Comprehensive Genomic and Transcriptomic Analysis for Guiding Therapeutic Decisions in Patients with Rare Cancers. Cancer Discov. 2021, 11, 2780–2795. [Google Scholar] [CrossRef]
- Bayerisches Landesamt für Gesundheit und Lebensmittelsicherheit. Krebsregister. Available online: https://www.lgl.bayern.de/gesundheit/krebsregister/auswertung_forschung/datenbank/index.htm (accessed on 9 March 2023).
- Eckstein, M.; Agaimy, A.; Woenckhaus, J.; Winter, A.; Bittmann, I.; Janzen, J.; Bertz, S.; Haller, F.; Hartmann, A. DICER1 mutation-positive giant botryoid fibroepithelial polyp of the urinary bladder mimicking embryonal rhabdomyosarcoma. Hum. Pathol. 2019, 84, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Meintker, L.; Haller, F.; Togel, L.; Schmidt, D.; Waibel, H.; Hartmann, A.; Mackensen, A.; Meidenbauer, N. Successful Targeting of BRAF V600E Mutation With Vemurafenib in a Treatment-Resistant Extragonadal Nonseminomatous Germ-Cell Tumor. JCO Precis. Oncol. 2020, 4, 233–238. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Den Dunnen, J.T.; Dalgleish, R.; Maglott, D.R.; Hart, R.K.; Greenblatt, M.S.; McGowan-Jordan, J.; Roux, A.F.; Smith, T.; Antonarakis, S.E.; Taschner, P.E. HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum. Mutat. 2016, 37, 564–569. [Google Scholar] [CrossRef]
- Fokkema, I.; Kroon, M.; Lopez Hernandez, J.A.; Asscheman, D.; Lugtenburg, I.; Hoogenboom, J.; den Dunnen, J.T. The LOVD3 platform: Efficient genome-wide sharing of genetic variants. Eur. J. Hum. Genet. 2021, 29, 1796–1803. [Google Scholar] [CrossRef]
- Tamborero, D.; Dienstmann, R.; Rachid, M.H.; Boekel, J.; Baird, R.; Brana, I.; De Petris, L.; Yachnin, J.; Massard, C.; Opdam, F.L.; et al. Support systems to guide clinical decision-making in precision oncology: The Cancer Core Europe Molecular Tumor Board Portal. Nat. Med. 2020, 26, 992–994. [Google Scholar] [CrossRef]
- De Bruijn, I.; Kundra, R.; Mastrogiacomo, B.; Tran, T.N.; Sikina, L.; Mazor, T.; Li, X.; Ochoa, A.; Zhao, G.; Lai, B.; et al. Analysis and Visualization of Longitudinal Genomic and Clinical Data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. 2023, 83, 3861–3867. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Lee, J.M.; Riley, G.R.; Jang, W.; Rubinstein, W.S.; Church, D.M.; Maglott, D.R. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014, 42, D980–D985. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Lee, K.; Yu, N.; Jang, I.; Choi, I.; Kim, P.; Jang, Y.E.; Kim, B.; Kim, S.; Lee, B.; et al. ChimerDB 3.0: An enhanced database for fusion genes from cancer transcriptome and literature data mining. Nucleic Acids Res. 2017, 45, D784–D789. [Google Scholar] [CrossRef] [PubMed]
- Mitelman, F.; Johansson, B.; Mertens, F. (Eds.) Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. Available online: https://mitelmandatabase.isb-cgc.org (accessed on 9 March 2023).
- Leichsenring, J.; Horak, P.; Kreutzfeldt, S.; Heining, C.; Christopoulos, P.; Volckmar, A.L.; Neumann, O.; Kirchner, M.; Ploeger, C.; Budczies, J.; et al. Variant classification in precision oncology. Int. J. Cancer 2019, 145, 2996–3010. [Google Scholar] [CrossRef] [PubMed]
- Horak, P.; Leichsenring, J.; Kreutzfeldt, S.; Kazdal, D.; Teleanu, V.; Endris, V.; Volckmar, A.L.; Renner, M.; Kirchner, M.; Heilig, C.E.; et al. Variant interpretation in molecular pathology and oncology: An introduction. Pathologe 2021, 42, 369–379. [Google Scholar] [CrossRef]
- Uhrig, S.; Ellermann, J.; Walther, T.; Burkhardt, P.; Frohlich, M.; Hutter, B.; Toprak, U.H.; Neumann, O.; Stenzinger, A.; Scholl, C.; et al. Accurate and efficient detection of gene fusions from RNA sequencing data. Genome Res. 2021, 31, 448–460. [Google Scholar] [CrossRef]
- Pavlidis, N.; Pentheroudakis, G. Cancer of unknown primary site. Lancet 2012, 379, 1428–1435. [Google Scholar] [CrossRef]
- Cobain, E.F.; Chinnaiyan, A.M. Is Universal Next-Generation Sequencing Testing of Patients With Advanced Cancer Ready for Prime Time?-Reply. JAMA Oncol. 2021, 7, 1246–1247. [Google Scholar] [CrossRef] [PubMed]
- Crimini, E.; Repetto, M.; Tarantino, P.; Ascione, L.; Antonarelli, G.; Rocco, E.G.; Barberis, M.; Mazzarella, L.; Curigliano, G. Challenges and Obstacles in Applying Therapeutical Indications Formulated in Molecular Tumor Boards. Cancers 2022, 14, 3193. [Google Scholar] [CrossRef] [PubMed]
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.B.; Barlesi, F.; Lolkema, M.P.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. Available online: https://gco.iarc.fr/today (accessed on 9 March 2023).
- Buttner, R.; Wolf, J.; Kron, A.; Nationales Netzwerk Genomische Medizin. The national Network Genomic Medicine (nNGM): Model for innovative diagnostics and therapy of lung cancer within a public healthcare system. Pathologe 2019, 40, 276–280. [Google Scholar] [CrossRef]
- Nguyen, L.; Martens, J.W.M.; Van Hoeck, A.; Cuppen, E. Pan-cancer landscape of homologous recombination deficiency. Nat. Commun. 2020, 11, 5584. [Google Scholar] [CrossRef]

| Characteristics | Value | |
|---|---|---|
| Period | May 2016–December 2020 | |
| All cases | N (%) | 590 (100) |
| Internal cases | N (%) | 489 (82.9) |
| External cases | N (%) | 101 (17.1) |
| Time diagnosis to MTB inclusion (months) | ||
| All cases | median (Min, Max) | 19 (0, 299) |
| Females | median (Min, Max) | 15 (0, 299) |
| Males | median (Min, Max) | 19 (0, 286) |
| Disease stage | ||
| Local | N (% of All cases) | |
| Relapse | N (% of All cases) | 61 (10.3) |
| Metastasis | N (% of All cases) | 410 (69.5) |
| Previous therapies | ||
| Yes | N (% of All cases) | 490 (83.1) |
| No | N (% of All cases) | 29 (4.9) |
| Not evaluable | N (% of All cases) | 71 (12.0) |
| Yes, females | N (% of All males) | 193 (85.4) |
| Yes, males | N (% of All females) | 297 (90.3) |
| Evaluable cases | ||
| Total | N (% of All cases) | 554 (93.9) |
| Females | N (%) | 226 (40.8) |
| Median age | Yrs (Min, Max) | 57 (16, 88) |
| Males | N (%) | 329 (59.4) |
| Median age | Yrs (Min, Max) | 63 (19, 88) |
| Localization primary tumor | ||
| Colorectal | N (% Evaluable Cases) | 87 (15.7) |
| Neuroendocrine | N (% Evaluable Cases) | 53 (9.6) |
| Lung, NSCLC | N (% Evaluable Cases) | 44 (7.9) |
| Pancreas | N (% Evaluable Cases) | 44 (7.9) |
| Head and neck | N (% Evaluable Cases) | 38 (6.9) |
| Soft tissue | N (% Evaluable Cases) | 31 (5.6) |
| Prostate | N (% Evaluable Cases) | 29 (5.2) |
| CUP | N (% Evaluable Cases) | 26 (4.7) |
| Esophagus | N (% Evaluable Cases) | 24 (4.3) |
| Biliary duct | N (% Evaluable Cases) | 24 (4.3) |
| Gynecological | N (% Evaluable Cases) | 22 (4.0) |
| Breast | N (% Evaluable Cases) | 21 (3.8) |
| CNS | N (% Evaluable Cases) | 19 (3.4) |
| Stomach | N (% Evaluable Cases) | 18 (3.2) |
| Salivary gland | N (% Evaluable Cases) | 14 (2.5) |
| Bladder | N (% Evaluable Cases) | 11 (2.0) |
| Gallbladder | N (% Evaluable Cases) | 11 (2.0) |
| Skin, non-melanoma | N (% Evaluable Cases) | 9 (1.6) |
| Kidney | N (% Evaluable Cases) | 7 (1.3) |
| Thyroid | N (% Evaluable Cases) | 6 (1.1) |
| Small intestinal | N (% Evaluable Cases) | 4 (0.7) |
| Germ cell | N (% Evaluable Cases) | 3 (0.5) |
| Skin, melanoma | N (% Evaluable Cases) | 3 (0.5) |
| Anus | N (% Evaluable Cases) | 3 (0.5) |
| Hematopoietic | N (% Evaluable Cases) | 2 (0.4) |
| Bone | N (% Evaluable Cases) | 2 (0.4) |
| Thymus | N (% Evaluable Cases) | 1 (0.2) |
| Clinical Translation: | Reasons for Non-Adherence (N): | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MTB Recommend 1 | Yes N (%) | No N (%) | OT | SD | PC | PD | NHR | PIR | DU | NA | |
| stomach | 3 | 0 | 3 (100) | 2 | - | 1 | - | - | - | - | - |
| gynecological | 5 | 0 | 5 (100) | 2 | 1 | 1 | - | - | - | 1 | - |
| pancreas | 8 | 2 (25) | 6 (75) | 3 | - | - | - | - | 1 | - | 2 |
| CUP | 4 | 1 (25) | 3 (75) | - | 2 | - | - | - | 1 | - | - |
| CNS | 11 | 3 (27) | 8 (73) | 3 | 2 | 1 | 1 | - | 1 | - | 3 |
| lung, NSCLC | 15 | 4 (27) | 11 (73) | 2 | 1 | 5 | - | - | - | - | - |
| colorectal | 17 | 5 (29) | 12 (71) | 3 | - | 5 | 2 | 2 | - | - | - |
| head and neck | 10 | 3 (30) | 7 (70) | 2 | - | 2 | - | 2 | - | - | 1 |
| neuroendocrine | 6 | 2 (33) | 4 (67) | 2 | - | 1 | - | - | - | 1 | - |
| esophagus | 9 | 3 (33) | 6 (67) | - | - | 1 | 2 | 1 | - | 1 | 1 |
| biliary duct | 8 | 3 (38) | 5 (62) | - | - | 3 | - | 1 | 1 | - | - |
| prostate | 5 | 2 (40) | 3 (60) | 2 | 1 | - | - | - | - | - | - |
| breast | 7 | 3 (43) | 4 (57) | 3 | - | - | 1 | - | - | - | - |
| soft tissue | 9 | 5 (56) | 4 (44) | 3 | - | - | - | - | 1 | - | - |
| esophagus | 9 | 3 (33) | 6 (67) | - | - | 1 | 2 | 1 | - | 1 | 1 |
| Patient | Entity | Clinical Evidence Level | Drug Class/Inhibitor | Targetable Alteration | TTF (Months) |
|---|---|---|---|---|---|
| UKER28 UKER22 UKER62 UKER64 UKER253 UKER335 UKER336 UKER363 UKER361 UKER415 UKER440 UKER462 UKER68 UKER72 UKER73 UKER161 UKER125 UKER86 UKER589 UKER398 UKER391 UKER185 UKER222 UKER231 UKER262 UKER458 UKER488 UKER490 UKER519 UKER533 UKER513 UKER565 UKER29 UKER373 UKER309 UKER61 UKER122 UKER166 UKER298 UKER502 UKER504 UKER523 UKER233 UKER514 | biliary duct biliary duct breast breast kidney lung, NSCLC lung, NSCLC lung, NSCLC lung, NSCLC pancreas pancreas prostate CNS CNS CNS colorectal colorectal colorectal CUP esophagus esophagus germ cell head and neck head and neck melanoma prostate salivary gland salivary gland soft tissue soft tissue soft tissue thyroid biliary duct esophagus neuroendocrine breast colorectal colorectal neuroendocrine skin, non-melanoma skin, non-melanoma soft tissue head and neck soft tissue | M1A M1A M1A M1A M1A M1A M1A M1A M1A M1A M1A M1A M2A M2A M2A M2A M2A M2A M2A M2A M2A M2A M2A M2A M2A M2A M2A M2A M2A M2A M2A M2A M2B M2B M2B M2C M2C M2C M2C M2C M2C M2C M3 M3 | Infigratinib, clinical study Ivosidenib PARP inhibitor Alpelisib Cabozantinib Alectinib Crizotinib Dabrafenib/Trametinib Alectinib Olaparib Olaparib Olaparib Osimertinib Dabrafenib/Trametinib Alpelisib Crizotinib Olaparib Rucaparib Trastuzumab Osimertinib Olaparib Vemurafenib Panitumumab Paclitaxel/Trastuzumab Imatinib Olaparib Trametinib Trastuzumab Emtansine Crizotinib Olaparib Olaparib, after 3 mo. + Pembrolizumab Dabrafenib/Trametinib Olaparib Alectinib Everolimus Ponatinib Palbociclib Panitumumab/Trametinib Olaparib Palbociclib Palbociclib Everolimus Olaparib Ponatinib | PDE3B::FGFR2 gene fusion IDH1 p.Arg132Cys BRCA2 p.Asn3124Ile PIK3CA p.His1047Arg MET p.Met1268Thr EML4::ALK gene fusion MET GCN: 21.2 BRAF p.Val600Glu EML4::ALK gene fusion BRCA2 p.Cys3222Trpfs BRCA2 p.Tyr1894Ter BRCA2 p.Asn3124Ile EGFR p.Leu62Arg/p.Thr263Pro/ EGFR GCN: 33.4 BRAF p.Gly466Glu/NRAS p.Gly12Asp PIK3CA p.Cys420Arg MET GCN: 11.4 BRCA1 p.Ser4LeufsTer18 BRCA2 p.Ser3366AsnfsTer5 ERBB2 p.Arg678Gln/ERBB2 GCN: 4.9 EGFR p.Gly719Ala/EGFR GCN: 19 Franconia Anemia (FA)/ CCND1 GCN: 22.8/CDK4 GCN: 5.5/CCNE1 GCN: 5.2 BRAF p.Val600Glu EGFR GCN: 9.3 ERBB2 GCN: 6.5 KIT p.Ala502_Tyr503dup NBEA::BRCA2 gene fusion BRAF p.Asp594Asn/ FBXW7 p.Val464Met ERBB3 GCN: 3.7 + ERBB2 GCN: 3.2 TNS1::ALK gene fusion BRCA2 deletion BRCA2 p.Asn3124Ile BRAF p.Val600Glu BAP1 p.Ser37ArgfsTer47 BRE::ALK gene fusion TSC2 p.Leu234SerfsTer60 FGFR1 GCN: 7.5/FGF3 GCN: 5.3/ FGF19 GCN: 4.8/FGF4 GCN: 4.5 CCND1 GCN: 10.3 MAP2K1 p.Lys57Glu ATM Splice Acceptor/ NBN p.Lys219AsnfsTer16 CDKN2A p.Asp84Val CDKN2A p.Val82ArgfsTer44/ CDKN2A p.Arg80Ter PTEN p.Met134del FANCL p.Tyr111Cys FGFR4 GCN: 4.5 | 11 11 8 5 4 1 11 24 20 38 1 8 3 0 19 5 1 4 7 20 13 3 14 5 1 2 13 1 1 1 14 8 9 1 0 11 1 22 1 1 0 1 12 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tögel, L.; Schubart, C.; Lettmaier, S.; Neufert, C.; Hoyer, J.; Wolff, K.; Moskalev, E.A.; Stöhr, R.; Agaimy, A.; Reis, A.; et al. Determinants Affecting the Clinical Implementation of a Molecularly Informed Molecular Tumor Board Recommendation: Experience from a Tertiary Cancer Center. Cancers 2023, 15, 5892. https://doi.org/10.3390/cancers15245892
Tögel L, Schubart C, Lettmaier S, Neufert C, Hoyer J, Wolff K, Moskalev EA, Stöhr R, Agaimy A, Reis A, et al. Determinants Affecting the Clinical Implementation of a Molecularly Informed Molecular Tumor Board Recommendation: Experience from a Tertiary Cancer Center. Cancers. 2023; 15(24):5892. https://doi.org/10.3390/cancers15245892
Chicago/Turabian StyleTögel, Lars, Christoph Schubart, Sebastian Lettmaier, Clemens Neufert, Juliane Hoyer, Kerstin Wolff, Evgeny A Moskalev, Robert Stöhr, Abbas Agaimy, André Reis, and et al. 2023. "Determinants Affecting the Clinical Implementation of a Molecularly Informed Molecular Tumor Board Recommendation: Experience from a Tertiary Cancer Center" Cancers 15, no. 24: 5892. https://doi.org/10.3390/cancers15245892





