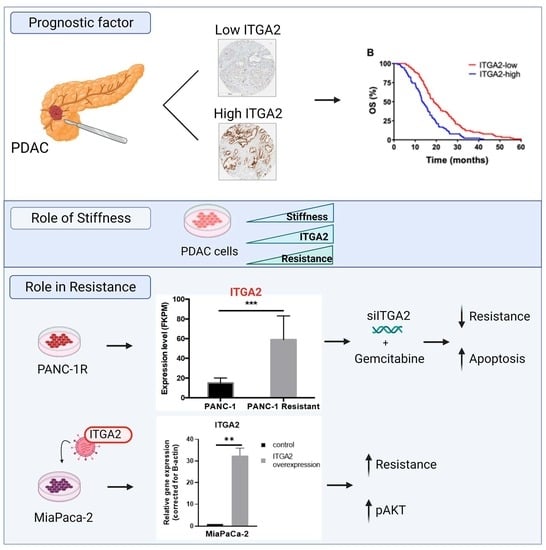
Prognostic Significance of Integrin Subunit Alpha 2 (ITGA2) and Role of Mechanical Cues in Resistance to Gemcitabine in Pancreatic Ductal Adenocarcinoma (PDAC)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Culture Conditions
2.2. Evaluation of ITGA2 mRNA Expression in Human Normal and Tumor Tissues and Correlation with Survival in PDAC
2.3. Evaluation of ITGA2 mRNA and Protein Expression in Two Independent Cohorts of PDAC Ppatients
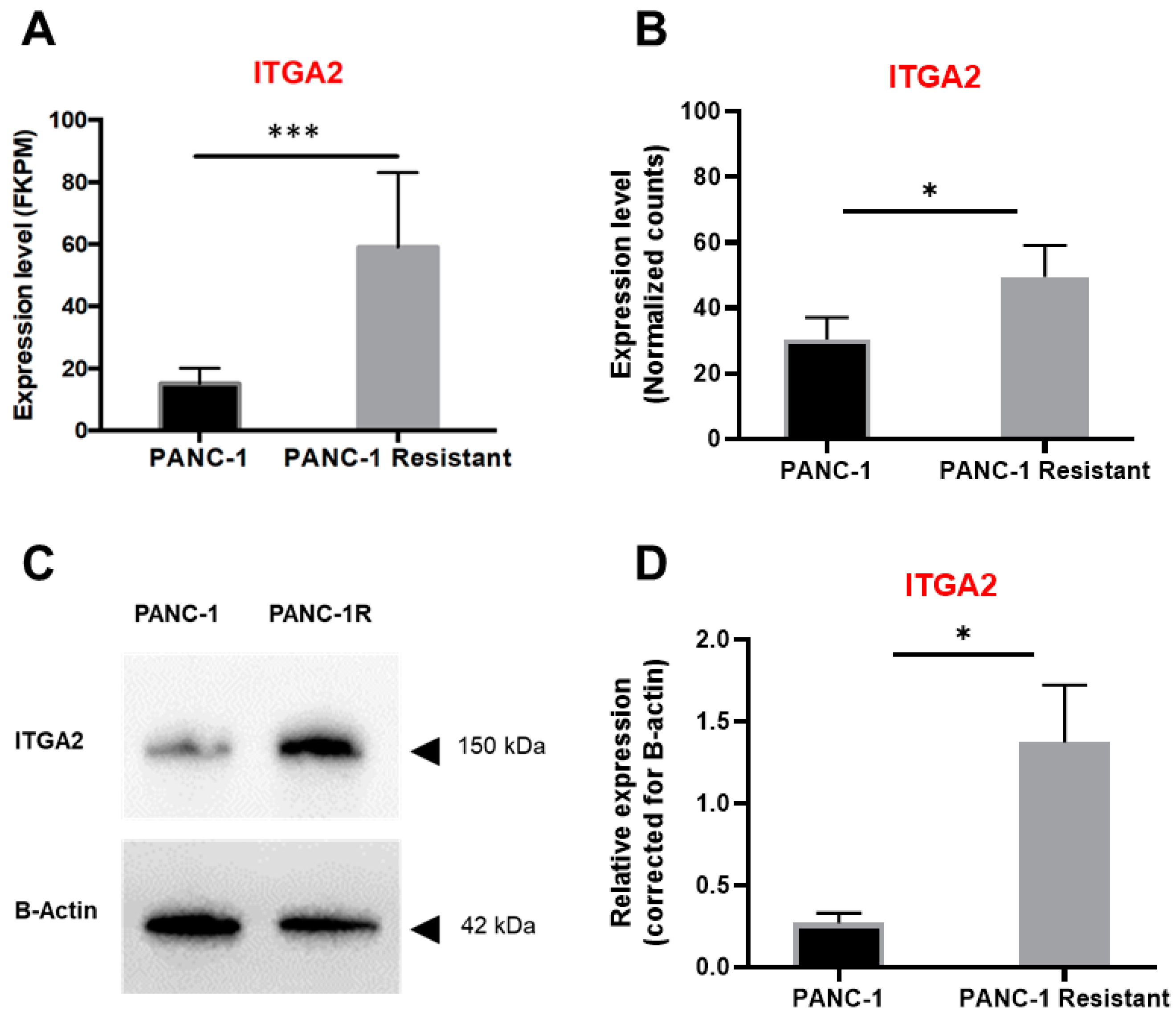
2.4. Evaluation of PANC-1 and Its Gemcitabine-Resistant Clone (PANC-1R) by RNA-Sequencing and Label-Free Proteomics
2.5. RT-qPCR, Western Blot and Immunocytochemistry of Cells Growing as Monolayer or in 3D
2.6. Drug Sensitivity, Genetic Knockdown and Overexpression Experiments
2.7. Wound-Healing and Invasion Assay in 3D Collagen Matrix
2.8. Apoptosis Assay
2.9. Analysis of Phospho-AKT by Enzyme Linked Immunosorbent (ELISA) Assay
2.10. In Vivo
2.11. Statistical Analysis
3. Results
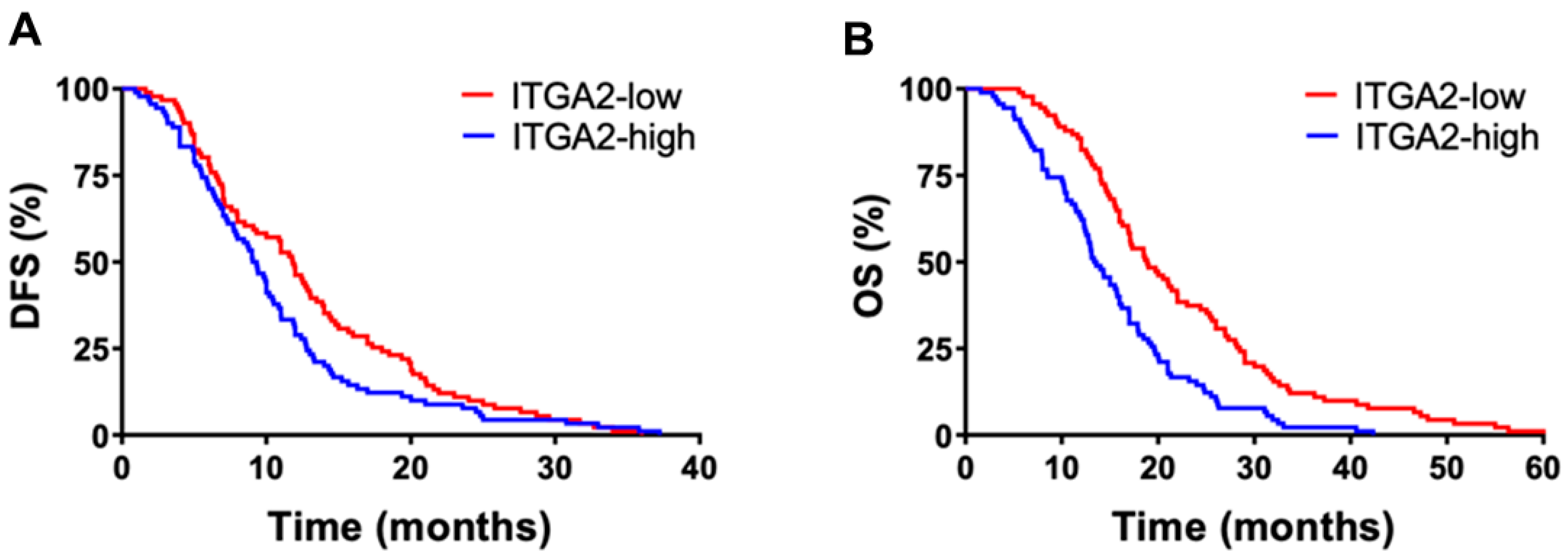
3.1. High Expression Levels of ITGA2 Are an Unfavorable Prognostic Factor in Gemcitabine-Treated Patients
3.2. High Expression Levels of ITGA2 Are Associated with Gemcitabine Resistance and Stiffness
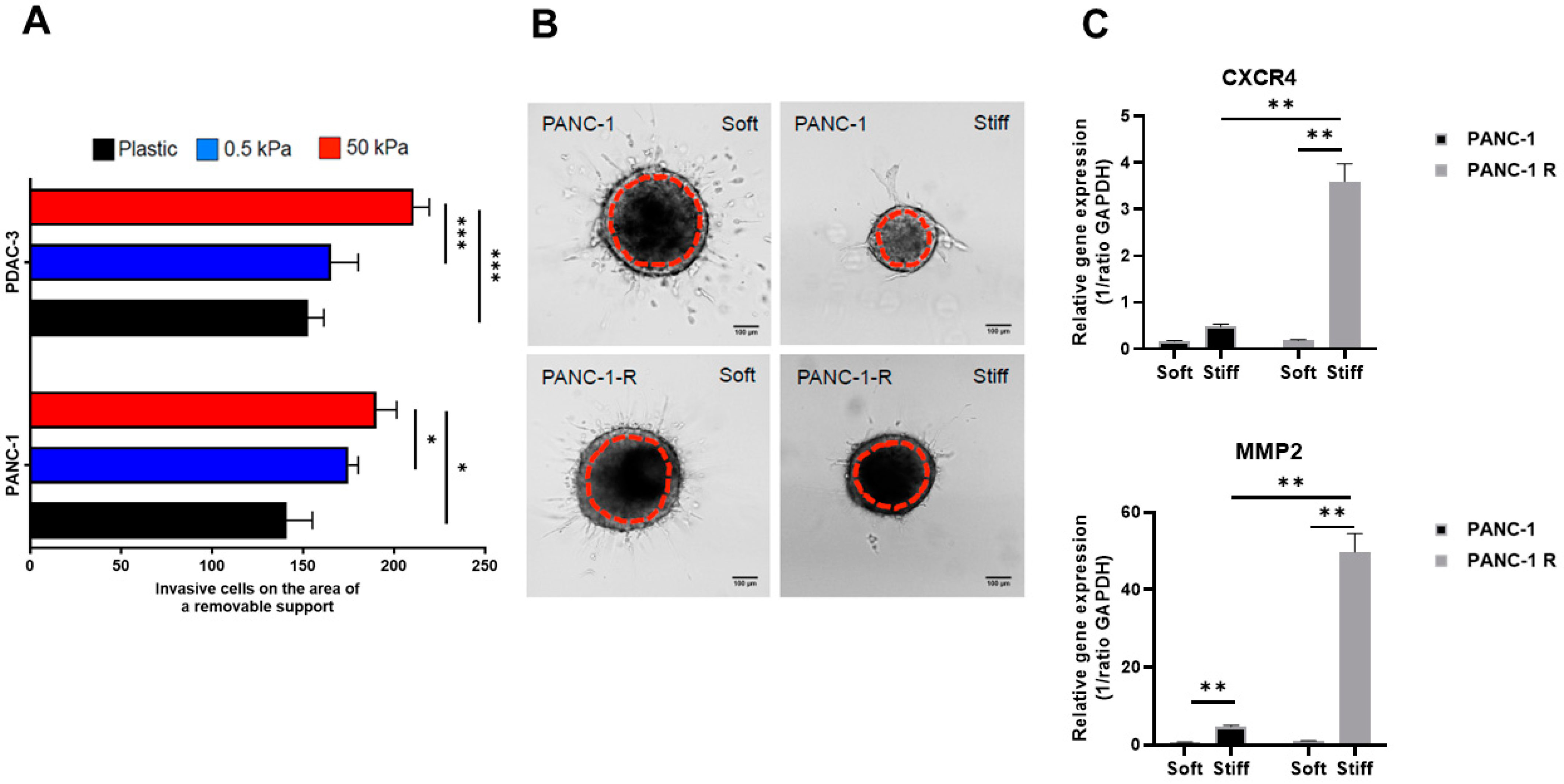
3.3. Substrate Stiffness Influences Invasive Behavior in 2D and 3D
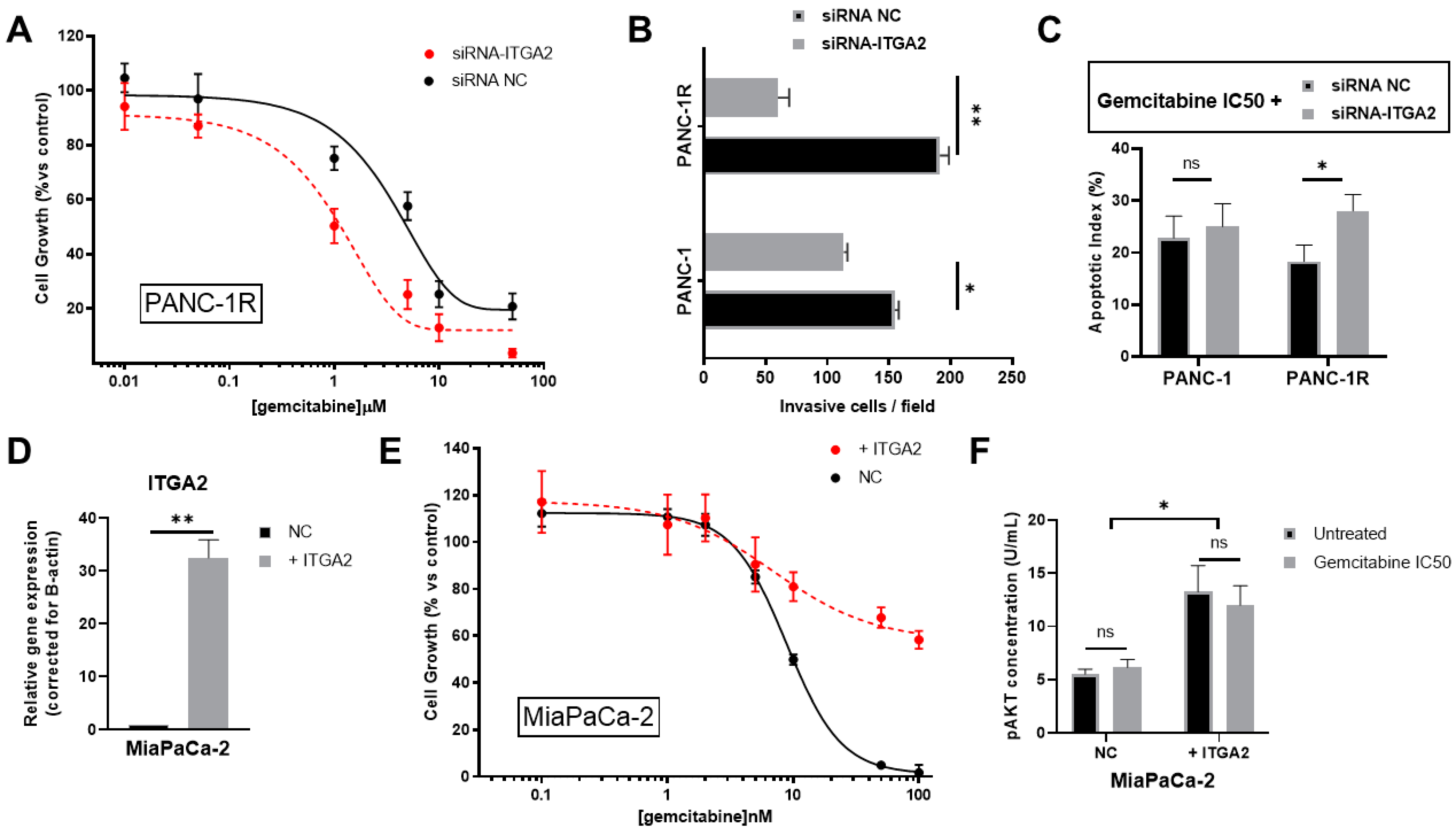
3.4. ITGA2 Expression Levels Modulate Gemcitabine Sensitivity In Vitro
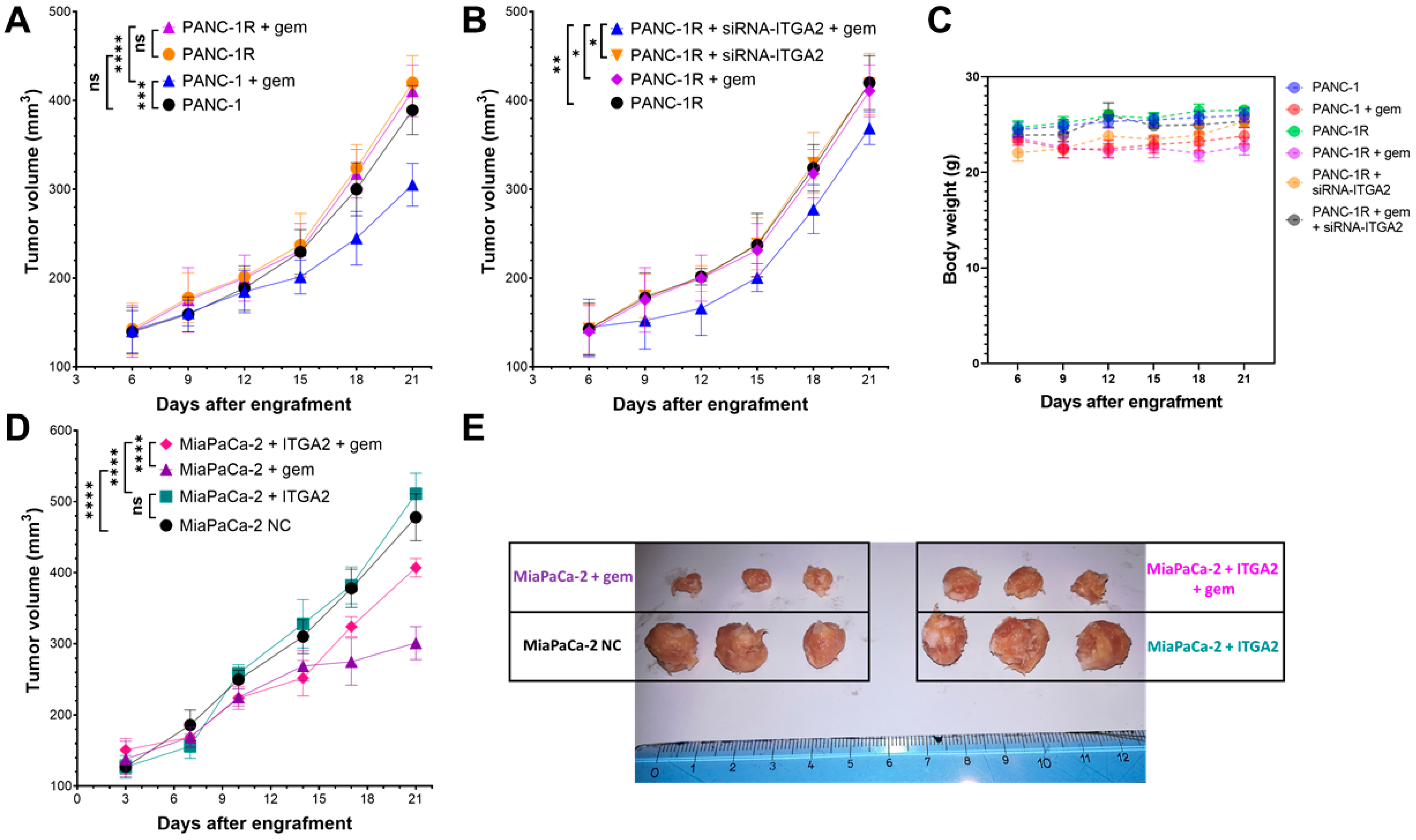
3.5. ITGA2 Expression Levels Modulate Gemcitabine Sensitivity In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miller, K.D.; Nogueira, L.; Devasia, T.; Mariotto, A.B.; Yabroff, K.R.; Jemal, A.; Kramer, J.; Siegel, R.L. Cancer Treatment and Survivorship Statistics, 2022. CA Cancer J. Clin. 2022, 72, 409–436. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Sarantis, P.; Koustas, E.; Papadimitropoulou, A.; Papavassiliou, A.G.; Karamouzis, M.V. Pancreatic Ductal Adenocarcinoma: Treatment Hurdles, Tumor Microenvironment and Immunotherapy. World J. Gastrointest. Oncol. 2020, 12, 173–181. [Google Scholar] [CrossRef]
- Milella, M.; Bassi, C.; Boggi, U.; Brunetti, O.; Cavaliere, A.; Crippa, S.; De Vita, F.; Falconi, M.; Frassineti, G.L.; Giommoni, E.; et al. Evolving Pancreatic Cancer Treatment: From Diagnosis to Healthcare Management. Crit. Rev. Oncol. Hematol. 2022, 169, 103571. [Google Scholar] [CrossRef]
- Caparello, C.; Meijer, L.L.; Garajova, I.; Falcone, A.; Le Large, T.Y.; Funel, N.; Kazemier, G.; Peters, G.J.; Vasile, E.; Giovannetti, E. FOLFIRINOX and Translational Studies: Towards Personalized Therapy in Pancreatic Cancer. World J. Gastroenterol. 2016, 22, 6987. [Google Scholar] [CrossRef]
- Coppola, S.; Carnevale, I.; Danen, E.H.J.; Peters, G.J.; Schmidt, T.; Assaraf, Y.G.; Giovannetti, E. A Mechanopharmacology Approach to Overcome Chemoresistance in Pancreatic Cancer. Drug Resist. Updat. 2017, 31, 43–51. [Google Scholar] [CrossRef]
- Beatty, G.L.; Werba, G.; Lyssiotis, C.A.; Simeone, D.M. The Biological Underpinnings of Therapeutic Resistance in Pancreatic Cancer. Genes Dev. 2021, 35, 940–962. [Google Scholar] [CrossRef]
- Liot, S.; Balas, J.; Aubert, A.; Prigent, L.; Mercier-Gouy, P.; Verrier, B.; Bertolino, P.; Hennino, A.; Valcourt, U.; Lambert, E. Stroma Involvement in Pancreatic Ductal Adenocarcinoma: An Overview Focusing on Extracellular Matrix Proteins. Front. Immunol. 2021, 12, 612271. [Google Scholar] [CrossRef]
- Nabavizadeh, A.; Payen, T.; Iuga, A.C.; Sagalovskiy, I.R.; Desrouilleres, D.; Saharkhiz, N.; Palermo, C.F.; Sastra, S.A.; Oberstein, P.E.; Rosario, V.; et al. Noninvasive Young’s Modulus Visualization of Fibrosis Progression and Delineation of Pancreatic Ductal Adenocarcinoma (PDAC) Tumors Using Harmonic Motion Elastography (HME) in Vivo. Theranostics 2020, 10, 4614–4626. [Google Scholar] [CrossRef]
- Zhu, L.; Guo, J.; Jin, Z.; Xue, H.; Dai, M.; Zhang, W.; Sun, Z.; Xu, J.; Marticorena Garcia, S.R.; Asbach, P.; et al. Distinguishing Pancreatic Cancer and Autoimmune Pancreatitis with in Vivo Tomoelastography. Eur. Radiol. 2021, 31, 3366–3374. [Google Scholar] [CrossRef]
- Whatcott, C.J.; Diep, C.H.; Jiang, P.; Watanabe, A.; LoBello, J.; Sima, C.; Hostetter, G.; Shepard, H.M.; Von Hoff, D.D.; Han, H. Desmoplasia in Primary Tumors and Metastatic Lesions of Pancreatic Cancer. Clin. Cancer Res. 2015, 21, 3561–3568. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.V.; Nyberg, K.D.; Scott, M.B.; Welsh, A.M.; Nguyen, A.H.; Wu, N.; Hohlbauch, S.V.; Geisse, N.A.; Gibb, E.A.; Robertson, A.G.; et al. Stiffness of Pancreatic Cancer Cells Is Associated with Increased Invasive Potential. Integr. Biol. 2016, 8, 1232–1245. [Google Scholar] [CrossRef] [PubMed]
- Rice, A.J.; Cortes, E.; Lachowski, D.; Cheung, B.C.H.; Karim, S.A.; Morton, J.P.; Del Río Hernández, A. Matrix Stiffness Induces Epithelial-Mesenchymal Transition and Promotes Chemoresistance in Pancreatic Cancer Cells. Oncogenesis 2017, 6, e352. [Google Scholar] [CrossRef] [PubMed]
- Kechagia, J.Z.; Ivaska, J.; Roca-Cusachs, P. Integrins as Biomechanical Sensors of the Microenvironment. Nat. Rev. Mol. Cell Biol. 2019, 20, 457–473. [Google Scholar] [CrossRef]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in Cancer: Biological Implications and Therapeutic Opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Hamidi, H.; Ivaska, J. Every Step of the Way: Integrins in Cancer Progression and Metastasis. Nat. Rev. Cancer 2018, 18, 533–548. [Google Scholar] [CrossRef]
- Huang, W.; Zhu, J.; Shi, H.; Wu, Q.; Zhang, C. ITGA2 Overexpression Promotes Esophageal Squamous Cell Carcinoma Aggression via FAK/AKT Signaling Pathway. OncoTargets Ther. 2021, 14, 3583–3596. [Google Scholar] [CrossRef]
- Ma, L.; Sun, Y.; Li, D.; Li, H.; Jin, X.; Ren, D. Overexpressed ITGA2 Contributes to Paclitaxel Resistance by Ovarian Cancer Cells through the Activation of the AKT/FoxO1 Pathway. Aging 2020, 12, 5336–5351. [Google Scholar] [CrossRef]
- Ren, D.; Zhao, J.; Sun, Y.; Li, D.; Meng, Z.; Wang, B.; Fan, P.; Liu, Z.; Jin, X.; Wu, H. Overexpressed ITGA2 Promotes Malignant Tumor Aggression by Up-Regulating PD-L1 Expression through the Activation of the STAT3 Signaling Pathway. J. Exp. Clin. Cancer Res. 2019, 38, 485. [Google Scholar] [CrossRef] [PubMed]
- Adorno-Cruz, V.; Liu, H. Regulation and Functions of Integrin A2 in Cell Adhesion and Disease. Genes Dis. 2019, 6, 16–24. [Google Scholar] [CrossRef]
- Lei, X.; Chen, G.; Li, J.; Wen, W.; Gong, J.; Fu, J. Comprehensive Analysis of Abnormal Expression, Prognostic Value and Oncogenic Role of the Hub Gene FN1 in Pancreatic Ductal Adenocarcinoma via Bioinformatic Analysis and in Vitro Experiments. PeerJ 2021, 9, e12141. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Kitagawa, T.; Baron, B.; Abiko, Y.; Chiba, I.; Kuramitsu, Y. ITGA2, LAMB3, and LAMC2 May Be the Potential Therapeutic Targets in Pancreatic Ductal Adenocarcinoma: An Integrated Bioinformatics Analysis. Sci. Rep. 2021, 11, 10563. [Google Scholar] [CrossRef] [PubMed]
- Massihnia, D.; Avan, A.; Funel, N.; Maftouh, M.; van Krieken, A.; Granchi, C.; Raktoe, R.; Boggi, U.; Aicher, B.; Minutolo, F.; et al. Phospho-Akt Overexpression Is Prognostic and Can Be Used to Tailor the Synergistic Interaction of Akt Inhibitors with Gemcitabine in Pancreatic Cancer. J. Hematol. Oncol. 2017, 10, 9. [Google Scholar] [CrossRef]
- Yang, G.; Guan, W.; Cao, Z.; Guo, W.; Xiong, G.; Zhao, F.; Feng, M.; Qiu, J.; Liu, Y.; Zhang, M.Q.; et al. Integrative Genomic Analysis of Gemcitabine Resistance in Pancreatic Cancer by Patient-Derived Xenograft Models. Clin. Cancer Res. 2021, 27, 3383–3396. [Google Scholar] [CrossRef]
- Quint, K.; Tonigold, M.; Di Fazio, P.; Montalbano, R.; Lingelbach, S.; Rückert, F.; Alinger, B.; Ocker, M.; Neureiter, D. Pancreatic Cancer Cells Surviving Gemcitabine Treatment Express Markers of Stem Cell Differentiation and Epithelial-Mesenchymal Transition. Int. J. Oncol. 2012, 41, 2093–2102. [Google Scholar] [CrossRef]
- Avan, A.; Caretti, V.; Funel, N.; Galvani, E.; Maftouh, M.; Honeywell, R.J.; Lagerweij, T.; Van Tellingen, O.; Campani, D.; Fuchs, D.; et al. Crizotinib Inhibits Metabolic Inactivation of Gemcitabine in C-Met–Driven Pancreatic Carcinoma. Cancer Res. 2013, 73, 6745. [Google Scholar] [CrossRef]
- Giovannetti, E.; Wang, Q.; Avan, A.; Funel, N.; Lagerweij, T.; Lee, J.-H.; Caretti, V.; van der Velde, A.; Boggi, U.; Wang, Y.; et al. Role of CYB5A in Pancreatic Cancer Prognosis and Autophagy Modulation. J. Natl. Cancer Inst. 2014, 106, djt346. [Google Scholar] [CrossRef]
- Sciarrillo, R.; Wojtuszkiewicz, A.; Kooi, I.E.; Gómez, V.E.; Boggi, U.; Jansen, G.; Kaspers, G.-J.; Cloos, J.; Giovannetti, E. Using RNA-Sequencing to Detect Novel Splice Variants Related to Drug Resistance in In Vitro Cancer Models. J. Vis. Exp. 2016, 118, 54714. [Google Scholar] [CrossRef]
- Sciarrillo, R.; Wojtuszkiewicz, A.; El Hassouni, B.; Funel, N.; Gandellini, P.; Lagerweij, T.; Buonamici, S.; Blijlevens, M.; Zeeuw van der Laan, E.A.; Zaffaroni, N.; et al. Splicing Modulation as Novel Therapeutic Strategy against Diffuse Malignant Peritoneal Mesothelioma. EBioMedicine 2019, 39, 215–225. [Google Scholar] [CrossRef]
- Firuzi, O.; Che, P.P.; El Hassouni, B.; Buijs, M.; Coppola, S.; Löhr, M.; Funel, N.; Heuchel, R.; Carnevale, I.; Schmidt, T.; et al. Role of C-MET Inhibitors in Overcoming Drug Resistance in Spheroid Models of Primary Human Pancreatic Cancer and Stellate Cells. Cancers 2019, 11, 638. [Google Scholar] [CrossRef]
- Le Large, T.Y.S.; El Hassouni, B.; Funel, N.; Kok, B.; Piersma, S.R.; Pham, T.V.; Olive, K.P.; Kazemier, G.; van Laarhoven, H.W.M.; Jimenez, C.R.; et al. Proteomic Analysis of Gemcitabine-Resistant Pancreatic Cancer Cells Reveals That Microtubule-Associated Protein 2 Upregulation Associates with Taxane Treatment. Ther. Adv. Med. Oncol. 2019, 11, 175883591984123. [Google Scholar] [CrossRef]
- Ying, X.; Jing, L.; Ma, S.; Li, Q.; Luo, X.; Pan, Z.; Feng, Y.; Feng, P. GSK3β Mediates Pancreatic Cancer Cell Invasion in Vitro via the CXCR4/MMP-2 Pathway. Cancer Cell Int. 2015, 15, 70. [Google Scholar] [CrossRef]
- El Hassouni, B.; Mantini, G.; Immordino, B.; Peters, G.J.; Giovannetti, E. CX-5461 Inhibits Pancreatic Ductal Adenocarcinoma Cell Growth, Migration and Induces DNA Damage. Molecules 2019, 24, 4445. [Google Scholar] [CrossRef]
- Hubeek, I.; Giovannetti, E.; Broekhuizen, A.J.F.; Pastor-Anglada, M.; Kaspers, G.J.L.; Peters, G.J. Immunocytochemical Detection of HENT1 and HCNT1 in Normal Tissues, Lung Cancer Cell Lines, and NSCLC Patient Samples. Nucleosides Nucleotides Nucleic Acids 2008, 27, 787–793. [Google Scholar] [CrossRef]
- Wolcott, K.M.; Woodard, G.E. CD34 Positive Cells Isolated from Traumatized Human Skeletal Muscle Require the CD34 Protein for Multi-Potential Differentiation. Cell. Signal. 2020, 74, 109711. [Google Scholar] [CrossRef]
- Truong, H.H.; de Sonneville, J.; Ghotra, V.P.S.; Xiong, J.; Price, L.; Hogendoorn, P.C.W.; Spaink, H.H.; van de Water, B.; Danen, E.H.J. Automated Microinjection of Cell-Polymer Suspensions in 3D ECM Scaffolds for High-Throughput Quantitative Cancer Invasion Screens. Biomaterials 2012, 33, 181–188. [Google Scholar] [CrossRef]
- Rajan, N.; Habermehl, J.; Coté, M.-F.; Doillon, C.J.; Mantovani, D. Preparation of Ready-to-Use, Storable and Reconstituted Type I Collagen from Rat Tail Tendon for Tissue Engineering Applications. Nat. Protoc. 2006, 1, 2753–2758. [Google Scholar] [CrossRef]
- Truong, H.H.; Xiong, J.; Ghotra, V.P.S.; Nirmala, E.; Haazen, L.; Le Dévédec, S.E.; Balcioğlu, H.E.; He, S.; Snaar-Jagalska, B.E.; Vreugdenhil, E.; et al. Β1 Integrin Inhibition Elicits a Prometastatic Switch Through the TGFβ–MiR-200–ZEB Network in E-Cadherin–Positive Triple-Negative Breast Cancer. Sci. Signal. 2014, 7, ra15. [Google Scholar] [CrossRef]
- Balcioglu, H.E.; van de Water, B.; Danen, E.H.J. Tumor-Induced Remote ECM Network Orientation Steers Angiogenesis. Sci. Rep. 2016, 6, 22580. [Google Scholar] [CrossRef]
- Giovannetti, E.; Mey, V.; Danesi, R.; Mosca, I.; Del Tacca, M. Synergistic Cytotoxicity and Pharmacogenetics of Gemcitabine and Pemetrexed Combination in Pancreatic Cancer Cell Lines. Clin. Cancer Res. 2004, 10, 2936–2943. [Google Scholar] [CrossRef]
- Bononi, G.; Di Stefano, M.; Poli, G.; Ortore, G.; Meier, P.; Masetto, F.; Caligiuri, I.; Rizzolio, F.; Macchia, M.; Chicca, A.; et al. Reversible Monoacylglycerol Lipase Inhibitors: Discovery of a New Class of Benzylpiperidine Derivatives. J. Med. Chem. 2022, 65, 7118–7140. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, O.; Papini, F.; Mantini, G.; Gregori, A.; Parrino, B.; Liu, D.S.K.; Cascioferro, S.; Carbone, D.; Peters, G.J.; Frampton, A.E.; et al. “Open Sesame?”: Biomarker Status of the Human Equilibrative Nucleoside Transporter-1 and Molecular Mechanisms Influencing Its Expression and Activity in the Uptake and Cytotoxicity of Gemcitabine in Pancreatic Cancer. Cancers 2020, 12, 3206. [Google Scholar] [CrossRef] [PubMed]
- Weiss, F.; Lauffenburger, D.; Friedl, P. Towards Targeting of Shared Mechanisms of Cancer Metastasis and Therapy Resistance. Nat. Rev. Cancer 2022, 22, 157–173. [Google Scholar] [CrossRef]
- Ellenrieder, V.; Alber, B.; Lacher, U.; Hendler, S.F.; Menke, A.; Boeck, W.; Wagner, M.; Wilda, M.; Friess, H.; Büchler, M.; et al. Role of MT-MMPs and MMP-2 in Pancreatic Cancer Progression. Int. J. Cancer 2000, 85, 14–20. [Google Scholar] [CrossRef]
- Marchesi, F.; Monti, P.; Leone, B.E.; Zerbi, A.; Vecchi, A.; Piemonti, L.; Mantovani, A.; Allavena, P. Increased Survival, Proliferation, and Migration in Metastatic Human Pancreatic Tumor Cells Expressing Functional CXCR4. Cancer Res. 2004, 64, 8420–8427. [Google Scholar] [CrossRef] [PubMed]
- Qin, A.; Liu, Q.; Wang, J. Ropivacaine Inhibits Proliferation, Invasion, Migration and Promotes Apoptosis of Papillary Thyroid Cancer Cells via Regulating ITGA2 Expression. Drug Dev. Res. 2020, 81, 700–707. [Google Scholar] [CrossRef]
- Juin, A.; Billottet, C.; Moreau, V.; Destaing, O.; Albiges-Rizo, C.; Rosenbaum, J.; Génot, E.; Saltel, F. Physiological Type I Collagen Organization Induces the Formation of a Novel Class of Linear Invadosomes. Mol. Biol. Cell 2012, 23, 297–309. [Google Scholar] [CrossRef]
- Sliker, B.H.; Goetz, B.T.; Barnes, R.; King, H.; Maurer, H.C.; Olive, K.P.; Solheim, J.C. HLA-B Influences Integrin Beta-1 Expression and Pancreatic Cancer Cell Migration. Exp. Cell Res. 2020, 390, 111960. [Google Scholar] [CrossRef]
- Bergonzini, C.; Kroese, K.; Zweemer, A.J.M.; Danen, E.H.J. Targeting Integrins for Cancer Therapy—Disappointments and Opportunities. Front. Cell Dev. Biol. 2022, 10, 863850. [Google Scholar] [CrossRef]
- Shih, Y.-R.V.; Tseng, K.-F.; Lai, H.-Y.; Lin, C.-H.; Lee, O.K. Matrix Stiffness Regulation of Integrin-Mediated Mechanotransduction during Osteogenic Differentiation of Human Mesenchymal Stem Cells. J. Bone Miner. Res. 2011, 26, 730–738. [Google Scholar] [CrossRef]
- Liu, L.; Wang, S.; Cen, C.; Peng, S.; Chen, Y.; Li, X.; Diao, N.; Li, Q.; Ma, L.; Han, P. Identification of Differentially Expressed Genes in Pancreatic Ductal Adenocarcinoma and Normal Pancreatic Tissues Based on Microarray Datasets. Mol. Med. Rep. 2019, 20, 1901–1914. [Google Scholar] [CrossRef] [PubMed]
- Deichmann, S.; Schindel, L.; Braun, R.; Bolm, L.; Taylor, M.; Deshpande, V.; Schilling, O.; Bronsert, P.; Keck, T.; Ferrone, C.; et al. Overexpression of Integrin Alpha 2 (ITGA2) Correlates with Poor Survival in Patients with Pancreatic Ductal Adenocarcinoma. J. Clin. Pathol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Kalli, M.; Stylianopoulos, T. Defining the Role of Solid Stress and Matrix Stiffness in Cancer Cell Proliferation and Metastasis. Front. Oncol. 2018, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Anlaş, A.A.; Nelson, C.M. Soft Microenvironments Induce Chemoresistance by Increasing Autophagy Downstream of Integrin-Linked Kinase. Cancer Res. 2020, 80, 4103–4113. [Google Scholar] [CrossRef]
- Waters, A.M.; Der, C.J. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a031435. [Google Scholar] [CrossRef]
- Rovithi, M.; Avan, A.; Funel, N.; Leon, L.G.; Gomez, V.E.; Wurdinger, T.; Griffioen, A.W.; Verheul, H.M.W.; Giovannetti, E. Development of Bioluminescent Chick Chorioallantoic Membrane (CAM) Models for Primary Pancreatic Cancer Cells: A Platform for Drug Testing. Sci. Rep. 2017, 7, 44686. [Google Scholar] [CrossRef]
- Ulrich, T.A.; de Juan Pardo, E.M.; Kumar, S. The Mechanical Rigidity of the Extracellular Matrix Regulates the Structure, Motility, and Proliferation of Glioma Cells. Cancer Res. 2009, 69, 4167–4174. [Google Scholar] [CrossRef]
- Chen, W.; Park, S.; Patel, C.; Bai, Y.; Henary, K.; Raha, A.; Mohammadi, S.; You, L.; Geng, F. The Migration of Metastatic Breast Cancer Cells Is Regulated by Matrix Stiffness via YAP Signalling. Heliyon 2021, 7, e06252. [Google Scholar] [CrossRef]
- Papalazarou, V.; Zhang, T.; Paul, N.R.; Juin, A.; Cantini, M.; Maddocks, O.D.K.; Salmeron-Sanchez, M.; Machesky, L.M. The Creatine–Phosphagen System Is Mechanoresponsive in Pancreatic Adenocarcinoma and Fuels Invasion and Metastasis. Nat. Metab. 2020, 2, 62–80. [Google Scholar] [CrossRef]
- Nia, H.T.; Liu, H.; Seano, G.; Datta, M.; Jones, D.; Rahbari, N.; Incio, J.; Chauhan, V.P.; Jung, K.; Martin, J.D.; et al. Solid Stress and Elastic Energy as Measures of Tumour Mechanopathology. Nat. Biomed. Eng. 2017, 1, 0004. [Google Scholar] [CrossRef]
- Wolf, K.; Wu, Y.I.; Liu, Y.; Geiger, J.; Tam, E.; Overall, C.; Stack, M.S.; Friedl, P. Multi-Step Pericellular Proteolysis Controls the Transition from Individual to Collective Cancer Cell Invasion. Nat. Cell Biol. 2007, 9, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, N.; Zhang, C.; Schwarz, A.M.; Hinz, S.; Wang, C.; Williams, N.S.; Schwarz, M.A.; Schwarz, R.E. Comparative Benefits of Nab-Paclitaxel over Gemcitabine or Polysorbate-Based Docetaxel in Experimental Pancreatic Cancer. Carcinogenesis 2013, 34, 2361–2369. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Huang, Z.; Chen, H.; Zhou, B.; Liao, Y.; Wang, Z. Emodin Reverses Gemcitabine Resistance of Pancreatic Cancer Cell Lines Through Inhibition of IKKβ/NF-ΚB Signaling Pathway. OncoTargets Ther. 2020, 13, 9839–9848. [Google Scholar] [CrossRef] [PubMed]
- Chuang, Y.-C.; Wu, H.-Y.; Lin, Y.-L.; Tzou, S.-C.; Chuang, C.-H.; Jian, T.-Y.; Chen, P.-R.; Chang, Y.-C.; Lin, C.-H.; Huang, T.-H.; et al. Blockade of ITGA2 Induces Apoptosis and Inhibits Cell Migration in Gastric Cancer. Biol. Proced. Online 2018, 20, 10. [Google Scholar] [CrossRef] [PubMed]
- Darvishi, B.; Eisavand, M.R.; Majidzadeh-A, K.; Farahmand, L. Matrix Stiffening and Acquired Resistance to Chemotherapy: Concepts and Clinical Significance. Br. J. Cancer 2022, 126, 1253–1263. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Clauser, K.R.; Öhlund, D.; Rickelt, S.; Huang, Y.; Gupta, M.; Mani, D.R.; Carr, S.A.; Tuveson, D.A.; Hynes, R.O. Proteomic Analyses of ECM during Pancreatic Ductal Adenocarcinoma Progression Reveal Different Contributions by Tumor and Stromal Cells. Proc. Natl. Acad. Sci. USA 2019, 116, 19609–19618. [Google Scholar] [CrossRef]
- Meng, Z.; Garrett, C.R.; Shen, Y.; Liu, L.; Yang, P.; Huo, Y.; Zhao, Q.; Spelman, A.R.; Ng, C.S.; Chang, D.Z.; et al. Prospective Randomised Evaluation of Traditional Chinese Medicine Combined with Chemotherapy: A Randomised Phase II Study of Wild Toad Extract plus Gemcitabine in Patients with Advanced Pancreatic Adenocarcinomas. Br. J. Cancer 2012, 107, 411–416. [Google Scholar] [CrossRef]
- Guo, P.; Moses-Gardner, A.; Huang, J.; Smith, E.R.; Moses, M.A. ITGA2 as a Potential Nanotherapeutic Target for Glioblastoma. Sci. Rep. 2019, 9, 6195. [Google Scholar] [CrossRef]

| Univariate Analysis | No, % | OS Months (95% CI) | p | DFS Months (95% CI) | p |
|---|---|---|---|---|---|
| No. Patients | 85 | 21.9 (17.9–25.9) | 14.5 (9.3–19.6) | ||
| Age, years | |||||
| ≤65 | 40 (47) | 17.9 (13.1–22.7) | 0.36 | 9.4 (5.2–13.7) | 0.35 |
| >65 | 44 (53) | 23.6 (18.1–29.0) | 12.5 (7.7–17.3) | ||
| Sex | |||||
| Male | 46 (54) | 17.7 (13.6–21.8) | 0.61 | 10.7 (6.8–14.5) | 0.22 |
| Female | 39 (46) | 24.5 (18.3–30.7) | 11.7 (5.9–17.5) | ||
| Grading | |||||
| 1–2 | 32 (38) | 22.9 (16.3–29.5) | 0.67 | 12.0 (7.4–16.6) | 0.69 |
| 3 | 53 (62) | 18.9 (14.9–22.9) | 10.3 (5.7–14.9) | ||
| Stage | |||||
| IIA | 42 (50) | 21.4 (15.7–27.1) | 0.60 | 12.0 (7.4–16.6) | 0.87 |
| IIB | 43 (50) | 19.8 (15.4–24.2) | 10.3 (5.7–14.9) | ||
| ITGA2 expression | |||||
| low | 43 (50) | 20.9 (16.5–25.1) | 0.042 | 10.7 (8.7–18.7) | 0.045 |
| high | 42 (50) | 13.6 (6.4–20.7) | 8.3 (2.6–14.1) |
| Univariate Analysis | No. % | OS Months (95% CI) | p | DFS Months (95% CI) | p |
|---|---|---|---|---|---|
| No. Patients | 96 | 18.3 (16.6–20.0) | 10.0 (8.8–11.3) | ||
| Age, years | |||||
| ≤65 | 45 (47) | 18.5 (15.9–21.1) | 0.98 | 10.4 (8.5–12.3) | 0.74 |
| >65 | 51 (53) | 18.2 (16.0–20.4) | 9.7 (8.0–11.3) | ||
| Sex | |||||
| Male | 49 (51) | 18.4 (16.3–20.5) | 0.80 | 9.9 (7.1–10.7) | 0.61 |
| Female | 47 (49) | 18.2 (15.6–21.0) | 11.2 (9.4–12.9) | ||
| Grading | |||||
| 1–2 | 52 (55) | 19.8 (17.4–22.2) | 0.06 | 10.8 (8.9–12.6) | 0.10 |
| 3 | 43 (45) | 16.2 (14.0–18.4) | 8.8 (7.3–10.3) | ||
| Stage | |||||
| IIA | 6 (6) | 18.7 (14.5–22.8) | 0.81 | 9.1 (6.0–12.2) | 0.57 |
| IIB | 90 (94) | 18.7 (16.9–20.5) | 10.1 (8.8–11.5) | ||
| ITGA2 expression | |||||
| low | 48 (50) | 20.6 (18.1–23.1) | 0.01 | 11.4 (9.5–13.4) | 0.04 |
| high | 48 (50) | 16.1 (13.9–18.2) | 8.7 (7.2–10.1) | ||
| Multivariate analysis | Covariates | Risk of death | p | Risk of progression | p |
| Grading (WHO) | 1 vs. 2 | 1.6 (0.9–2.3) | 0.06 | 1.3 (0.8–1.9) | 0.19 |
| ITGA2 expression | Low vs. high | 1.7 (1.1–2.6) | 0.01 | 1.5 (0.9–2.1) | 0.07 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gregori, A.; Bergonzini, C.; Capula, M.; Mantini, G.; Khojasteh-Leylakoohi, F.; Comandatore, A.; Khalili-Tanha, G.; Khooei, A.; Morelli, L.; Avan, A.; et al. Prognostic Significance of Integrin Subunit Alpha 2 (ITGA2) and Role of Mechanical Cues in Resistance to Gemcitabine in Pancreatic Ductal Adenocarcinoma (PDAC). Cancers 2023, 15, 628. https://doi.org/10.3390/cancers15030628
Gregori A, Bergonzini C, Capula M, Mantini G, Khojasteh-Leylakoohi F, Comandatore A, Khalili-Tanha G, Khooei A, Morelli L, Avan A, et al. Prognostic Significance of Integrin Subunit Alpha 2 (ITGA2) and Role of Mechanical Cues in Resistance to Gemcitabine in Pancreatic Ductal Adenocarcinoma (PDAC). Cancers. 2023; 15(3):628. https://doi.org/10.3390/cancers15030628
Chicago/Turabian StyleGregori, Alessandro, Cecilia Bergonzini, Mjriam Capula, Giulia Mantini, Fatemeh Khojasteh-Leylakoohi, Annalisa Comandatore, Ghazaleh Khalili-Tanha, Alireza Khooei, Luca Morelli, Amir Avan, and et al. 2023. "Prognostic Significance of Integrin Subunit Alpha 2 (ITGA2) and Role of Mechanical Cues in Resistance to Gemcitabine in Pancreatic Ductal Adenocarcinoma (PDAC)" Cancers 15, no. 3: 628. https://doi.org/10.3390/cancers15030628
APA StyleGregori, A., Bergonzini, C., Capula, M., Mantini, G., Khojasteh-Leylakoohi, F., Comandatore, A., Khalili-Tanha, G., Khooei, A., Morelli, L., Avan, A., Danen, E. H., Schmidt, T., & Giovannetti, E. (2023). Prognostic Significance of Integrin Subunit Alpha 2 (ITGA2) and Role of Mechanical Cues in Resistance to Gemcitabine in Pancreatic Ductal Adenocarcinoma (PDAC). Cancers, 15(3), 628. https://doi.org/10.3390/cancers15030628