Circulating Chromosome Conformation Signatures Significantly Enhance PSA Positive Predicting Value and Overall Accuracy for Prostate Cancer Detection
Abstract
:Simple Summary
Abstract
1. Background
2. Methods
2.1. Patient Population
2.2. Sample Preparation
2.3. EpiSwitch PCR
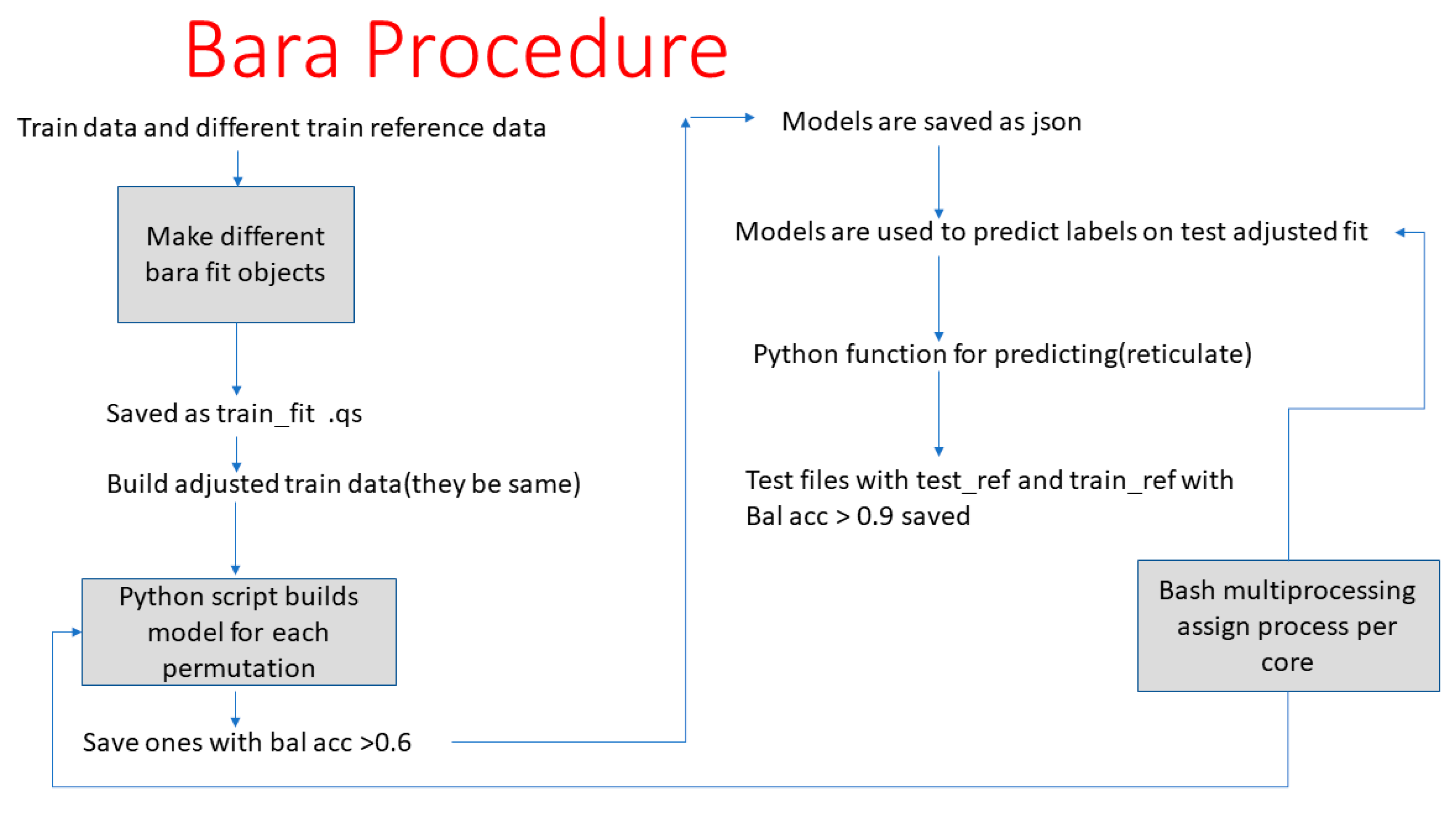
2.4. Statistical Analysis
3. Results
3.1. Bayesian Test Performance for Individual and Combined Cohorts
3.2. Test Optimization
3.3. Evaluation of Test Performance in the Combined Cohort Comparing Continuous and Dichotomous PSA
4. Discussion
4.1. Clinical Importance
4.2. Future Implementation of the PSE Test
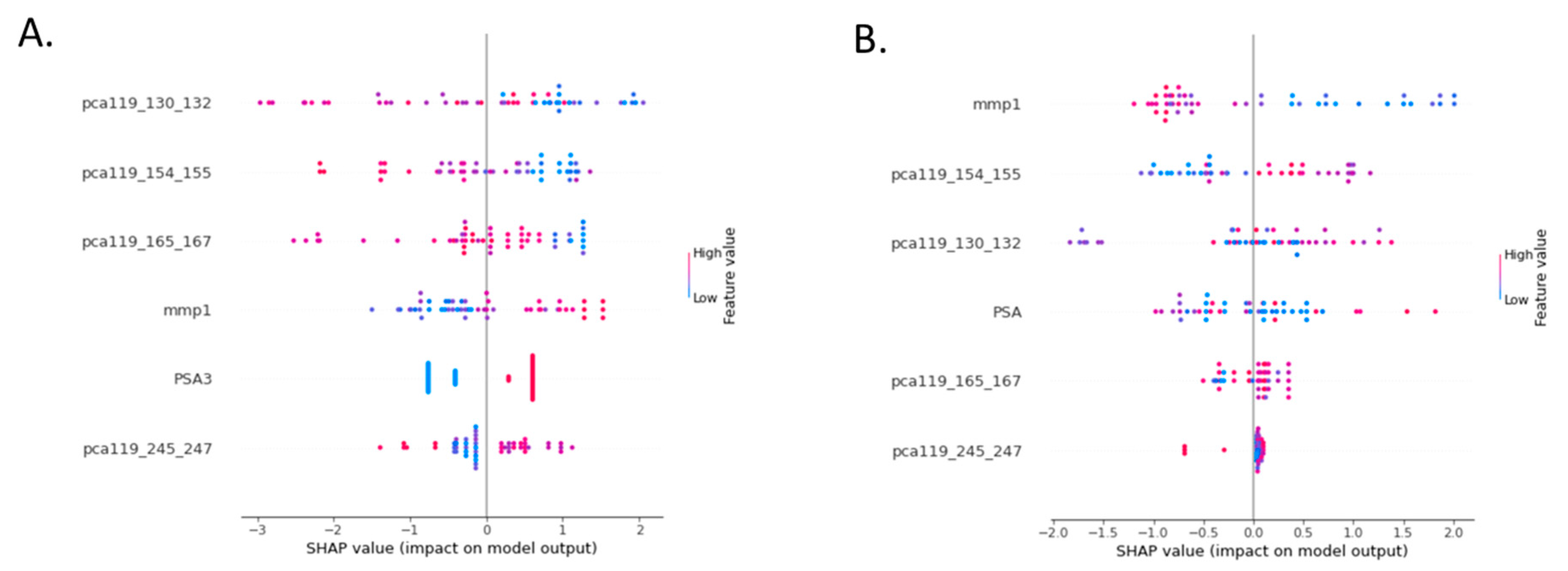
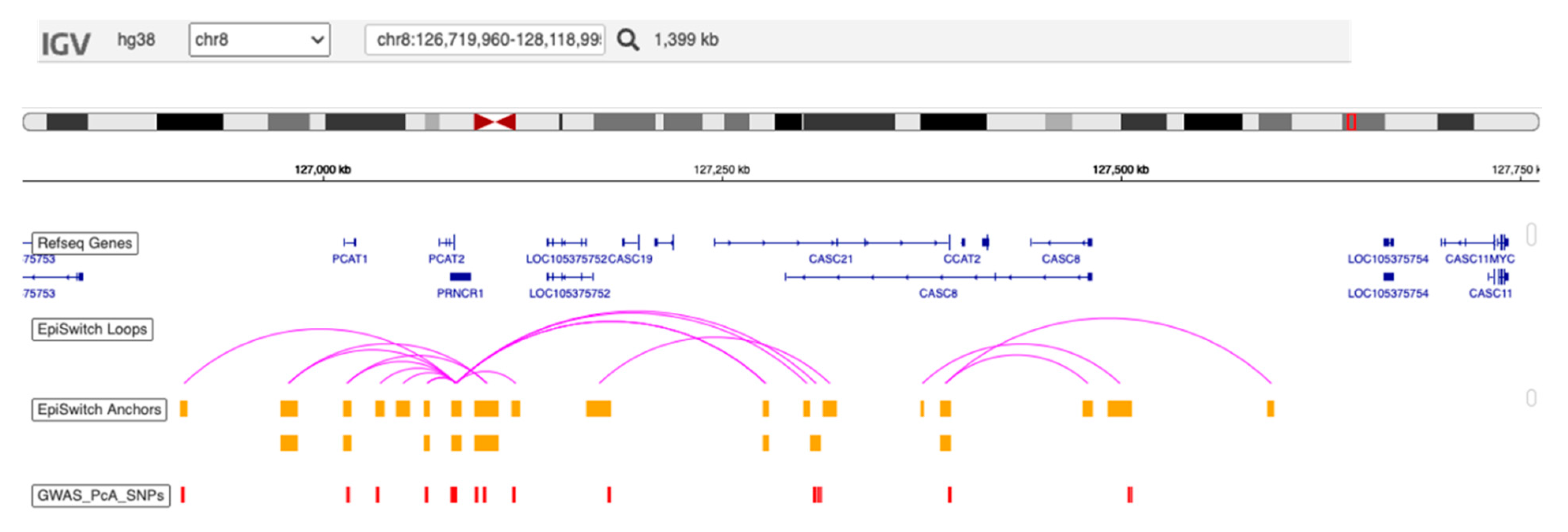
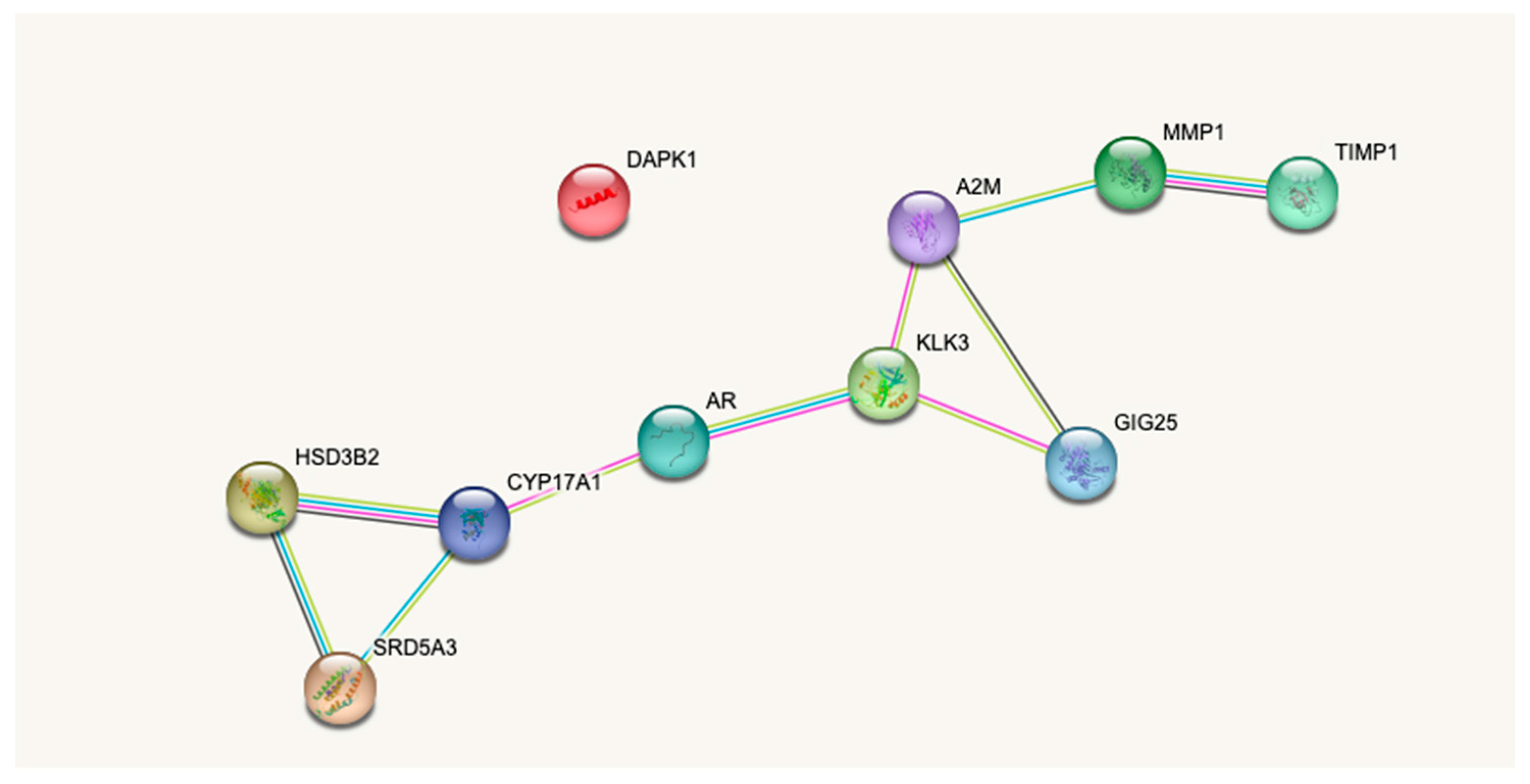
4.3. Pathophysiological Relevance of the Assay
4.4. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- ACS. American Cancer Society. 2020. Available online: http://www.cancer.org/cancer/prostatecancer/overviewguide/prostate-cancer-overview-key-statistics (accessed on 1 October 2022).
- CRUK. Cancer Research UK. 2020. Available online: http://www.cancerresearchuk.org/home/ (accessed on 1 October 2022).
- Sakr, W.; Haas, G.; Cassin, B.; Pontes, J.; Crissman, J. The Frequency of Carcinoma and Intraepithelial Neoplasia of the Prostate in Young Male Patients. J. Urol. 1993, 150, 379–385. [Google Scholar] [CrossRef]
- Sanchez-Chapado, M.; Olmedilla, G.; Cabeza, M.; Donat, E.; Ruiz, A. Prevalence of PCa and prostatic intraepithelial neoplasia in Caucasian Mediterranean males: An autopsy study. Prostate 2003, 54, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, P.A.; Moch, H.; Cubilla, A.L.; Ulbright, T.M.; Reuter, V.E. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs—Part B: Prostate and Bladder Tumours. Eur. Urol. 2016, 70, 106–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mottet, N.; van den Bergh, R.C.; Briers, E.; Van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer-2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2021, 79, 243–262. [Google Scholar] [CrossRef] [PubMed]
- Holmstrom, B.; Johansson, M.; Bergh, A.; Stenman, U.H.; Hallmans, G.; Stattin, P. Prostate specific antigen for early detection of PCa: Longitudinal study. BMJ 2009, 339, b3537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falchook, A.D.; Martin, N.E.; Basak, R.; Smith, A.B.; Milowsky, M.I.; Chen, R.C. Stage at presentation and survival outcomes of patients with Gleason 8-10 PCa and low prostate-specific antigen. Urol. Oncol. 2016, 34, 119.e19–119.e26. [Google Scholar] [CrossRef] [PubMed]
- Punnen, S.; Pavan, N.; Parekh, D.J. Finding the Wolf in Sheep’s Clothing: The 4Kscore Is a Novel Blood Test That Can Accurately Identify the Risk of Aggressive PCa. Rev. Urol. 2015, 17, 3–13. [Google Scholar]
- Hugosson, J.; Roobol, M.J.; Mansson, M.; Tammela, T.L.J.; Zappa, M.; Nelen, V.; Kwiatkowski, M.; Lujan, M.; Carlsson, S.V.; Talala, K.M.; et al. A 16-yr Follow-up of the European Randomized study of Screening for PCa. Eur. Urol. 2019, 76, 43–51. [Google Scholar] [CrossRef] [Green Version]
- Eldred-Evans, D.; Burak, P.; Connor, M.J.; Day, E.; Evans, M.; Fiorentino, F.; Gammon, M.; Hosking-Jervis, F.; Klimowska-Nassar, N.; McGuire, W.; et al. Population-Based PCa Screening with Magnetic Resonance Imaging or Ultrasonography: The IP1-PROSTAGRAM Study. JAMA Oncol. 2021, 7, 395–402. [Google Scholar] [CrossRef]
- Yu, J.; Boo, Y.; Kang, M.; Sung, H.H.; Jeong, B.C.; Seo, S.; Jeon, S.S.; Lee, H.; Jeon, H.J. Can Prostate-Specific Antigen Density Be an Index to Distinguish Patients Who Can Omit Repeat Prostate Biopsy in Patients with Negative Magnetic Resonance Imaging? Cancer Manag. Res. 2021, 13, 5467–5475. [Google Scholar] [CrossRef]
- Schulz, W.A.; Elo, J.P.; Florl, A.R.; Pennanen, S.; Santourlidis, S.; Engers, R.; Buchardt, M.; Seifert, H.-H.; Visakorpi, T. Genomewide DNA hypomethylation is associated with alterations on chromosome 8 in prostate carcinoma. Genes Chromosom. Cancer 2002, 35, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Yegnasubramanian, S.; Kowalski, J.; Gonzalgo, M.L.; Zahurak, M.; Piantadosi, S.; Walsh, P.C.; Bova, G.S.; De Marzo, A.M.; Isaacs, W.B.; Nelson, W.G. Hypermethylation of CpG islands in primary and metastatic human PCa. Cancer Res. 2004, 64, 1975–1986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Tu, S.W.; Hsieh, J.T. Down-regulation of human DAB2IP gene expression mediated by polycomb Ezh2 complex and histone deacetylase in PCa. J. Biol. Chem. 2005, 280, 22437–22444. [Google Scholar] [CrossRef] [Green Version]
- Iljin, K.; Wolf, M.; Edgren, H.; Gupta, S.; Kilpinen, S.; Skotheim, R.I.; Peltola, M.; Smit, F.; Verhaegh, G.; Schalken, J.; et al. TMPRSS2 fusions with oncogenic ETS factors in PCa involve unbalanced genomic rearrangements and are associated with HDAC1 and epigenetic reprogramming. Cancer Res. 2006, 66, 10242–10246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukhopadhyay, S.; Ramadass, A.S.; Akoulitchev, A.; Gordon, S. Formation of distinct chromatin conformation signatures epigenetically regulate macrophage activation. Int. Immunopharmacol. 2014, 18, 7–11. [Google Scholar] [CrossRef]
- Cortese, R.; Kwan, A.; Lalonde, E.; Bryzgunova, O.; Bondar, A.; Wu, Y.; Gordevicius, J.; Park, M.; Oh, G.; Kaminsky, Z.; et al. Epigenetic markers of PCa in plasma circulating DNA. Hum. Mol. Genet. 2012, 21, 3619–3631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dekker, J.; Rippe, K.; Dekker, M.; Kleckner, N. Capturing Chromosome Conformation. Science 2002, 295, 1306–1311. [Google Scholar] [CrossRef] [Green Version]
- Bastonini, E.; Jeznach, M.; Field, M.; Juszczyk, K.; Corfield, E.; Dezfouli, M.; Ahmat, N.; Smith, A.; Womersley, H.; Jordan, P.; et al. Chromatin barcodes as biomarkers for melanoma. Pigment. Cell Melanoma Res. 2014, 27, 788–800. [Google Scholar] [CrossRef]
- Jakub, J.W.; Grotz, T.E.; Jordan, P.; Hunter, E.; Pittelkow, M.; Ramadass, A.; Akoulitchev, A.; Markovic, S. A pilot study of chromosomal aberrations and epigenetic changes in peripheral blood samples to identify patients with melanoma. Melanoma Res. 2015, 25, 406–411. [Google Scholar] [CrossRef]
- Alshaker, H.; Mills, R.; Hunter, E.; Salter, M.; Ramadass, A.; Skinner, B.M.; Westra, W.; Green, J.; Akoulitchev, A.; Winkler, M.; et al. Chromatin conformation changes in peripheral blood can detect PCa and stratify disease risk groups. J. Transl. Med. 2021, 19, 46. [Google Scholar] [CrossRef] [PubMed]
- Alshaker, H.; Hunter, E.; Salter, M.; Ramadass, A.; Westra, W.; Winkler, M.; Green, J.; Akoulitchev, A.; Pchejetski, D. Monocytes acquire PCa specific chromatin conformations upon indirect co-culture with PCa cells. Front. Oncol. 2022, 12, 990842. [Google Scholar] [CrossRef] [PubMed]
- Salter, M.; Corfield, E.; Ramadass, A.; Grand, F.; Green, J.; Westra, J.; Lim, C.R.; Farrimond, L.; Feneberg, E.; Scaber, J.; et al. Initial Identification of a Blood-Based Chromosome Conformation Signature for Aiding in the Diagnosis of Amyotrophic Lateral Sclerosis. Ebiomedicine 2018, 33, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Carini, C.; Hunter, E.; Scottish Early Rheumatoid Arthritis Inception cohort Investigators; Ramadass, A.S.; Green, J.; Akoulitchev, A.; McInnes, I.B.; Goodyear, C.S.; Porter, D. Chromosome conformation signatures define predictive markers of inadequate response to methotrexate in early rheumatoid arthritis. J. Transl. Med. 2018, 16, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Guestrin, C. XGBoost. 2016, pp. 785–794. Available online: https://arxiv.org/abs/1603.02754 (accessed on 15 October 2022).
- Biecek, P.; Burzykowski, T. Shapley Additive Explanations (SHAP) for Average Attributions. In Explanatory Model Analysis; Chapman and Hall/CRC: Boca Raton, FL, USA, 2021; pp. 95–106. [Google Scholar] [CrossRef]
- Gradin, R.; Lindstedt, M.; Johansson, H. Batch adjustment by reference alignment (BARA): Improved prediction performance in biological test sets with batch effects. PLoS ONE 2019, 14, e0212669. [Google Scholar] [CrossRef]
- Schröder, F.H.; Hugosson, J.; Roobol-Bouts, M.J.; Tammela, T.L.J.; Ciatto, S.; Nelen, V.; Kwiatkowski, M.; Lujan, M.; Lilja, H.; Zappa, M.; et al. Screening and Prostate-Cancer Mortality in a Randomized European Study. N. Engl. J. Med. 2009, 360, 1320–1328. [Google Scholar] [CrossRef] [Green Version]
- Andriole, G.L.; Crawford, E.D.; Grubb, R.L., 3rd; Buys, S.S.; Chia, D.; Church, T.R.; Fouad, M.N.; Gelmann, E.P.; Kvale, P.A.; Reding, D.J.; et al. Mortality results from a randomized prostate-cancer screening trial. N. Engl. J. Med. 2009, 360, 1310–1319. [Google Scholar] [CrossRef] [Green Version]
- Oesterling, J.E.; Jacobsen, S.J.; Chute, C.G.; Guess, H.A.; Girman, C.J.; Panser, L.A.; Lieber, M.M. Serum prostate-specific antigen in a community-based population of healthy men. Establishment of age-specific reference ranges. JAMA 1993, 270, 860–864. [Google Scholar] [CrossRef]
- Sun, Y.-W.; Chen, K.-M.; Kawasawa, Y.I.; Salzberg, A.C.; Cooper, T.K.; Caruso, C.; Aliaga, C.; Zhu, J.; Gowda, K.; Amin, S.; et al. Hypomethylated Fgf3 is a potential biomarker for early detection of oral cancer in mice treated with the tobacco carcinogen dibenzo[def,p]chrysene. PLoS ONE 2017, 12, e0186873. [Google Scholar] [CrossRef] [Green Version]
- Christova, R.; Jones, T.; Wu, P.J.; Bolzer, A.; Costa-Pereira, A.P.; Watling, D.; Kerr, I.M.; Sheer, D. P-STAT1 mediates higher-order chromatin remodelling of the human MHC in response to IFNgamma. J. Cell Sci. 2007, 120 Pt 18, 3262–3270. [Google Scholar] [CrossRef] [Green Version]
- Tordini, F.; Aldinucci, M.; Milanesi, L.; Liò, P.; Merelli, I. The Genome Conformation as an Integrator of Multi-Omic Data: The Example of Damage Spreading in Cancer. Front. Genet. 2016, 7, 194. [Google Scholar] [CrossRef]
- di Iulio, J.; Bartha, I.; Wong, E.H.M.; Yu, H.C.; Lavrenko, V.; Yang, D.; Jung, I.; Hicks, M.A.; Shah, N.; Kirkness, E.F.; et al. The human noncoding genome defined by genetic diversity. Nat. Genet. 2018, 50, 333–337. [Google Scholar] [CrossRef] [PubMed]

| PSA | ||||
| Accuracy | 0.87 | |||
| Cancer present | Cancer absent | Sensitivity | 0.35 | |
| Test positive | 13 | 27 | Specificity | 0.93 |
| Test negative | 24 | 342 | PPV | 0.32 |
| NPV | 0.93 | |||
| MRI | ||||
| Accuracy | 0.83 | |||
| Cancer present | Cancer absent | Sensitivity | 0.57 | |
| Test positive | 21 | 51 | Specificity | 0.86 |
| Test negative | 16 | 318 | PPV | 0.29 |
| NPV | 0.95 | |||
| Ultrasound | ||||
| Accuracy | 0.78 | |||
| Cancer present | Cancer absent | Sensitivity | 0.59 | |
| Test positive | 22 | 74 | Specificity | 0.79 |
| Test negative | 15 | 295 | PPV | 0.23 |
| NPV | 0.95 |
| PROSTAGRAM Cohort | 109 |
|---|---|
| Control | 88 |
| Insignificant cancer | 8 |
| Significant cancer | 13 |
| PSA < 3 | 86 |
| PSA > 3 | 23 |
| Gleason sum = 0 | 88 |
| Gleason sum = 6 | 13 |
| Gleason sum = 7 | 8 |
| Imperial Cohort | 38 |
| Control | 9 |
| Organ confined | 9 |
| Locally advanced | 12 |
| Metastatic | 8 |
| PSA < 3 | 4 |
| PSA > 3 | 34 |
| Gleason sum = 0 | 10 |
| Gleason sum = 6 | 10 |
| Gleason sum = 7 | 8 |
| Gleason sum = 8 | 6 |
| Gleason sum = 9 | 4 |
| Binary PSA alone (across both cohorts) | ||||
| Accuracy | 0.79 | |||
| Cancer present | Cancer absent | Sensitivity | 0.33 | |
| Test positive | 2 | 12 | Specificity | 0.83 |
| Test negative | 4 | 61 | PPV | 0.14 |
| NPV | 0.93 | |||
| Continuous PSA alone (across both cohorts) | ||||
| Accuracy | 0.43 | |||
| Cancer present | Cancer absent | Sensitivity | 0.66 | |
| Test positive | 4 | 43 | Specificity | 0.41 |
| Test negative | 2 | 30 | PPV | 0.08 |
| NPV | 0.93 | |||
| Episwitch alone (across both cohorts) | ||||
| Accuracy | 0.64 | |||
| Cancer present | Cancer absent | Sensitivity | 0.61 | |
| Test positive | 16 | 34 | Specificity | 0.76 |
| Test negative | 5 | 54 | PPV | 0.91 |
| NPV | 0.32 | |||
| PSE with binary PSA (across both cohorts) | ||||
| Accuracy | 0.79 | |||
| Cancer present | Cancer absent | Sensitivity | 0.53 | |
| Test positive | 18 | 4 | Specificity | 0.93 |
| Test negative | 16 | 58 | PPV | 0.81 |
| NPV | 0.78 | |||
| PSE with continuous PSA (across both cohorts) | ||||
| Accuracy | 0.94 | |||
| Cancer present | Cancer absent | Sensitivity | 0.86 | |
| Test positive | 25 | 2 | Specificity | 0.97 |
| Test negative | 4 | 70 | PPV | 0.93 |
| NPV | 0.95 | |||
| PSE with continuous PSA (Prostagram cohort) | ||||
| Accuracy | 0.92 | |||
| Cancer present | Cancer absent | Sensitivity | 0.5 | |
| Test positive | 4 | 2 | Specificity | 0.97 |
| Test negative | 4 | 69 | PPV | 0.67 |
| NPV | 0.95 | |||
| PSE with continuous PSA (Imperial cohort) | ||||
| Accuracy | 1 | |||
| Cancer present | Cancer absent | Sensitivity | 1 | |
| Test positive | 21 | 0 | Specificity | 1 |
| Test negative | 0 | 1 | PPV | 1 |
| NPV | 1 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pchejetski, D.; Hunter, E.; Dezfouli, M.; Salter, M.; Powell, R.; Green, J.; Naithani, T.; Koutsothanasi, C.; Alshaker, H.; Jaipuria, J.; et al. Circulating Chromosome Conformation Signatures Significantly Enhance PSA Positive Predicting Value and Overall Accuracy for Prostate Cancer Detection. Cancers 2023, 15, 821. https://doi.org/10.3390/cancers15030821
Pchejetski D, Hunter E, Dezfouli M, Salter M, Powell R, Green J, Naithani T, Koutsothanasi C, Alshaker H, Jaipuria J, et al. Circulating Chromosome Conformation Signatures Significantly Enhance PSA Positive Predicting Value and Overall Accuracy for Prostate Cancer Detection. Cancers. 2023; 15(3):821. https://doi.org/10.3390/cancers15030821
Chicago/Turabian StylePchejetski, Dmitri, Ewan Hunter, Mehrnoush Dezfouli, Matthew Salter, Ryan Powell, Jayne Green, Tarun Naithani, Christina Koutsothanasi, Heba Alshaker, Jiten Jaipuria, and et al. 2023. "Circulating Chromosome Conformation Signatures Significantly Enhance PSA Positive Predicting Value and Overall Accuracy for Prostate Cancer Detection" Cancers 15, no. 3: 821. https://doi.org/10.3390/cancers15030821
APA StylePchejetski, D., Hunter, E., Dezfouli, M., Salter, M., Powell, R., Green, J., Naithani, T., Koutsothanasi, C., Alshaker, H., Jaipuria, J., Connor, M. J., Eldred-Evans, D., Fiorentino, F., Ahmed, H., Akoulitchev, A., & Winkler, M. (2023). Circulating Chromosome Conformation Signatures Significantly Enhance PSA Positive Predicting Value and Overall Accuracy for Prostate Cancer Detection. Cancers, 15(3), 821. https://doi.org/10.3390/cancers15030821






