The Rochester Modified Magee Algorithm (RoMMa): An Outcomes Based Strategy for Clinical Risk-Assessment and Risk-Stratification in ER Positive, HER2 Negative Breast Cancer Patients Being Considered for Oncotype DX® Testing
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Turner, B.M.; Katerji, H.; Zhang Hicks, D.G. Biomarker and Multigene Assay Testing in ER Positive HER-2 Negative Breast Carcinomas: An International Guidelines-Based Approach. Hum. Pathol. Rep. 2021, 26, 300574. [Google Scholar] [CrossRef]
- Paik, S.; Shak, S.; Tang, G.; Kim, C.; Baker, J.; Cronin, M.; Baehner, F.L.; Walker, M.G.; Watson, D.; Park, T.; et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004, 351, 2817–2826. [Google Scholar] [CrossRef] [PubMed]
- Blok, E.J.; Bastiaannet, E.; Van den Hout, W.B.; Liefers, G.J.; Smit, V.T.H.B.M.; Kroep, J.R.; van de Velde, C.J.H. Systematic review of the clinical and economic value of gene expression profiles for invasive early breast cancer available in Europe. Cancer Treat Rev. 2018, 62, 74–90. [Google Scholar] [CrossRef]
- Sparano, J.A.; Gray, R.J.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E., Jr.; Dees, E.C.; Perez, E.A.; Olson, J.A., Jr.; et al. Prospective Validation of a 21-Gene Expression Assay in Breast Cancer. N. Engl. J. Med. 2015, 373, 2005–2014. [Google Scholar] [CrossRef]
- Kalinsky, K.; Barlow, W.E.; Gralow, J.R.; Meric-Bernstam, F.; Albain, K.S.; Hayes, D.F.; Lin, N.U.; Perez, E.A.; Goldstein, L.J.; Chia, S.K.; et al. 21-Gene Assay to Inform Chemotherapy Benefit in Node-Positive Breast Cancer. N. Engl. J. Med. 2021, 385, 2336–2347. [Google Scholar] [CrossRef]
- Gluz, O.; Nitz, U.A.; Christgen, M.; Kates, R.E.; Shak, S.; Clemens, M.; Kraemer, S.; Aktas, B.; Kuemmel, S.; Reimer, T.; et al. West German Study Group Phase III PlanB Trial: First Prospective Outcome Data for the 21-Gene Recurrence Score Assay and Concordance of Prognostic Markers by Central and Local Pathology Assessment. J. Clin. Oncol. 2016, 34, 2341–2349. [Google Scholar] [CrossRef]
- Mamounas, E.P.; Tang, G.; Fisher, B.; Paik, S.; Shak, S.; Costantino, J.P.; Watson, D.; Geyer, C.E., Jr.; Wickerham, D.L.; Wolmark, N. Association between the 21-gene recurrence score assay and risk of locoregional recurrence in node-negative, estrogen receptor-positive breast cancer: Results from NSABP B-14 and NSABP B-20. J. Clin. Oncol. 2010, 28, 1677–1683. [Google Scholar] [CrossRef]
- Sgroi, D.C.; Sestak, I.; Cuzick, J.; Zhang, Y.; Schnabel, C.A.; Schroeder, B.; Erlander, M.G.; Dunbier, A.; Sidhu, K.; Lopez-Knowles, E.; et al. Prediction of late distant recurrence in patients with oestrogen-receptor-positive breast cancer: A prospective comparison of the breast-cancer index (BCI) assay, 21-gene recurrence score, and IHC4 in the TransATAC study population. Lancet Oncol. 2013, 14, 1067–1076. [Google Scholar] [CrossRef]
- Paik, S.; Tang, G.; Shak, S.; Kim, C.; Baker, J.; Kim, W.; Cronin, M.; Baehner, F.L.; Watson, D.; Bryant, J.; et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J. Clin. Oncol. 2006, 24, 3726–3734. [Google Scholar] [CrossRef]
- Tang, G.; Cuzick, J.; Costantino, J.P.; Dowsett, M.; Forbes, J.F.; Crager, M.; Mamounas, E.P.; Shak, S.; Wolmark, N. Risk of recurrence and chemotherapy benefit for patients with node-negative, estrogen receptor-positive breast cancer: Recurrence score alone and integrated with pathologic and clinical factors. J. Clin. Oncol. 2011, 29, 4365–4372. [Google Scholar] [CrossRef]
- Albain, K.S.; Barlow, W.E.; Shak, S.; Hortobagyi, G.N.; Livingston, R.B.; Yeh, I.T.; Ravdin, P.; Bugarini, R.; Baehner, F.L.; Davidson, N.E.; et al. Breast Cancer Intergroup of North America. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: A retrospective analysis of a randomised trial. Lancet Oncol. 2010, 11, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Dowsett, M.; Cuzick, J.; Wale, C.; Forbes, J.; Mallon, E.A.; Salter, J.; Quinn, E.; Dunbier, A.; Baum, M.; Buzdar, A.; et al. Prediction of risk of distant recurrence using the 21-gene recurrence score in node-negative and node-positive postmenopausal patients with breast cancer treated with anastrozole or tamoxifen: A TransATAC study. J. Clin. Oncol. 2010, 28, 1829–1834. [Google Scholar] [CrossRef] [PubMed]
- Cuzick, J.; Dowsett, M.; Pineda, S.; Wale, C.; Salter, J.; Quinn, E.; Zabaglo, L.; Mallon, E.; Green, A.R.; Ellis, I.O.; et al. Prognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the Genomic Health recurrence score in early breast cancer. J. Clin. Oncol. 2011, 29, 4273–4278. [Google Scholar] [CrossRef]
- Tang, G.; Shak, S.; Paik, S.; Anderson, S.J.; Costantino, J.P.; Geyer, C.E.; Mamounas, E.P.; Wickerham, D.L.; Wolmark, N. Comparison of the prognostic and predictive utilities of the 21-gene Recurrence Score assay and Adjuvant! for women with node-negative, ER-positive breast cancer: Results from NSABP B-14 and NSABP B-20. Breast Cancer Res. Treat. 2011, 127, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Toi, M.; Iwata, H.; Yamanaka, T.; Masuda, N.; Ohno, S.; Nakamura, S.; Nakayama, T.; Kashiwaba, M.; Kamigaki, S.; Kuroi, K. Japan Breast Cancer Research Group-Translational Research Group. Clinical significance of the 21-gene signature (Oncotype DX) in hormone receptor-positive early stage primary breast cancer in the Japanese population. Cancer 2010, 116, 3112–3118. [Google Scholar] [CrossRef]
- Naoi, Y.; Kishi, K.; Tsunashima, R.; Shimazu, K.; Shimomura, A.; Maruyama, N.; Shimoda, M.; Kagara, N.; Baba, Y.; Kim, S.J.; et al. Comparison of efficacy of 95-gene and 21-gene classifier (Oncotype DX) for prediction of recurrence in ER-positive and node-negative breast cancer patients. Breast Cancer Res. Treat. 2013, 140, 299–306. [Google Scholar] [CrossRef]
- Yorozuya, K.; Takeuchi, T.; Yoshida, M.; Mouri, Y.; Kousaka, J.; Fujii, K.; Nakano, S.; Fukutomi, T.; Hara, K.; Ichihara, S.; et al. Evaluation of Oncotype DX Recurrence Score as a prognostic factor in Japanese women with estrogen receptor-positive, node-negative primary Stage I or IIA breast cancer. J. Cancer Res. Clin. Oncol. 2010, 136, 939–944. [Google Scholar] [CrossRef]
- Goldstein, L.J.; Gray, R.; Badve, S.; Childs, B.H.; Yoshizawa, C.; Rowley, S.; Shak, S.; Baehner, F.L.; Ravdin, P.M.; Davidson, N.E.; et al. Prognostic utility of the 21-gene assay in hormone receptor-positive operable breast cancer compared with classical clinicopathologic features. J. Clin. Oncol. 2008, 26, 4063–4071. [Google Scholar] [CrossRef]
- Le Du, F.; Gonzalez-Angulo, A.M.; Park, M.; Liu, D.D.; Hortobagyi, G.N.; Ueno, N.T. Effect of 21-Gene RT-PCR Assay on Adjuvant Therapy and Outcomes in Patients with Stage I Breast Cancer. Clin. Breast Cancer 2015, 15, 458–466. [Google Scholar] [CrossRef]
- Freitas, M.R.; Simon, S.D. Comparison between Oncotype DX test and standard prognostic criteria in estrogen receptor positive early-stage breast cancer. Einstein 2011, 9, 354–358. [Google Scholar] [CrossRef]
- Aktas, B.; Bankfalvi, A.; Heubner, M.; Kimmig, R.; Kasimir-Bauer, S. Evaluation and correlation of risk recurrence in early breast cancer assessed by Oncotype DX®, clinicopathological markers and tumor cell dissemination in the blood and bone marrow. Mol. Clin. Oncol. 2013, 1, 1049–1054. [Google Scholar] [CrossRef] [PubMed]
- Acs, G.; Kiluk, J.; Loftus, L.; Laronga, C. Comparison of Oncotype DX and Mammostrat risk estimations and correlations with histologic tumor features in low-grade, estrogen receptor-positive invasive breast carcinomas. Mod. Pathol. 2013, 26, 1451–1460. [Google Scholar] [CrossRef] [PubMed]
- Kok, M.; Linn, S.C.; Van Laar, R.K.; Jansen, M.P.; Van den Berg, T.M.; Delahaye, L.J.; Glas, A.M.; Peterse, J.L.; Hauptmann, M.; Foekens, J.A.; et al. Comparison of gene expression profiles predicting progression in breast cancer patients treated with tamoxifen. Breast Cancer Res. Treat. 2009, 113, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Özmen, V.; Çakar, B.; Gökmen, E.; Özdoğan, M.; Güler, N.; Uras, C.; Ok, E.; Demircan, O.; Işıkdoğan, A.; Saip, P. Cost effectiveness of Gene Expression Profiling in Patients with Early-Stage Breast Cancer in a Middle-Income Country, Turkey: Results of a Prospective Multicenter Study. Eur. J. Breast Health 2019, 15, 183–190. [Google Scholar] [CrossRef]
- Levine, M.N.; Julian, J.A.; Bedard, P.L.; Eisen, A.; Trudeau, M.E.; Higgins, B.; Bordeleau, L.; Pritchard, K.I. Prospective Evaluation of the 21-Gene Recurrence Score Assay for Breast Cancer Decision-Making in Ontario. J. Clin. Oncol. 2016, 34, 1065–1071. [Google Scholar] [CrossRef]
- Leung, R.C.; Yau, T.C.; Chan, M.C.; Chan, S.W.; Chan, T.W.; Tsang, Y.Y.; Wong, T.T.; Chao, C.; Yoshizawa, C.; Soong, I.S.; et al. The Impact of the Oncotype DX Breast Cancer Assay on Treatment Decisions for Women with Estrogen Receptor-Positive, Node-Negative Breast Carcinoma in Hong Kong. Clin. Breast Cancer 2016, 16, 372–378. [Google Scholar] [CrossRef]
- Gligorov, J.; Pivot, X.B.; Jacot, W.; Naman, H.L.; Spaeth, D.; Misset, J.L.; Largillier, R.; Sautiere, J.L.; de Roquancourt, A.; Pomel, C.; et al. Prospective Clinical Utility Study of the Use of the 21-Gene Assay in Adjuvant Clinical Decision Making in Women with Estrogen Receptor-Positive Early Invasive Breast Cancer: Results from the SWITCH Study. Oncologist 2015, 20, 873–879. [Google Scholar] [CrossRef]
- Lee, M.H.; Han, W.; Lee, J.E.; Kim, K.S.; Park, H.; Kim, J.; Bae, S.Y.; Shin, H.J.; Lee, J.W.; Lee, E.S. The clinical impact of 21-gene recurrence score on treatment decisions for patients with hormone receptor-positive early breast cancer in Korea. Cancer Res. Treat 2015, 47, 208–214. [Google Scholar] [CrossRef]
- Jaafar, H.; Bashir, M.A.; Taher, A.; Qawasmeh, K.; Jaloudi, M. Impact of Oncotype DX testing on adjuvant treatment decisions in patients with early breast cancer: A single-center study in the United Arab Emirates. Asia Pac. J. Clin. Oncol. 2014, 10, 354–360. [Google Scholar] [CrossRef]
- Davidson, J.A.; Cromwell, I.; Ellard, S.L.; Lohrisch, C.; Gelmon, K.A.; Shenkier, T.; Villa, D.; Lim, H.; Sun, S.; Taylor, S.; et al. A prospective clinical utility and pharmacoeconomic study of the impact of the 21-gene Recurrence Score® assay in oestrogen receptor positive node negative breast cancer. Eur. J. Cancer 2013, 49, 2469–2475. [Google Scholar] [CrossRef]
- Holt, S.; Bertelli, G.; Humphreys, I.; Valentine, W.; Durrani, S.; Pudney, D.; Rolles, M.; Moe, M.; Khawaja, S.; Sharaiha, Y.; et al. A decision impact, decision conflict and economic assessment of routine Oncotype DX testing of 146 women with node-negative or pNImi, ER-positive breast cancer in the U.K. Br. J. Cancer 2013, 108, 2250–2258. [Google Scholar] [CrossRef] [PubMed]
- Biroschak, J.R.; Schwartz, G.F.; Palazzo, J.P.; Toll, A.D.; Brill, K.L.; Jaslow, R.J.; Lee, S.Y. Impact of Oncotype DX on treatment decisions in ER-positive, node-negative breast cancer with histologic correlation. Breast J. 2013, 19, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Ademuyiwa, F.O.; Miller, A.; O’Connor, T.; Edge, S.B.; Thorat, M.A.; Sledge, G.W.; Levine, E.; Badve, S. The effects of oncotype DX recurrence scores on chemotherapy utilization in a multi-institutional breast cancer cohort. Breast Cancer Res Treat 2011, 126, 797–802. [Google Scholar] [CrossRef] [PubMed]
- Albanell, J.; Gonzalez, A.; Ruiz-Borrego, M.; Alba, E.; Garcia-Saenz, J.A.; Corominas, J.M.; Burgues, O.; Furio, V.; Rojo, A.; Palacios, J.; et al. Prospective transGEICAM study of the impact of the 21-gene Recurrence Score assay and traditional clinicopathological factors on adjuvant clinical decision making in women with estrogen receptor-positive (ER+) node-negative breast cancer. Ann. Oncol. 2012, 23, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.S.; Mumby, P.B.; Norton, J.; Rychlik, K.; Smerage, J.; Kash, J.; Chew, H.K.; Gaynor, E.R.; Hayes, D.F.; Epstein, A.; et al. Prospective multicenter study of the impact of the 21-gene recurrence score assay on medical oncologist and patient adjuvant breast cancer treatment selection. J. Clin. Oncol. 2010, 28, 1671–1676. [Google Scholar] [CrossRef]
- Henry, L.R.; Stojadinovic, A.; Swain, S.M.; Prindiville, S.; Cordes, R.; Soballe, P.W. The influence of a gene expression profile on breast cancer decisions. J. Surg. Oncol. 2009, 99, 319–323. [Google Scholar] [CrossRef]
- Oratz, R.; Paul, D.; Cohn, A.L.; Sedlacek, S.M. Impact of a commercial reference laboratory test recurrence score on decision making in early-stage breast cancer. J. Oncol. Pract. 2007, 3, 182–186. [Google Scholar] [CrossRef]
- Kuchel, A.; Robinson, T.; Comins, C.; Shere, M.; Varughese, M.; Sparrow, G.; Sahu, A.; Saunders, L.; Bahl, A.; Cawthorn, S.J.; et al. The impact of the 21-gene assay on adjuvant treatment decisions in oestrogen receptor-positive early breast cancer: A prospective study. Br. J. Cancer 2016, 114, 731–736. [Google Scholar] [CrossRef]
- Bargallo, J.E.; Lara, F.; Shaw-Dulin, R.; Perez-Sánchez, V.; Villarreal-Garza, C.; Maldonado-Martinez, H.; Mohar-Betancourt, A.; Yoshizawa, C.; Burke, E.; Decker, T.; et al. A study of the impact of the 21-gene breast cancer assay on the use of adjuvant chemotherapy in women with breast cancer in a Mexican public hospital. J. Surg. Oncol. 2015, 111, 203–207. [Google Scholar] [CrossRef]
- Yamauchi, H.; Nakagawa, C.; Takei, H.; Chao, C.; Yoshizawa, C.; Yagata, H.; Yoshida, A.; Hayashi, N.; Hell, S.; Nakamura, S. Prospective study of the effect of the 21-gene assay on adjuvant clinical decision-making in Japanese women with estrogen receptor-positive, node-negative, and node-positive breast cancer. Clin. Breast Cancer 2014, 14, 191–197. [Google Scholar] [CrossRef]
- Fried, G.; Moskovitz, M. Treatment decisions in estrogen receptor-positive early breast cancer patients with intermediate oncotype DX recurrence score results. SpringerPlus 2014, 3, 71. [Google Scholar] [CrossRef] [PubMed]
- Cheung, P.S.; Tong, A.C.; Leung, R.C.; Kwan, W.H.; Yau, T.C. Initial experience with the Oncotype DX assay in decision-making for adjuvant therapy of early oestrogen receptor-positive breast cancer in Hong Kong. Hong Kong Med. J. 2014, 20, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Eiermann, W.; Rezai, M.; Kümmel, S.; Kühn, T.; Warm, M.; Friedrichs, K.; Schneeweiss, A.; Markmann, S.; Eggemann, H.; Hilfrich, J.; et al. The 21-gene recurrence score assay impacts adjuvant therapy recommendations for ER-positive, node-negative and node-positive early breast cancer resulting in a risk-adapted change in chemotherapy use. Ann. Oncol. 2013, 24, 618–624. [Google Scholar] [CrossRef] [PubMed]
- de Boer, R.H.; Baker, C.; Speakman, D.; Chao, C.Y.; Yoshizawa, C.; Mann, G.B. The impact of a genomic assay (Oncotype DX) on adjuvant treatment recommendations in early breast cancer. Med. J. Aust. 2013, 199, 205–208. [Google Scholar] [CrossRef]
- Geffen, D.B.; Abu-Ghanem, S.; Sion-Vardy, N.; Braunstein, R.; Tokar, M.; Ariad, S.; Delgado, B.; Bayme, M.; Koretz, M. The impact of the 21-gene recurrence score assay on decision making about adjuvant chemotherapy in early-stage estrogen-receptor-positive breast cancer in an oncology practice with a unified treatment policy. Ann. Oncol. 2011, 22, 2381–2386. [Google Scholar] [CrossRef]
- Hussey, P.S.; Wertheimer, S.; Mehrotra, A. The association between health care quality and cost: A systematic review. Ann. Intern. Med. 2013, 158, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, M.C.; Skinner, J.A. Comparative effectiveness and health care spending--implications for reform. N. Engl. J. Med. 2010, 362, 460–465. [Google Scholar] [CrossRef]
- Wang, S.Y.; Dang, W.; Richman, I.; Mougalian, S.S.; Evans, S.B.; Gross, C.P. Cost-Effectiveness Analyses of the 21-Gene Assay in Breast Cancer: Systematic Review and Critical Appraisal. J. Clin. Oncol. 2018, 36, 1619–1627. [Google Scholar] [CrossRef]
- Turner, B.M.; Gimenez-Sanders, M.A.; Soukiazian, A.; Breaux, A.C.; Skinner, K.; Shayne, M.; Soukiazian, N.; Ling, M.; Hicks, D.G. A multi-institutional validation and outcome study of the Rochester Modified Magee algorithm (RoMMa) and prediction of an Oncotype DX® recurrence score <26. Cancer Med. 2019, 8, 4176–4188. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Wishart, G.C.; Azzato, E.M.; Greenberg, D.C.; Rashbass, J.; Kearins, O.; Lawrence, G.; Caldas, C.; Pharoah, P.D. PREDICT: A new UK prognostic model that predicts survival following surgery for invasive breast cancer. Breast Cancer Res. 2010, 12, 1–10. [Google Scholar] [CrossRef]
- Candido dos Reis, F.J.; Wishart, G.C.; Dicks, E.M.; Greenberg, D.; Rashbass, J.; Schmidt, M.K.; van den Broek, A.J.; Ellis, I.O.; Green, A.; Rakha, E.; et al. An updated PREDICT breast cancer prognostication and treatment benefit prediction model with independent validation. Breast Cancer Res. 2017, 19, 58. [Google Scholar] [CrossRef]
- Engelhardt, E.G.; van den Broek, A.J.; Linn, S.C.; Wishart, G.C.; Emiel, J.T.; van de Velde, A.O.; Smit, V.T.; Voogd, A.C.; Siesling, S.; Brinkhuis, M.; et al. Accuracy of the online prognostication tools PREDICT and Adjuvant! for early-stage breast cancer patients younger than 50 years. Eur. J. Cancer 2017, 78, 37–44. [Google Scholar] [CrossRef]
- Available online: https://webapp.oncoassist.com/public/index.php/adjuvant_tools/breast_cancer_survival (accessed on 12 February 2022).
- Flanagan, M.B.; Dabbs, D.J.; Brufsky, A.M.; Beriwal, S.; Bhargava, R. Histopathologic variables predict Oncotype DX recurrence score. Mod. Pathol. 2008, 21, 1255–1261. [Google Scholar] [CrossRef]
- Klein, M.E.; Dabbs, D.J.; Shuai, Y.; Brufsky, A.M.; Jankowitz, R.; Puhalla, S.L.; Bhargava, R. Prediction of the Oncotype DX® recurrence score: Use of pathology-generated equations derived by linear regression analysis. Mod. Pathol. 2013, 26, 658–664. [Google Scholar] [CrossRef] [PubMed]
- Clark, B.Z.; Dabbs, D.J.; Cooper, K.L.; Bhargava, R. Impact of progesterone receptor semiquantitative immunohistochemical result on Oncotype DX® recurrence score: A quality assurance study of 1074 cases. Appl. Immunohistochem. Mol. Morphol. 2013, 21, 287–291. [Google Scholar] [CrossRef]
- Zbytek, B.; Cohen, C.; Wang, J.; Page, A.; Williams, D.J.; Adams, A.L. Nottingham-defined mitotic score: Comparison with visual and image cytometric phosphohistone H3 labeling indices and correlation with Oncotype DX® recurrence score. Appl. Immunohistochem. Mol. Morphol. 2013, 21, 48–53. [Google Scholar] [CrossRef]
- Kraus, J.A.; Dabbs, D.J.; Beriwal, S.; Bhargava, R. Semi-quantitative immunohistochemical assay versus Oncotype DX® qRT-PCR assay for estrogen and progesterone receptors: An independent quality assurance study. Mod. Pathol. 2012, 25, 869–876. [Google Scholar] [CrossRef]
- Lee, J.J.; Shen, J. Is the Oncotype DX® assay necessary in strongly estrogen receptor-positive breast cancers? Am. Surg. 2011, 77, 1364–1367. [Google Scholar] [CrossRef]
- Williams, D.J.; Cohen, C.; Darrow, M.; Page, A.J.; Chastain, B.; Adams, A.L. Proliferation (Ki-67 and phosphohistone H3) and Oncotype DX® recurrence score in estrogen receptor-positive breast cancer. Appl. Immunohistochem. Mol. Morphol. 2011, 19, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Sahebjam, S.; Aloyz, R.; Pilavdzic, D.; Brisson, M.L.; Ferrario, C.; Bouganim, N.; Cohen, V.; Miller, W.H.; Panasci, L.C. Ki-67 is a major, but not the sole determinant of Oncotype DX® recurrence score. Br. J. Cancer 2011, 105, 1342–1345. [Google Scholar] [CrossRef] [PubMed]
- Auerbach, J.; Kim, M.; Fineberg, S. Can features evaluated in the routine pathologic assessment of lymph node-negative estrogen receptor-positive stage I or II invasive breast cancer be used to predict the Oncotype DX® recurrence score? Arch. Pathol. Lab. Med. 2010, 134, 1697–1701. [Google Scholar] [CrossRef] [PubMed]
- Allison, K.H.; Kandalaft, P.L.; Sitlani, C.M.; Dintzis, S.M.; Gown, A.M. Routine pathologic parameters can predict Oncotype DX® recurrence scores in subsets of ER positive patients: Who does not always need testing? Breast Cancer Res. Treat. 2012, 131, 413–424. [Google Scholar] [CrossRef]
- Tang, P.; Wang, J.; Hicks, D.G.; Wang, X.; Schiffhauer, L.; McMahon, L.; Yang, Q.; Shayne, M.; Huston, A.; Skinner, K.A.; et al. A lower Allred score for progesterone receptor is strongly associated with a higher recurrence score of 21-gene assay in breast cancer. Cancer Investig. 2010, 28, 978–982. [Google Scholar] [CrossRef]
- Turner, B.M.; Skinner, K.A.; Tang, P.; Jackson, M.C.; Soukiazian, N.; Shayne, M.; Huston, A.; Ling, M.; Hicks, D.G. Use of modified Magee equations and histologic criteria to predict the Oncotype DX recurrence score. Mod. Pathol. 2015, 28, 921–931. [Google Scholar] [CrossRef]
- Bhargava, R.; Clark, B.Z.; Carter, G.J.; Brufsky, A.M.; Dabbs, D.J. The healthcare value of the Magee Decision Algorithm™: Use of Magee Equations™ and mitosis score to safely forgo molecular testing in breast cancer. Mod. Pathol. 2020, 33, 1563–1570. [Google Scholar] [CrossRef]
- Bhargava, R.; Clark, B.Z.; Dabbs, D.J. Breast Cancers with Magee Equation Score of Less Than 18, or 18-25 and Mitosis Score of 1, Do Not Require Oncotype DX Testing: A Value Study. Am. J. Clin. Pathol. 2019, 151, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, J.M.; Christiansen, J.; Gustavson, M.; Rimm, D.L.; Piper, T.; de Velde, C.J.V.; Hasenburg, A.; Kieback, D.G.; Putter, H.; Markopoulos, C.J.; et al. Validation of the IHC4 Breast Cancer Prognostic Algorithm Using Multiple Approaches on the Multinational TEAM Clinical Trial. Arch Pathol. Lab. Med. 2016, 140, 66–74. [Google Scholar] [CrossRef]
- Bhargava, R.; Dabbs, D.J. The Story of the Magee Equations: The Ultimate in Applied Immunohistochemistry. Appl. Immunohistochem. Mol. Morphol. 2022, 1097. [Google Scholar] [CrossRef]
- Sparano, J.A.; Gray, R.J.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E., Jr.; Dees, E.C.; Goetz, M.P.; Olson, J.A., Jr.; et al. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N. Engl. J. Med. 2018, 379, 111–121. [Google Scholar] [CrossRef]
- Elston, C.W.; Ellis, I.O. Pathological prognostic factors in breast cancer. I, the value of histological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology 1991, 19, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, T.O.; Leung, S.C.Y.; Rimm, D.L.; Dodson, A.; Acs, B.; Badve, S.; Denkert, C.; Ellis, M.J.; Fineberg, S.; Flowers, M.; et al. Assessment of Ki67 in Breast Cancer: Updated recommendations from the International Ki67 in Breast Cancer Working Group. J. Natl. Cancer Inst. 2021, 113, 808–819. [Google Scholar] [CrossRef]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the Areas under Two or More Correlated Receiver Operating Characteris-tic Curves: A Nonparametric Approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Hicks, D.G.; Turner, B. Pathologic diagnosis, immunohistochemistry, multigene assays and breast cancer treatment: Progress toward “precision” cancer therapy. Biotech. Histochem. 2015, 90, 81–92. [Google Scholar] [CrossRef]
- Turner, B.M.; Hicks, D.G. Pathologic diagnosis of breast cancer patients: Evolution of the traditional clinical-pathologic paradigm toward “precision” cancer therapy. Biotech. Histochem. 2017, 92, 175–200. [Google Scholar] [CrossRef] [PubMed]
- Dannehl, D.; Engler, T.; Volmer, L.L.; Staebler, A.; Fischer, A.K.; Weiss, M.; Hahn, M.; Walter, C.B.; Grischke, E.M.; Fend, F.; et al. Recurrence Score® Result Impacts Treatment Decisions in Hormone Receptor-Positive, HER2-Negative Patients with Early Breast Cancer in a Real-World Setting-Results of the IRMA Trial. Cancers 2022, 14, 5365. [Google Scholar] [CrossRef]
- Mittmann, N.; Earle, C.C.; Cheng, S.Y.; Julian, J.A.; Rahman, F.; Seung, S.J.; Levine, M.N. Population-Based Study to Determine the Health System Costs of Using the 21-Gene Assay. J. Clin. Oncol. 2018, 36, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Ishibe, N.; Schully, S.; Freedman, A.; Ramsey, S.D. Use of Oncotype DX® in Women with node-positive breast cancer. PLoS Curr. 2011, 3, RRN1249. [Google Scholar] [CrossRef]
- Ward, S.; Scope, A.; Rafia, R.; Pandor, A.; Harnan, S.; Evans, P.; Wyld, L. Gene expression profiling and expanded immunohistochemistry tests to guide the use of adjuvant chemotherapy in breast cancer management: A systematic review and cost-effectiveness analysis. Health Technol. Assess. 2013, 17, 1–302. [Google Scholar] [CrossRef]
- Rouzier, R.; Pronzato, P.; Chéreau, E.; Carlson, J.; Hunt, B.; Valentine, W.J. Multigene assays and molecular markers in breast cancer: Systematic review of health economic analyses. Breast Cancer Res. Treat. 2013, 139, 621–637. [Google Scholar] [CrossRef]
- Marrone, M.; Stewart, A.; Dotson, W.D. Clinical utility of gene-expression profiling in women with early breast cancer: An overview of systematic reviews. Genet Med. 2015, 17, 519–532. [Google Scholar] [PubMed]
- Klang, S.H.; Hammerman, A.; Liebermann, N.; Efrat, N.; Doberne, J.; Hornberger, J. Economic implications of 21-gene breast cancer risk assay from the perspective of an Israeli-managed health-care organization. Value Health 2010, 13, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Hornberger, J.; Cosler, L.E.; Lyman, G.H. Economic analysis of targeting chemotherapy using a 21-gene RT-PCR assay in lymph-node-negative, estrogen-receptor-positive, early-stage breast cancer. Am. J. Manag. Care. 2005, 11, 313–324. [Google Scholar]
- Lyman, G.H.; Cosler, L.E.; Kuderer, N.M.; Hornberger, J. Impact of a 21-gene RT-PCR assay on treatment decisions in early-stage breast cancer: An economic analysis based on prognostic and predictive validation studies. Cancer 2007, 109, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Kondo, M.; Hoshi, S.L.; Yamanaka, T.; Ishiguro, H.; Toi, M. Economic evaluation of the 21-gene signature (Oncotype DX) in lymph node-negative/positive, hormone receptor-positive early-stage breast cancer based on Japanese validation study (JBCRG-TR03). Breast Cancer Res. Treat. 2011, 127, 739–749. [Google Scholar] [PubMed]
- de Lima, M.A.; Clemons, M.; Van Katwyk, S.; Stober, C.; Robertson, S.J.; Vandermeer, L.; Fergusson, D.; Thavorn, K. Cost analysis of using Magee scores as a surrogate of Oncotype DX for adjuvant treatment decisions in women with early breast cancer. J. Eval. Clin. Pract. 2020, 26, 889–892. [Google Scholar] [CrossRef]
- Hall, P.S.; McCabe, C.; Stein, R.C.; Cameron, D. Economic evaluation of genomic test-directed chemotherapy for early-stage lymph node-positive breast cancer. J. Natl. Cancer Inst. 2012, 104, 56–66. [Google Scholar]
- Kim, H.S.; Umbricht, C.B.; Illei, P.B.; Cimino-Mathews, A.; Cho, S.; Chowdhury, N.; Figueroa-Magalhaes, M.C.; Pesce, C.; Jeter, S.C.; Mylander, C.; et al. Optimizing the Use of Gene Expression Profiling in Early Stage Breast Cancer. J. Clin. Oncol. 2016, 34, 4390–4397. [Google Scholar]
- Marazzi, F.; Barone, R.; Masiello, V.; Magri, V.; Mulè, A.; Santoro, A.; Cacciatori, F.; Boldrini, L.; Franceschini, G.; Moschella, F.; et al. Oncotype DX Predictive Nomogram for Recurrence Score Output: The Novel System ADAPTED01 Based on Quantitative Immunochemistry Analysis. Clin. Breast Cancer 2020, 20, e600–e611. [Google Scholar] [CrossRef]
- Turner, B.M.; Tang, P.; Hicks, D.G. Letter to the Editor. The value of algorithms predicting the Oncotype DX recurrence score should not be underestimated! Breast Cancer Res. Treat. 2017, 164, 249–250. [Google Scholar] [CrossRef]
- Soukiazian, A.; Hicks, D.G.; Turner, B.M. Reconsidering “low risk” criteria for breast cancer recurrence in hormone positive patients. Breast J. 2019. preprint. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Tozbikian, G.; Zynger, D.L.; Li, Z. Using the Modified Magee equation to identify patiens unlikely to benefit from the 21-gene recurrence score assay (Oncotype DX assay). Am. J. Clin. Pathol. 2017, 147, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Zynger, D.L.; Li, X.; Li, Z. Comparison of Oncotype DX with Modified Magee equation recurrence in low-grade invasive carcinoma of breast. Am. J. Clin. Pathol. 2017, 148, 5167–5172. [Google Scholar] [CrossRef]
- Acs, G.; Esposito, N.N.; Kiluk, J.; Loftus, L.; Laronga, C. A mitotically active, cellular tumor stroma and/or inflammatory cells associated with tumor cells may contribute to intermediate or high Oncotype DX Recurrence Scores in low-grade invasive breast carcinomas. Mod. Pathol. 2012, 25, 556–566. [Google Scholar] [CrossRef] [PubMed]
- Weigelt, B.; Baehner, F.L.; Reis-Filho, J.S. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: A retrospective of the last decade. J. Pathol. 2010, 220, 263–280. [Google Scholar] [CrossRef]
- Allison, K.H.; Hammond, M.E.H.; Dowsett, M.; McKernin, S.E.; Carey, L.A.; Fitzgibbons, P.L.; Hayes, D.F.; Lakhani, S.R.; Chavez-MacGregor, M.; Perlmutter, J.; et al. Estrogen and Progesterone Receptor Testing in Breast Cancer: ASCO/CAP Guideline Update. J. Clin. Oncol. 2020, 38, 1346–1366. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W.; et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2018, 36, 2105–2122. [Google Scholar] [CrossRef]



| N (%) | Mean Months of Follow-Up (Range) | |
|---|---|---|
| TOTAL POPULATION | 355 (100) | 93.2 (20–60) |
| White | 310 (87.3) | 92.8 (20–160) |
| Black | 32 (9.0) | 90.7 (62–41) |
| Hispanic | 6 (1.7) | 117.3 (91–146) |
| Asian | 5 (1.4) | 107 (96–119) |
| Unknown | 2 (0.6) | 94 (90–98) |
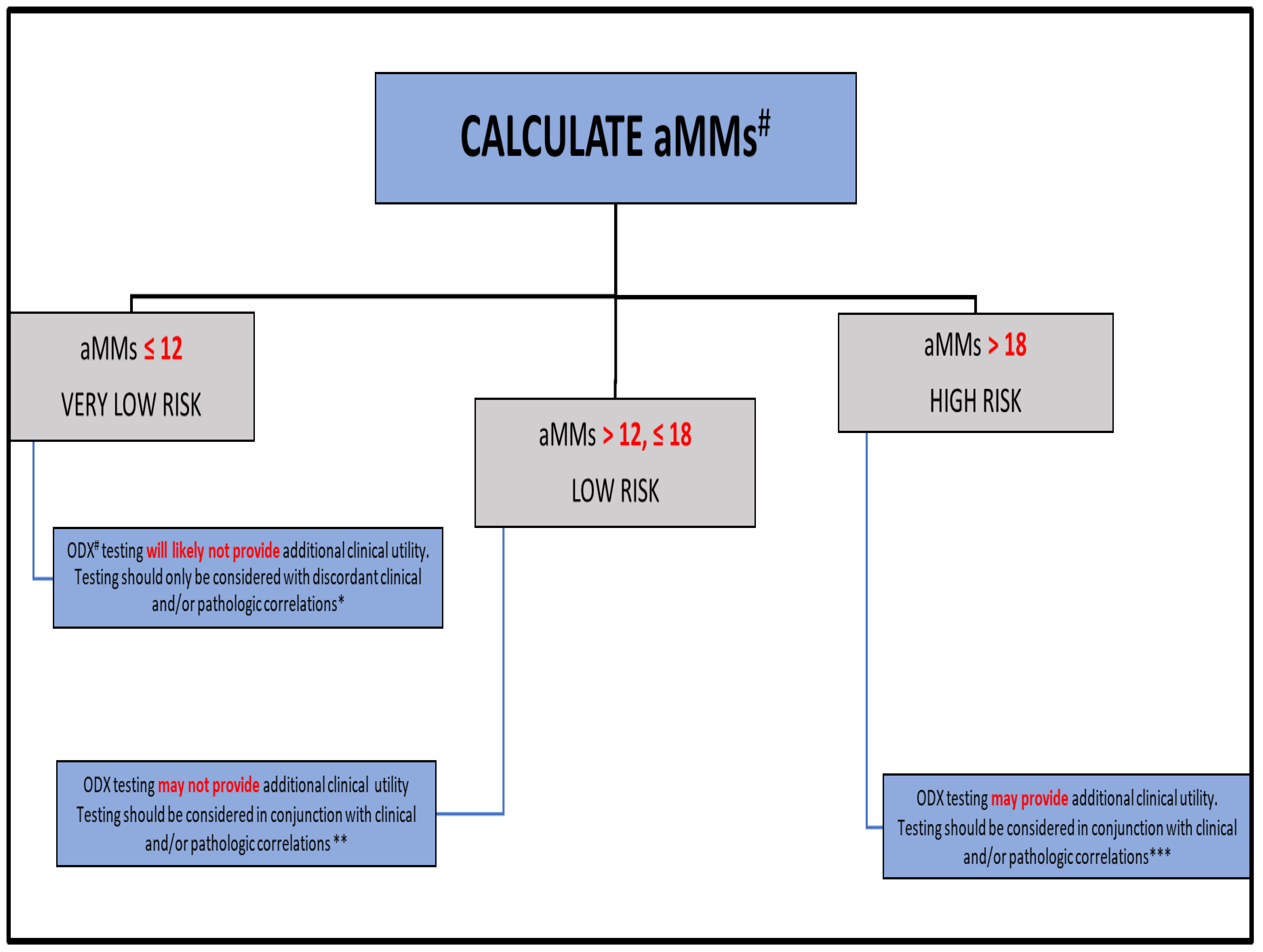
| aMMs * ≤ 12 | 62 (17.5) | 102.2 (63–150) |
| aMMs >12 & ≤18 | 173 (48.7) | 94.3 (59–160) |
| aMMs > 18 | 120 (33.8) | 87.1 (20–142) |
| ODX * < 11 | 94 (26.5) | 91.9 (62–157) |
| ODX 11–25 ** | 208 (58.6) | 93.9 (24–154) |
| ODX > 25 ** | 53 (14.9) | 90.1 (20–160) |
| RECURRENCES | 31 (8.7) | 59.3 (3–144) *** |
| MEAN (RANGE) | ||
| AGE | 59 (33–84) | |
| AGE OF RECURRENCE | 65 (43–83) | |
| aMMs | 16.7 (6.5–32.3) | |
| ODX | 14.6 (0–44) | |
| NS * | 5.7 (3–9) | |
| ER-H SCORE **** | 261.1 (40–300) | |
| PR-H SCORE **** | 175.0 (0–300) | |
| Ki-67 | 14.0 (0–70) | |
| Size (cm) | 2.2 (0.4–14) |
| OUTCOME | |||
|---|---|---|---|
| Risk Category | Recurrence N (%) | No Recurrence N (%) | p-Value |
| VERY LOW (N) | |||
| Average Modified Magee score ≤12 (62) | 2 (3.2) | 60 (96.8) | 0.27 |
| Oncotype DX <11 (94) | 7 (7.4) | 87 (92.6) | |
| LOW (N) | |||
| Average Modified Magee score >12, ≤18 (173) | 14 (8.1) | 159 (91.9) | 0.84 |
| Oncotype DX 11–25 (208) * | 18 (8.7) | 190 (91.3) | |
| HIGH (N) | |||
| Average Modified Magee score >18 (120) | 15(12.5) | 105 (87.5) | 0.83 |
| Oncotype DX ≥16–25 (32) ** and Oncotype DX >25 (21) *** | 6 (11.3) | 47 (88.7) | |
| ODX Risk Category ** | ||||
|---|---|---|---|---|
| aMMs Risk Category * | Very Low | Low | High | TOTAL |
| Very low | 0 | 2 | 0 | 2 |
| Low | 5 | 7 | 2 *** | 14 |
| High | 2 | 9 | 4 | 15 |
| TOTAL | 7 | 18 | 6 | 31 |
| ODX Risk Category ** | ||||
|---|---|---|---|---|
| aMMs Risk Category * | Very Low | Low | High | TOTAL |
| Very low | 26 | 30 | 6 | 62 |
| Low | 53 | 101 | 19 *** | 173 |
| High | 15 | 77 | 28 | 120 |
| TOTAL | 94 | 208 | 53 | 355 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turner, B.M.; Finkelman, B.S.; Hicks, D.G.; Numbereye, N.; Moisini, I.; Dhakal, A.; Skinner, K.; Sanders, M.A.G.; Wang, X.; Shayne, M.; et al. The Rochester Modified Magee Algorithm (RoMMa): An Outcomes Based Strategy for Clinical Risk-Assessment and Risk-Stratification in ER Positive, HER2 Negative Breast Cancer Patients Being Considered for Oncotype DX® Testing. Cancers 2023, 15, 903. https://doi.org/10.3390/cancers15030903
Turner BM, Finkelman BS, Hicks DG, Numbereye N, Moisini I, Dhakal A, Skinner K, Sanders MAG, Wang X, Shayne M, et al. The Rochester Modified Magee Algorithm (RoMMa): An Outcomes Based Strategy for Clinical Risk-Assessment and Risk-Stratification in ER Positive, HER2 Negative Breast Cancer Patients Being Considered for Oncotype DX® Testing. Cancers. 2023; 15(3):903. https://doi.org/10.3390/cancers15030903
Chicago/Turabian StyleTurner, Bradley M., Brian S. Finkelman, David G. Hicks, Numbere Numbereye, Ioana Moisini, Ajay Dhakal, Kristin Skinner, Mary Ann G. Sanders, Xi Wang, Michelle Shayne, and et al. 2023. "The Rochester Modified Magee Algorithm (RoMMa): An Outcomes Based Strategy for Clinical Risk-Assessment and Risk-Stratification in ER Positive, HER2 Negative Breast Cancer Patients Being Considered for Oncotype DX® Testing" Cancers 15, no. 3: 903. https://doi.org/10.3390/cancers15030903
APA StyleTurner, B. M., Finkelman, B. S., Hicks, D. G., Numbereye, N., Moisini, I., Dhakal, A., Skinner, K., Sanders, M. A. G., Wang, X., Shayne, M., Schiffhauer, L., Katerji, H., & Zhang, H. (2023). The Rochester Modified Magee Algorithm (RoMMa): An Outcomes Based Strategy for Clinical Risk-Assessment and Risk-Stratification in ER Positive, HER2 Negative Breast Cancer Patients Being Considered for Oncotype DX® Testing. Cancers, 15(3), 903. https://doi.org/10.3390/cancers15030903





