Practical Application of Circulating Tumor-Related DNA of Human Papillomavirus in Liquid Biopsy to Evaluate the Molecular Response in Patients with Oropharyngeal Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Clinical Aspects of HPV Genotypes and Prognostic Role of HPV16
3. Clinical Factors Affecting the Quantity of ctHPV16
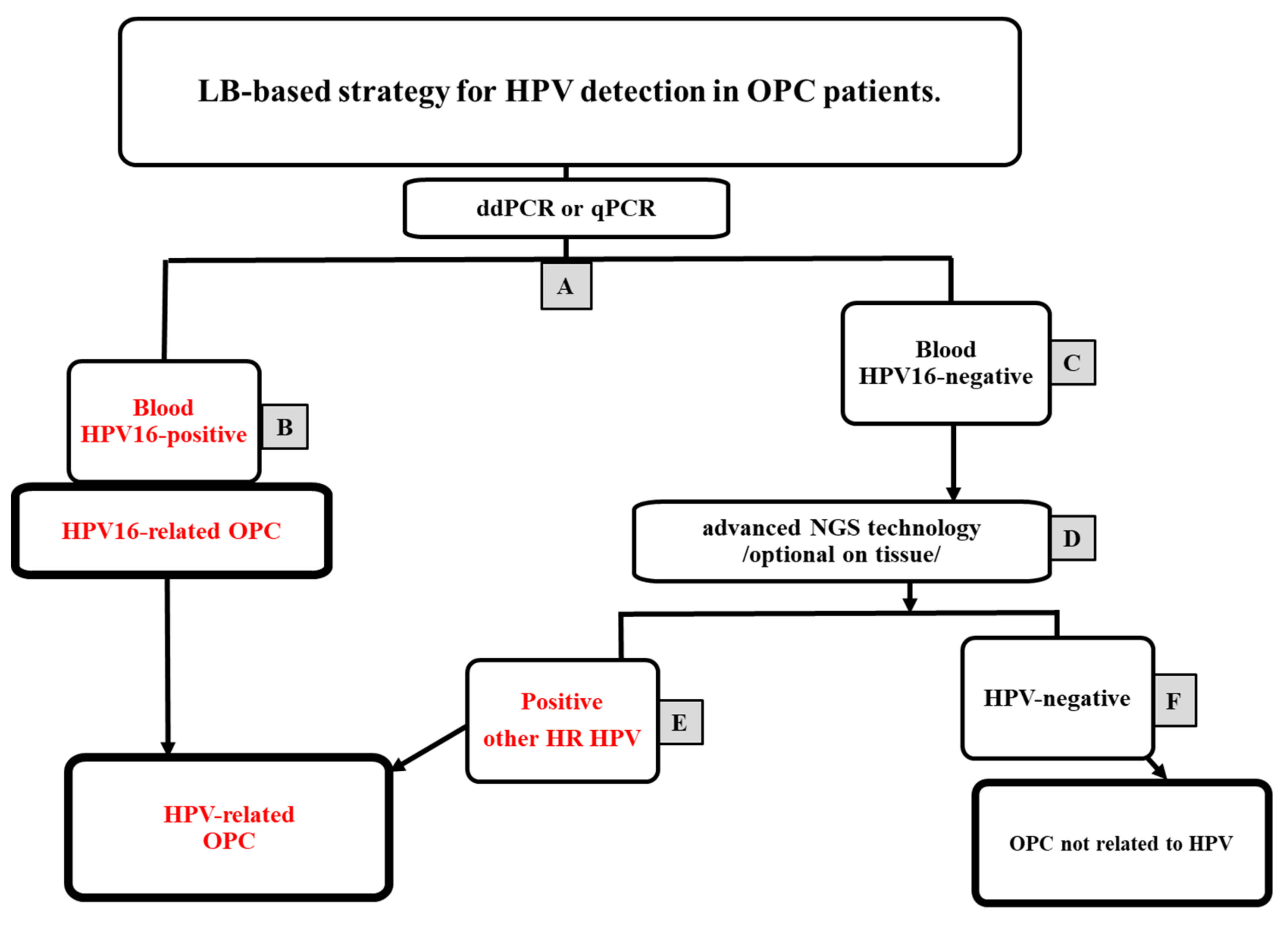
4. Proposal for an LB-Based HPV Diagnostics Regimen
4.1. E6/E7 Sequence as the ctDNA Marker
4.2. ddPCR or qPCR in the First Line of OPC Diagnostics
4.3. NGS in the Second Line of OPC Diagnostics
5. ctHPV16-Based Molecular Response in LB
5.1. ctHPV16 Assay Prior to Treatment—Complementation of the Primary Diagnosis
5.2. ctHPV16 Assay during Treatment—Prediction of Good Response
5.3. ctHPV16 Assay after Treatment Completion—Early Detection of Treatment Failure
5.4. ctHPV16 during Surveillance—Detection of Recurrence or Metastatic Disease
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar] [PubMed]
- Mouliere, F.; El Messaoudi, S.; Gongora, C.; Guedj, A.S.; Robert, B.; Del Rio, M.; Molina, F.; Lamy, P.J.; Lopez-Crapez, E.; Mathonnet, M.; et al. Circulating Cell-Free DNA from Colorectal Cancer Patients May Reveal High KRAS or BRAF Mutation Load. Transl. Oncol. 2013, 6, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Li, M.; Dressman, D.; He, Y.; Shen, D.; Szabo, S.; Diaz, L.A., Jr.; Goodman, S.N.; David, K.A.; Juhl, H.; et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. USA 2005, 102, 16368–16373. [Google Scholar] [CrossRef] [PubMed]
- Anker, P.; Stroun, M. Circulating DNA in plasma or serum. Medicina 2000, 60, 699–702. [Google Scholar]
- Jen, J.; Wu, L.; Sidransky, D. An overview on the isolation and analysis of circulating tumor DNA in plasma and serum. Ann. N. Y. Acad. Sci. 2000, 906, 8–12. [Google Scholar] [CrossRef]
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol. Detect. Quantif. 2019, 17, 100087. [Google Scholar] [CrossRef]
- Cai, X.; Janku, F.; Zhan, Q.; Fan, J.B. Accessing Genetic Information with Liquid Biopsies. Trends Genet. TIG 2015, 31, 564–575. [Google Scholar] [CrossRef]
- Esposito, A.; Bardelli, A.; Criscitiello, C.; Colombo, N.; Gelao, L.; Fumagalli, L.; Minchella, I.; Locatelli, M.; Goldhirsch, A.; Curigliano, G. Monitoring tumor-derived cell-free DNA in patients with solid tumors: Clinical perspectives and research opportunities. Cancer Treat. Rev. 2014, 40, 648–655. [Google Scholar] [CrossRef]
- Gingras, I.; Salgado, R.; Ignatiadis, M. Liquid biopsy: Will it be the ‘magic tool’ for monitoring response of solid tumors to anticancer therapies? Curr. Opin. Oncol. 2015, 27, 560–567. [Google Scholar] [CrossRef]
- Sivars, L.; Palsdottir, K.; Crona Guterstam, Y.; Falconer, H.; Hellman, K.; Tham, E. The current status of cell-free human papillomavirus DNA as a biomarker in cervical cancer and other HPV-associated tumors: A review. Int. J. Cancer 2022, 1–11. [Google Scholar] [CrossRef]
- Mehanna, H.; Beech, T.; Nicholson, T.; El-Hariry, I.; McConkey, C.; Paleri, V.; Roberts, S. Prevalence of human papillomavirus in oropharyngeal and nonoropharyngeal head and neck cancer--systematic review and meta-analysis of trends by time and region. Head Neck 2013, 35, 747–755. [Google Scholar] [CrossRef]
- Del Mistro, A.; Frayle, H.; Menegaldo, A.; Favaretto, N.; Gori, S.; Nicolai, P.; Spinato, G.; Romeo, S.; Tirelli, G.; da Mosto, M.C.; et al. Age-independent increasing prevalence of Human Papillomavirus-driven oropharyngeal carcinomas in North-East Italy. Sci. Rep. 2020, 10, 9320. [Google Scholar] [CrossRef]
- Wittekindt, C.; Wagner, S.; Bushnak, A.; Prigge, E.S.; von Knebel Doeberitz, M.; Würdemann, N.; Bernhardt, K.; Pons-Kühnemann, J.; Maulbecker-Armstrong, C.; Klussmann, J.P. Increasing Incidence rates of Oropharyngeal Squamous Cell Carcinoma in Germany and Significance of Disease Burden Attributed to Human Papillomavirus. Cancer Prev. Res. 2019, 12, 375–382. [Google Scholar] [CrossRef]
- Zamani, M.; Grønhøj, C.; Jensen, D.H.; Carlander, A.F.; Agander, T.; Kiss, K.; Olsen, C.; Baandrup, L.; Nielsen, F.C.; Andersen, E.; et al. The current epidemic of HPV-associated oropharyngeal cancer: An 18-year Danish population-based study with 2,169 patients. Eur. J. Cancer 2020, 134, 52–59. [Google Scholar] [CrossRef]
- Bzhalava, D.; Eklund, C.; Dillner, J. International standardization and classification of human papillomavirus types. Virology 2015, 476, 341–344. [Google Scholar] [CrossRef]
- Bzhalava, D.; Guan, P.; Franceschi, S.; Dillner, J.; Clifford, G. A systematic review of the prevalence of mucosal and cutaneous human papillomavirus types. Virology 2013, 445, 224–231. [Google Scholar] [CrossRef]
- Choi, Y.J.; Park, J.S. Clinical significance of human papillomavirus genotyping. J. Gynecol. Oncol. 2016, 27, e21. [Google Scholar] [CrossRef]
- Lewis, J.S., Jr.; Mirabello, L.; Liu, P.; Wang, X.; Dupont, W.D.; Plummer, W.D.; Pinheiro, M.; Yeager, M.; Boland, J.F.; Cullen, M.; et al. Oropharyngeal Squamous Cell Carcinoma Morphology and Subtypes by Human Papillomavirus Type and by 16 Lineages and Sublineages. Head Neck Pathol. 2021, 15, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Schache, A.G.; Powell, N.G.; Cuschieri, K.S.; Robinson, M.; Leary, S.; Mehanna, H.; Rapozo, D.; Long, A.; Cubie, H.; Junor, E.; et al. HPV-Related Oropharynx Cancer in the United Kingdom: An Evolution in the Understanding of Disease Etiology. Cancer Res. 2016, 76, 6598–6606. [Google Scholar] [CrossRef]
- Wagner, S.; Prigge, E.S.; Wuerdemann, N.; Reder, H.; Bushnak, A.; Sharma, S.J.; Obermueller, T.; von Knebel Doeberitz, M.; Dreyer, T.; Gattenlöhner, S.; et al. Evaluation of p16(INK4a) expression as a single marker to select patients with HPV-driven oropharyngeal cancers for treatment de-escalation. Br. J. Cancer 2020, 123, 1114–1122. [Google Scholar] [CrossRef] [PubMed]
- Simoens, C.; Gorbaslieva, I.; Gheit, T.; Holzinger, D.; Lucas, E.; Ridder, R.; Rehm, S.; Vermeulen, P.; Lammens, M.; Vanderveken, O.M.; et al. HPV DNA genotyping, HPV E6*I mRNA detection, and p16(INK4a)/Ki-67 staining in Belgian head and neck cancer patient specimens, collected within the HPV-AHEAD study. Cancer Epidemiol. 2021, 72, 101925. [Google Scholar] [CrossRef]
- Guo, M.; Khanna, A.; Tinnirello, A.A.; Hwang, J.; Zhang, P.; Xu, L.; Li, G.; Dahlstrom, K.R.; Sturgis, E.M.; Stewart, J. Detection accuracy of the Cobas HPV assay for high-risk HPV in head and neck FNA biopsy specimens. Cancer Cytopathol. 2022, 130, 523–530. [Google Scholar] [CrossRef] [PubMed]
- El-Salem, F.; Mansour, M.; Gitman, M.; Miles, B.A.; Posner, M.R.; Bakst, R.L.; Genden, E.M.; Westra, W.H. Real-time PCR HPV genotyping in fine needle aspirations of metastatic head and neck squamous cell carcinoma: Exposing the limitations of conventional p16 immunostaining. Oral Oncol. 2019, 90, 74–79. [Google Scholar] [CrossRef]
- Shenker, R.F.; May, N.H.; Waltonen, J.D.; Yang, J.P.; O’Neill, S.S.; Frizzell, B.A.; Greven, K.M.; Hughes, R.T. Comparing Outcomes for Patients with Human Papillomavirus (HPV) Type 16 versus Other High-Risk HPV Types in Oropharyngeal Squamous Cell Carcinoma. Head Neck Pathol. 2021, 15, 866–874. [Google Scholar] [CrossRef]
- Mazul, A.L.; Rodriguez-Ormaza, N.; Taylor, J.M.; Desai, D.D.; Brennan, P.; Anantharaman, D.; Gheit, T.; Tommasino, M.; Abedi-Ardekani, B.; Olshan, A.F.; et al. Prognostic significance of non-HPV16 genotypes in oropharyngeal squamous cell carcinoma. Oral Oncol. 2016, 61, 98–103. [Google Scholar] [CrossRef]
- Mashiana, S.S.; Navale, P.; Khandakar, B.; Sobotka, S.; Posner, M.R.; Miles, B.A.; Zhang, W.; Gitman, M.; Bakst, R.L.; Genden, E.M.; et al. Human papillomavirus genotype distribution in head and neck cancer: Informing developing strategies for cancer prevention, diagnosis, treatment and surveillance. Oral Oncol. 2021, 113, 105109. [Google Scholar] [CrossRef]
- Garset-Zamani, M.; Carlander, A.F.; Jakobsen, K.K.; Friborg, J.; Kiss, K.; Marvig, R.L.; Olsen, C.; Nielsen, F.C.; Andersen, E.; Grønhøj, C.; et al. Impact of specific high-risk human papillomavirus genotypes on survival in oropharyngeal cancer. Int. J. Cancer 2022, 150, 1174–1183. [Google Scholar] [CrossRef]
- Kim, Y.; Joo, Y.H.; Kim, M.S.; Lee, Y.S. Prevalence of high-risk human papillomavirus and its genotype distribution in head and neck squamous cell carcinomas. J. Pathol. Transl. Med. 2020, 54, 411–418. [Google Scholar] [CrossRef]
- Ziai, H.; Warner, A.; Mundi, N.; Patel, K.; Chung, E.J.; Howlett, C.J.; Plantinga, P.; Yoo, J.; MacNeil, S.D.; Fung, K.; et al. Does HPV Subtype Predict Outcomes in Head and Neck Cancers? Int. J. Otolaryngol. 2021, 2021, 6672373. [Google Scholar] [CrossRef]
- Varier, I.; Keeley, B.R.; Krupar, R.; Patsias, A.; Dong, J.; Gupta, N.; Parasher, A.K.; Genden, E.M.; Miles, B.A.; Teng, M.; et al. Clinical characteristics and outcomes of oropharyngeal carcinoma related to high-risk non-human papillomavirus16 viral subtypes. Head Neck 2016, 38, 1330–1337. [Google Scholar] [CrossRef] [PubMed]
- Bratman, S.V.; Bruce, J.P.; O’Sullivan, B.; Pugh, T.J.; Xu, W.; Yip, K.W.; Liu, F.F. Human Papillomavirus Genotype Association With Survival in Head and Neck Squamous Cell Carcinoma. JAMA Oncol. 2016, 2, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Petrelli, F.; Sarti, E.; Barni, S. Predictive value of human papillomavirus in oropharyngeal carcinoma treated with radiotherapy: An updated systematic review and meta-analysis of 30 trials. Head Neck 2014, 36, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Rainsbury, J.W.; Ahmed, W.; Williams, H.K.; Roberts, S.; Paleri, V.; Mehanna, H. Prognostic biomarkers of survival in oropharyngeal squamous cell carcinoma: Systematic review and meta-analysis. Head Neck 2013, 35, 1048–1055. [Google Scholar] [CrossRef] [PubMed]
- No, J.H.; Sung, M.W.; Hah, J.H.; Choi, S.H.; Lee, M.C.; Kim, H.S.; Song, Y.S. Prevalence and prognostic value of human papillomavirus genotypes in tonsillar squamous cell carcinoma: A Korean multicenter study. Cancer 2015, 121, 535–544. [Google Scholar] [CrossRef]
- Yoo, S.H.; Ock, C.Y.; Keam, B.; Park, S.J.; Kim, T.M.; Kim, J.H.; Jeon, Y.K.; Chung, E.J.; Kwon, S.K.; Hah, J.H.; et al. Poor prognostic factors in human papillomavirus-positive head and neck cancer: Who might not be candidates for de-escalation treatment? Korean J. Intern. Med. 2019, 34, 1313–1323. [Google Scholar] [CrossRef]
- Chatfield-Reed, K.; Gui, S.; O’Neill, W.Q.; Teknos, T.N.; Pan, Q. HPV33+ HNSCC is associated with poor prognosis and has unique genomic and immunologic landscapes. Oral Oncol. 2020, 100, 104488. [Google Scholar] [CrossRef]
- Roberts, S.A.; Lawrence, M.S.; Klimczak, L.J.; Grimm, S.A.; Fargo, D.; Stojanov, P.; Kiezun, A.; Kryukov, G.V.; Carter, S.L.; Saksena, G.; et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013, 45, 970–976. [Google Scholar] [CrossRef]
- Saber, C.N.; Grønhøj Larsen, C.; Dalianis, T.; von Buchwald, C. Immune cells and prognosis in HPV-associated oropharyngeal squamous cell carcinomas: Review of the literature. Oral Oncol. 2016, 58, 8–13. [Google Scholar] [CrossRef]
- Dahlstrom, K.R.; Li, G.; Hussey, C.S.; Vo, J.T.; Wei, Q.; Zhao, C.; Sturgis, E.M. Circulating human papillomavirus DNA as a marker for disease extent and recurrence among patients with oropharyngeal cancer. Cancer 2015, 121, 3455–3464. [Google Scholar] [CrossRef]
- Cao, H.; Banh, A.; Kwok, S.; Shi, X.; Wu, S.; Krakow, T.; Khong, B.; Bavan, B.; Bala, R.; Pinsky, B.A.; et al. Quantitation of human papillomavirus DNA in plasma of oropharyngeal carcinoma patients. Int. J. Radiat. Oncol. Biol. Phys. 2012, 82, e351–e358. [Google Scholar] [CrossRef] [PubMed]
- Mazurek, A.M.; Rutkowski, T.; Śnietura, M.; Pigłowski, W.; Suwiński, R.; Składowski, K. Detection of circulating HPV16 DNA as a biomarker in the blood of patients with human papillomavirus-positive oropharyngeal squamous cell carcinoma. Head Neck 2019, 41, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Hanna, G.J.; Supplee, J.G.; Kuang, Y.; Mahmood, U.; Lau, C.J.; Haddad, R.I.; Jänne, P.A.; Paweletz, C.P. Plasma HPV cell-free DNA monitoring in advanced HPV-associated oropharyngeal cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018, 29, 1980–1986. [Google Scholar] [CrossRef] [PubMed]
- Damerla, R.R.; Lee, N.Y.; You, D.; Soni, R.; Shah, R.; Reyngold, M.; Katabi, N.; Wu, V.; McBride, S.M.; Tsai, C.J.; et al. Detection of Early Human Papillomavirus-Associated Cancers by Liquid Biopsy. JCO Precis. Oncol. 2019, 3, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Jeannot, E.; Becette, V.; Campitelli, M.; Calméjane, M.A.; Lappartient, E.; Ruff, E.; Saada, S.; Holmes, A.; Bellet, D.; Sastre-Garau, X. Circulating human papillomavirus DNA detected using droplet digital PCR in the serum of patients diagnosed with early stage human papillomavirus-associated invasive carcinoma. J. Pathol. Clin. Res. 2016, 2, 201–209. [Google Scholar] [CrossRef]
- Chera, B.S.; Kumar, S.; Beaty, B.T.; Marron, D.; Jefferys, S.; Green, R.; Goldman, E.C.; Amdur, R.; Sheets, N.; Dagan, R.; et al. Rapid Clearance Profile of Plasma Circulating Tumor HPV Type 16 DNA during Chemoradiotherapy Correlates with Disease Control in HPV-Associated Oropharyngeal Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 4682–4690. [Google Scholar] [CrossRef]
- Piccini, A.; Storey, A.; Romanos, M.; Banks, L. Regulation of human papillomavirus type 16 DNA replication by E2, glucocorticoid hormone and epidermal growth factor. J. Gen. Virol. 1997, 78, 1963–1970. [Google Scholar] [CrossRef]
- Reder, H.; Taferner, V.F.; Wittekindt, C.; Bräuninger, A.; Speel, E.M.; Gattenlöhner, S.; Wolf, G.; Klussmann, J.P.; Wuerdemann, N.; Wagner, S. Plasma Cell-Free Human Papillomavirus Oncogene E6 and E7 DNA Predicts Outcome in Oropharyngeal Squamous Cell Carcinoma. J. Mol. Diagn. JMD 2020, 22, 1333–1343. [Google Scholar] [CrossRef]
- Pinatti, L.M.; Walline, H.M.; Carey, T.E. Human Papillomavirus Genome Integration and Head and Neck Cancer. J. Dent. Res. 2018, 97, 691–700. [Google Scholar] [CrossRef]
- Bodily, J.; Laimins, L.A. Persistence of human papillomavirus infection: Keys to malignant progression. Trends Microbiol. 2011, 19, 33–39. [Google Scholar] [CrossRef]
- Tuna, M.; Amos, C.I. Next generation sequencing and its applications in HPV-associated cancers. Oncotarget 2017, 8, 8877–8889. [Google Scholar] [CrossRef]
- Araldi, R.P.; Sant’Ana, T.A.; Módolo, D.G.; de Melo, T.C.; Spadacci-Morena, D.D.; de Cassia Stocco, R.; Cerutti, J.M.; de Souza, E.B. The human papillomavirus (HPV)-related cancer biology: An overview. Biomed. Pharmacother. 2018, 106, 1537–1556. [Google Scholar] [CrossRef]
- Parfenov, M.; Pedamallu, C.S.; Gehlenborg, N.; Freeman, S.S.; Danilova, L.; Bristow, C.A.; Lee, S.; Hadjipanayis, A.G.; Ivanova, E.V.; Wilkerson, M.D.; et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 15544–15549. [Google Scholar] [CrossRef]
- Feller, L.; Wood, N.H.; Khammissa, R.A.; Lemmer, J. Human papillomavirus-mediated carcinogenesis and HPV-associated oral and oropharyngeal squamous cell carcinoma. Part 1: Human papillomavirus-mediated carcinogenesis. Head Face Med. 2010, 6, 15. [Google Scholar] [CrossRef]
- Scarth, J.A.; Patterson, M.R.; Morgan, E.L.; Macdonald, A. The human papillomavirus oncoproteins: A review of the host pathways targeted on the road to transformation. J. Gen. Virol. 2021, 102, 001540. [Google Scholar] [CrossRef]
- Narisawa-Saito, M.; Kiyono, T. Basic mechanisms of high-risk human papillomavirus-induced carcinogenesis: Roles of E6 and E7 proteins. Cancer Sci. 2007, 98, 1505–1511. [Google Scholar] [CrossRef]
- Snijders, P.J.; Steenbergen, R.D.; Heideman, D.A.; Meijer, C.J. HPV-mediated cervical carcinogenesis: Concepts and clinical implications. J. Pathol. 2006, 208, 152–164. [Google Scholar] [CrossRef]
- Akagi, K.; Li, J.; Broutian, T.R.; Padilla-Nash, H.; Xiao, W.; Jiang, B.; Rocco, J.W.; Teknos, T.N.; Kumar, B.; Wangsa, D.; et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2014, 24, 185–199. [Google Scholar] [CrossRef]
- Krasniqi, E.; Barba, M.; Venuti, A.; Pizzuti, L.; Cappuzzo, F.; Landi, L.; Carpano, S.; Marchetti, P.; Villa, A.; Vizza, E.; et al. Circulating HPV DNA in the Management of Oropharyngeal and Cervical Cancers: Current Knowledge and Future Perspectives. J. Clin. Med. 2021, 10, 1525. [Google Scholar] [CrossRef]
- van Ginkel, J.H.; Slieker, F.J.B.; de Bree, R.; van Es, R.J.J.; Van Cann, E.M.; Willems, S.M. Cell-free nucleic acids in body fluids as biomarkers for the prediction and early detection of recurrent head and neck cancer: A systematic review of the literature. Oral Oncol. 2017, 75, 8–15. [Google Scholar] [CrossRef]
- Keller, L.; Belloum, Y.; Wikman, H.; Pantel, K. Clinical relevance of blood-based ctDNA analysis: Mutation detection and beyond. Br. J. Cancer 2021, 124, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Swiecicki, P.L.; Brennan, J.R.; Mierzwa, M.; Spector, M.E.; Brenner, J.C. Head and Neck Squamous Cell Carcinoma Detection and Surveillance: Advances of Liquid Biomarkers. Laryngoscope 2019, 129, 1836–1843. [Google Scholar] [CrossRef] [PubMed]
- Fauzi, F.H.; Hamzan, N.I.; Rahman, N.A.; Suraiya, S.; Mohamad, S. Detection of human papillomavirus in oropharyngeal squamous cell carcinoma. J. Zhejiang Univ. Sci. B 2020, 21, 961–976. [Google Scholar] [CrossRef] [PubMed]
- Augustin, J.G.; Lepine, C.; Morini, A.; Brunet, A.; Veyer, D.; Brochard, C.; Mirghani, H.; Péré, H.; Badoual, C. HPV Detection in Head and Neck Squamous Cell Carcinomas: What Is the Issue? Front. Oncol. 2020, 10, 1751. [Google Scholar] [CrossRef] [PubMed]
- Mattox, A.K.; D’Souza, G.; Khan, Z.; Allen, H.; Henson, S.; Seiwert, T.Y.; Koch, W.; Pardoll, D.M.; Fakhry, C. Comparison of next generation sequencing, droplet digital PCR, and quantitative real-time PCR for the earlier detection and quantification of HPV in HPV-positive oropharyngeal cancer. Oral Oncol. 2022, 128, 105805. [Google Scholar] [CrossRef]
- Lo, Y.M. Quantitative analysis of Epstein-Barr virus DNA in plasma and serum: Applications to tumor detection and monitoring. Ann. N. Y. Acad. Sci. 2001, 945, 68–72. [Google Scholar] [CrossRef]
- Kojabad, A.A.; Farzanehpour, M.; Galeh, H.E.G.; Dorostkar, R.; Jafarpour, A.; Bolandian, M.; Nodooshan, M.M. Droplet digital PCR of viral DNA/RNA, current progress, challenges, and future perspectives. J. Med. Virol. 2021, 93, 4182–4197. [Google Scholar] [CrossRef]
- Wuerdemann, N.; Jain, R.; Adams, A.; Speel, E.M.; Wagner, S.; Joosse, S.A.; Klussmann, J.P. Cell-Free HPV-DNA as a Biomarker for Oropharyngeal Squamous Cell Carcinoma-A Step Towards Personalized Medicine? Cancers 2020, 12, 2997. [Google Scholar] [CrossRef]
- Jakobsen, K.K.; Carlander, A.F.; Bendtsen, S.K.; Garset-Zamani, M.; Lynggaard, C.D.; Grønhøj, C.; von Buchwald, C. Diagnostic Accuracy of HPV Detection in Patients with Oropharyngeal Squamous Cell Carcinomas: A Systematic Review and Meta-Analysis. Viruses 2021, 13, 1692. [Google Scholar] [CrossRef]
- Campo, F.; Zocchi, J.; Moretto, S.; Mazzola, F.; Petruzzi, G.; Donà, M.G.; Benevolo, M.; Iocca, O.; De Virgilio, A.; Pichi, B.; et al. Cell-Free Human Papillomavirus-DNA for Monitoring Treatment Response of Head and Neck Squamous Cell Carcinoma: Systematic Review and Meta-Analysis. Laryngoscope 2022, 132, 560–568. [Google Scholar] [CrossRef]
- Burd, E.M. Human Papillomavirus Laboratory Testing: The Changing Paradigm. Clin. Microbiol. Rev. 2016, 29, 291–319. [Google Scholar] [CrossRef]
- Burd, E.M. Validation of laboratory-developed molecular assays for infectious diseases. Clin. Microbiol. Rev. 2010, 23, 550–576. [Google Scholar] [CrossRef]
- Leung, E.; Han, K.; Zou, J.; Zhao, Z.; Zheng, Y.; Wang, T.T.; Rostami, A.; Siu, L.L.; Pugh, T.J.; Bratman, S.V. HPV Sequencing Facilitates Ultrasensitive Detection of HPV Circulating Tumor DNA. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2021, 27, 5857–5868. [Google Scholar] [CrossRef]
- Sastre-Garau, X.; Diop, M.; Martin, F.; Dolivet, G.; Marchal, F.; Charra-Brunaud, C.; Peiffert, D.; Leufflen, L.; Dembélé, B.; Demange, J.; et al. A NGS-based Blood Test For the Diagnosis of Invasive HPV-associated Carcinomas with Extensive Viral Genomic Characterization. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2021, 27, 5307–5316. [Google Scholar] [CrossRef]
- Speel, E.J. HPV Integration in Head and Neck Squamous Cell Carcinomas: Cause and Consequence. Recent Results Cancer Res. Der Krebsforsch. Prog. Dans Les. Rech. Sur Le Cancer 2017, 206, 57–72. [Google Scholar] [CrossRef]
- Chung, C.H.; Guthrie, V.B.; Masica, D.L.; Tokheim, C.; Kang, H.; Richmon, J.; Agrawal, N.; Fakhry, C.; Quon, H.; Subramaniam, R.M.; et al. Genomic alterations in head and neck squamous cell carcinoma determined by cancer gene-targeted sequencing. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2015, 26, 1216–1223. [Google Scholar] [CrossRef]
- Cirillo, M.; Craig, A.F.M.; Borchmann, S.; Kurtz, D.M. Liquid biopsy in lymphoma: Molecular methods and clinical applications. Cancer Treat. Rev. 2020, 91, 102106. [Google Scholar] [CrossRef]
- Rutkowski, T.W.; Mazurek, A.M.; Śnietura, M.; Hejduk, B.; Jędrzejewska, M.; Bobek-Billewicz, B.; d’Amico, A.; Pigłowski, W.; Wygoda, A.; Składowski, K.; et al. Circulating HPV16 DNA may complement imaging assessment of early treatment efficacy in patients with HPV-positive oropharyngeal cancer. J. Transl. Med. 2020, 18, 167. [Google Scholar] [CrossRef]
- Ragin, C.C.; Taioli, E. Survival of squamous cell carcinoma of the head and neck in relation to human papillomavirus infection: Review and meta-analysis. Int. J. Cancer 2007, 121, 1813–1820. [Google Scholar] [CrossRef]
- Fakhry, C.; Westra, W.H.; Li, S.; Cmelak, A.; Ridge, J.A.; Pinto, H.; Forastiere, A.; Gillison, M.L. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J. Natl. Cancer Inst. 2008, 100, 261–269. [Google Scholar] [CrossRef]
- Prigge, E.S.; Arbyn, M.; von Knebel Doeberitz, M.; Reuschenbach, M. Diagnostic accuracy of p16(INK4a) immunohistochemistry in oropharyngeal squamous cell carcinomas: A systematic review and meta-analysis. Int. J. Cancer 2017, 140, 1186–1198. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Quabius, E.S. Relevance of Human Papillomaviruses in Head and Neck Cancer-What Remains in 2021 from a Clinician’s Point of View? Viruses 2021, 13, 1173. [Google Scholar] [CrossRef]
- Molimard, C.; L’Huillier, V.; Overs, A.; Soret, C.; Algros, M.P.; Mougin, C.; Guenat, D.; Mauvais, O.; Prétet, J.L. Human papillomavirus DNA and p16 expression in head and neck squamous cell carcinoma in young French patients. J. Int. Med. Res. 2021, 49, 3000605211022534. [Google Scholar] [CrossRef] [PubMed]
- Husain, N.; Neyaz, A. Human papillomavirus associated head and neck squamous cell carcinoma: Controversies and new concepts. J. Oral Biol. Craniofacial Res. 2017, 7, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Hashida, Y.; Higuchi, T.; Matsumoto, S.; Iguchi, M.; Murakami, I.; Hyodo, M.; Daibata, M. Prognostic significance of human papillomavirus 16 viral load level in patients with oropharyngeal cancer. Cancer Sci. 2021, 112, 4404–4417. [Google Scholar] [CrossRef]
- Sabatini, M.E.; Chiocca, S. Human papillomavirus as a driver of head and neck cancers. Br. J. Cancer 2020, 122, 306–314. [Google Scholar] [CrossRef]
- Nauta, I.H.; Rietbergen, M.M.; van Bokhoven, A.; Bloemena, E.; Lissenberg-Witte, B.I.; Heideman, D.A.M.; Baatenburg de Jong, R.J.; Brakenhoff, R.H.; Leemans, C.R. Evaluation of the eighth TNM classification on p16-positive oropharyngeal squamous cell carcinomas in the Netherlands and the importance of additional HPV DNA testing. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018, 29, 1273–1279. [Google Scholar] [CrossRef]
- Bates, J.E.; Steuer, C.E. HPV as a Carcinomic Driver in Head and Neck Cancer: A De-escalated Future? Curr. Treat. Options Oncol. 2022, 23, 325–332. [Google Scholar] [CrossRef]
- Machiels, J.P.; René Leemans, C.; Golusinski, W.; Grau, C.; Licitra, L.; Gregoire, V. Squamous cell carcinoma of the oral cavity, larynx, oropharynx and hypopharynx: EHNS-ESMO-ESTRO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2020, 31, 1462–1475. [Google Scholar] [CrossRef]
- Cohen, E.E.; Karrison, T.G.; Kocherginsky, M.; Mueller, J.; Egan, R.; Huang, C.H.; Brockstein, B.E.; Agulnik, M.B.; Mittal, B.B.; Yunus, F.; et al. Phase III randomized trial of induction chemotherapy in patients with N2 or N3 locally advanced head and neck cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2014, 32, 2735–2743. [Google Scholar] [CrossRef]
- Haddad, R.; O’Neill, A.; Rabinowits, G.; Tishler, R.; Khuri, F.; Adkins, D.; Clark, J.; Sarlis, N.; Lorch, J.; Beitler, J.J.; et al. Induction chemotherapy followed by concurrent chemoradiotherapy (sequential chemoradiotherapy) versus concurrent chemoradiotherapy alone in locally advanced head and neck cancer (PARADIGM): A randomised phase 3 trial. Lancet Oncol. 2013, 14, 257–264. [Google Scholar] [CrossRef]
- Hitt, R.; Grau, J.J.; López-Pousa, A.; Berrocal, A.; García-Girón, C.; Irigoyen, A.; Sastre, J.; Martínez-Trufero, J.; Brandariz Castelo, J.A.; Verger, E.; et al. A randomized phase III trial comparing induction chemotherapy followed by chemoradiotherapy versus chemoradiotherapy alone as treatment of unresectable head and neck cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2014, 25, 25–216. [Google Scholar] [CrossRef]
- O’Sullivan, B.; Huang, S.H.; Siu, L.L.; Waldron, J.; Zhao, H.; Perez-Ordonez, B.; Weinreb, I.; Kim, J.; Ringash, J.; Bayley, A.; et al. Deintensification candidate subgroups in human papillomavirus-related oropharyngeal cancer according to minimal risk of distant metastasis. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013, 31, 543–550. [Google Scholar] [CrossRef]
- Marur, S.; Li, S.; Cmelak, A.J.; Gillison, M.L.; Zhao, W.J.; Ferris, R.L.; Westra, W.H.; Gilbert, J.; Bauman, J.E.; Wagner, L.I.; et al. E1308: Phase II Trial of Induction Chemotherapy Followed by Reduced-Dose Radiation and Weekly Cetuximab in Patients With HPV-Associated Resectable Squamous Cell Carcinoma of the Oropharynx- ECOG-ACRIN Cancer Research Group. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2017, 35, 490–497. [Google Scholar] [CrossRef]
- Spreafico, A.; Huang, S.H.; Xu, W.; Granata, R.; Liu, C.S.; Waldron, J.N.; Chen, E.; Ringash, J.; Bayley, A.; Chan, K.K.; et al. Impact of cisplatin dose intensity on human papillomavirus-related and -unrelated locally advanced head and neck squamous cell carcinoma. Eur. J. Cancer 2016, 67, 174–182. [Google Scholar] [CrossRef]
- Sadeghi, N.; Khalife, S.; Mascarella, M.A.; Ramanakumar, A.V.; Richardson, K.; Joshi, A.S.; Bouganim, N.; Taheri, R.; Fuson, A.; Siegel, R. Pathologic response to neoadjuvant chemotherapy in HPV-associated oropharynx cancer. Head Neck 2020, 42, 417–425. [Google Scholar] [CrossRef]
- Inukai, D.; Kan, T.; Yamanaka, S.; Okamoto, H.; Fujimoto, Y.; Ito, T.; Taniguchi, N.; Yamamoto, Y.; Tsuzuki, T.; Takami, A.; et al. Pathological and Virological Studies of p16-Positive Oropharyngeal Carcinoma with a Good Response to Neoadjuvant Chemotherapy. Microorganisms 2020, 8, 1497. [Google Scholar] [CrossRef]
- O’Boyle, C.J.; Siravegna, G.; Varmeh, S.; Queenan, N.; Michel, A.; Pang, K.C.S.; Stein, J.; Thierauf, J.C.; Sadow, P.M.; Faquin, W.C.; et al. Cell-free human papillomavirus DNA kinetics after surgery for human papillomavirus-associated oropharyngeal cancer. Cancer 2022, 128, 2193–2204. [Google Scholar] [CrossRef]
- Mehanna, H.; Rischin, D.; Wong, S.J.; Gregoire, V.; Ferris, R.; Waldron, J.; Le, Q.T.; Forster, M.; Gillison, M.; Laskar, S.; et al. De-Escalation After DE-ESCALATE and RTOG 1016: A Head and Neck Cancer InterGroup Framework for Future De-Escalation Studies. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2020, 38, 2552–2557. [Google Scholar] [CrossRef]
- Hamoir, M.; Ferlito, A.; Schmitz, S.; Hanin, F.X.; Thariat, J.; Weynand, B.; Machiels, J.P.; Grégoire, V.; Robbins, K.T.; Silver, C.E.; et al. The role of neck dissection in the setting of chemoradiation therapy for head and neck squamous cell carcinoma with advanced neck disease. Oral Oncol. 2012, 48, 203–210. [Google Scholar] [CrossRef]
- Andrade, R.S.; Heron, D.E.; Degirmenci, B.; Filho, P.A.; Branstetter, B.F.; Seethala, R.R.; Ferris, R.L.; Avril, N. Posttreatment assessment of response using FDG-PET/CT for patients treated with definitive radiation therapy for head and neck cancers. Int. J. Radiat. Oncol. Biol. Phys. 2006, 65, 1315–1322. [Google Scholar] [CrossRef] [PubMed]
- Manikantan, K.; Khode, S.; Dwivedi, R.C.; Palav, R.; Nutting, C.M.; Rhys-Evans, P.; Harrington, K.J.; Kazi, R. Making sense of post-treatment surveillance in head and neck cancer: When and what of follow-up. Cancer Treat. Rev. 2009, 35, 744–753. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, W.J., Jr. Salvage surgery for patients with recurrent squamous cell carcinoma of the upper aerodigestive tract: When do the ends justify the means? Laryngoscope 2000, 110, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Bledsoe, T.J.; Noble, A.R.; Hunter, G.K.; Rybicki, L.A.; Hoschar, A.; Chute, D.J.; Saxton, J.P.; Greskovich, J.F.; Adelstein, D.J.; Koyfman, S.A. Oropharyngeal squamous cell carcinoma with known human papillomavirus status treated with definitive chemoradiotherapy: Patterns of failure and toxicity outcomes. Radiat. Oncol. 2013, 8, 174. [Google Scholar] [CrossRef]
- Vedrine, P.O.; Thariat, J.; Hitier, M.; Janot, F.; Kaminsky, M.C.; Makeieff, M.; De Raucourt, D.; Lapeyre, M.; Toussaint, B. Need for neck dissection after radiochemotherapy? A study of the French GETTEC Group. Laryngoscope 2008, 118, 1775–1780. [Google Scholar] [CrossRef]
- van der Putten, L.; van den Broek, G.B.; de Bree, R.; van den Brekel, M.W.; Balm, A.J.; Hoebers, F.J.; Doornaert, P.; Leemans, C.R.; Rasch, C.R. Effectiveness of salvage selective and modified radical neck dissection for regional pathologic lymphadenopathy after chemoradiation. Head Neck 2009, 31, 593–603. [Google Scholar] [CrossRef]
- Lango, M.N.; Egleston, B.; Ende, K.; Feigenberg, S.; D’Ambrosio, D.J.; Cohen, R.B.; Ahmad, S.; Nicolaou, N.; Ridge, J.A. Impact of neck dissection on long-term feeding tube dependence in patients with head and neck cancer treated with primary radiation or chemoradiation. Head Neck 2010, 32, 341–347. [Google Scholar] [CrossRef]
- McCabe, K.J.; Rubinstein, D. Advances in head and neck imaging. Otolaryngol. Clin. N. Am. 2005, 38, 307–319. [Google Scholar] [CrossRef]
- Huang, S.H.; Perez-Ordonez, B.; Weinreb, I.; Hope, A.; Massey, C.; Waldron, J.N.; Kim, J.; Bayley, A.J.; Cummings, B.; Cho, B.C.; et al. Natural course of distant metastases following radiotherapy or chemoradiotherapy in HPV-related oropharyngeal cancer. Oral Oncol. 2013, 49, 79–85. [Google Scholar] [CrossRef]
- Wong, L.Y.; Wei, W.I.; Lam, L.K.; Yuen, A.P. Salvage of recurrent head and neck squamous cell carcinoma after primary curative surgery. Head Neck 2003, 25, 953–959. [Google Scholar] [CrossRef]
- Lee, J.Y.; Garcia-Murillas, I.; Cutts, R.J.; De Castro, D.G.; Grove, L.; Hurley, T.; Wang, F.; Nutting, C.; Newbold, K.; Harrington, K.; et al. Predicting response to radical (chemo)radiotherapy with circulating HPV DNA in locally advanced head and neck squamous carcinoma. Br. J. Cancer 2017, 117, 876–883. [Google Scholar] [CrossRef]
- Tanaka, H.; Takemoto, N.; Horie, M.; Takai, E.; Fukusumi, T.; Suzuki, M.; Eguchi, H.; Komukai, S.; Tatsumi, M.; Isohashi, F.; et al. Circulating tumor HPV DNA complements PET-CT in guiding management after radiotherapy in HPV-related squamous cell carcinoma of the head and neck. Int. J. Cancer 2021, 148, 995–1005. [Google Scholar] [CrossRef]
- Partlová, S.; Bouček, J.; Kloudová, K.; Lukešová, E.; Zábrodský, M.; Grega, M.; Fučíková, J.; Truxová, I.; Tachezy, R.; Špíšek, R.; et al. Distinct patterns of intratumoral immune cell infiltrates in patients with HPV-associated compared to non-virally induced head and neck squamous cell carcinoma. Oncoimmunology 2015, 4, e965570. [Google Scholar] [CrossRef]
- Vainshtein, J.M.; Spector, M.E.; Stenmark, M.H.; Bradford, C.R.; Wolf, G.T.; Worden, F.P.; Chepeha, D.B.; McHugh, J.B.; Carey, T.; Wong, K.K.; et al. Reliability of post-chemoradiotherapy F-18-FDG PET/CT for prediction of locoregional failure in human papillomavirus-associated oropharyngeal cancer. Oral Oncol. 2014, 50, 234–239. [Google Scholar] [CrossRef]
- Rulach, R.; Zhou, S.; Hendry, F.; Stobo, D.; James, A.; Dempsey, M.F.; Grose, D.; Lamb, C.; Schipani, S.; Rizwanullah, M.; et al. 12 week PET-CT has low positive predictive value for nodal residual disease in human papillomavirus-positive oropharyngeal cancers. Oral Oncol. 2019, 97, 76–81. [Google Scholar] [CrossRef]
- Liu, H.Y.; Milne, R.; Lock, G.; Panizza, B.J.; Bernard, A.; Foote, M.; McGrath, M.; Brown, E.; Gandhi, M.; Porceddu, S.V. Utility of a repeat PET/CT scan in HPV-associated Oropharyngeal Cancer following incomplete nodal response from (chemo)radiotherapy. Oral Oncol. 2019, 88, 153–159. [Google Scholar] [CrossRef]
- Posner, M.R.; Lorch, J.H.; Goloubeva, O.; Tan, M.; Schumaker, L.M.; Sarlis, N.J.; Haddad, R.I.; Cullen, K.J. Survival and human papillomavirus in oropharynx cancer in TAX 324: A subset analysis from an international phase III trial. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2011, 22, 1071–1077. [Google Scholar] [CrossRef]
- Ang, K.K.; Harris, J.; Wheeler, R.; Weber, R.; Rosenthal, D.I.; Nguyen-Tân, P.F.; Westra, W.H.; Chung, C.H.; Jordan, R.C.; Lu, C.; et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 2010, 363, 24–35. [Google Scholar] [CrossRef]
- Ley, J.; Wildes, T.; El-Mofty, S.; Adkins, D. Metastasis occurring eleven years after diagnosis of human papilloma virus-related oropharyngeal squamous cell carcinoma. Ecancermedicalscience 2014, 8, 480. [Google Scholar] [CrossRef]
- Trosman, S.J.; Koyfman, S.A.; Ward, M.C.; Al-Khudari, S.; Nwizu, T.; Greskovich, J.F.; Lamarre, E.D.; Scharpf, J.; Khan, M.J.; Lorenz, R.R.; et al. Effect of human papillomavirus on patterns of distant metastatic failure in oropharyngeal squamous cell carcinoma treated with chemoradiotherapy. JAMA Otolaryngol.—Head Neck Surg. 2015, 141, 457–462. [Google Scholar] [CrossRef]
- Fakhry, C.; Zhang, Q.; Nguyen-Tan, P.F.; Rosenthal, D.; El-Naggar, A.; Garden, A.S.; Soulieres, D.; Trotti, A.; Avizonis, V.; Ridge, J.A.; et al. Human papillomavirus and overall survival after progression of oropharyngeal squamous cell carcinoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2014, 32, 3365–3373. [Google Scholar] [CrossRef] [PubMed]
- Chera, B.S.; Kumar, S.; Shen, C.; Amdur, R.; Dagan, R.; Green, R.; Goldman, E.; Weiss, J.; Grilley-Olson, J.; Patel, S.; et al. Plasma Circulating Tumor HPV DNA for the Surveillance of Cancer Recurrence in HPV-Associated Oropharyngeal Cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2020, 38, 1050–1058. [Google Scholar] [CrossRef] [PubMed]
| Mean ± SD | Median | Min–Max | References | |
|---|---|---|---|---|
| HPV16 | 89.9 ± 3.7 | 89.0 | 84.3–96.3 | (N = 11) 96.3% [20]; 95% [21]; 92 % [22]; 92% [23]; 90% [24]; 89% [25]; 88% [13,26,27], 86% [28]; 84.3% [29]. |
| HPV35 | 4.1 ± 1.7 | 3.5 | 2.2–6.9 | (N = 6) 6.9% [29], 5.1% [27], 3.6% [30], 3.4% [15], 3.1% [31], 2.2% [19] |
| HPV33 | 3.7 ± 2.5 | 3.1 | 1.1–7.4 | (N = 8) 7.4% [15], 7% [13], 4.4% [29], 3.7% [27], 2.5% [31], 2.2% [19], 1.1% [20], 1.1% [30]. |
| HPV18 | 3.0 ± 2.1 | 2.3 | 1.4–5.9 | (N = 4) 5.9% [30], 3.1% [31], 1.5% [20], 1.4% [27] |
| Phase | ctHPV16 in LB | Radiological Response | Molecular Response | Clinical Options | |
|---|---|---|---|---|---|
| A | Prior treatment | Primary diagnosis | - | - | - |
| B | Ongoing treatment | Prediction of good response | cRR or pRR | cMR | Consider de-escalation * |
| C | After treatment completion | Assessment of early response to treatment (12 week) | cRR | cMR | Surveillance |
| pRR | cMR | Another radiological evaluation | |||
| cRR | pMR | Another LB:
| |||
| pRR | pMR | Salvage surgery | |||
| D | Surveillance | Detection of recurrence | cRR | cMR | Surveillance |
| recurrence | cMR | Biopsy or another radiological evaluation | |||
| recurrence | Mrec | Salvage surgery | |||
| cRR | Mrec | Consider PET-CT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mazurek, A.M.; Rutkowski, T.W. Practical Application of Circulating Tumor-Related DNA of Human Papillomavirus in Liquid Biopsy to Evaluate the Molecular Response in Patients with Oropharyngeal Cancer. Cancers 2023, 15, 1047. https://doi.org/10.3390/cancers15041047
Mazurek AM, Rutkowski TW. Practical Application of Circulating Tumor-Related DNA of Human Papillomavirus in Liquid Biopsy to Evaluate the Molecular Response in Patients with Oropharyngeal Cancer. Cancers. 2023; 15(4):1047. https://doi.org/10.3390/cancers15041047
Chicago/Turabian StyleMazurek, Agnieszka M., and Tomasz W. Rutkowski. 2023. "Practical Application of Circulating Tumor-Related DNA of Human Papillomavirus in Liquid Biopsy to Evaluate the Molecular Response in Patients with Oropharyngeal Cancer" Cancers 15, no. 4: 1047. https://doi.org/10.3390/cancers15041047
APA StyleMazurek, A. M., & Rutkowski, T. W. (2023). Practical Application of Circulating Tumor-Related DNA of Human Papillomavirus in Liquid Biopsy to Evaluate the Molecular Response in Patients with Oropharyngeal Cancer. Cancers, 15(4), 1047. https://doi.org/10.3390/cancers15041047







