Drug Resistance in Medulloblastoma Is Driven by YB-1, ABCB1 and a Seven-Gene Drug Signature
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Standard Culture Conditions
2.2. Cytotoxicity Assays
2.3. Establishment of Drug-Tolerant Cell Lines
2.4. Quantitative Real-Time PCR Analysis
2.5. Western Blotting
2.6. Modified Transwell Invasion Assay
2.7. shRNA Transduction
2.8. Chromatin Immunoprecipitation
2.9. Whole Transcriptome Sequencing
2.10. 3′mRNA Sequencing
2.11. Statistical Analysis
2.12. Bioinformatic Analysis of Published Datasets
3. Results
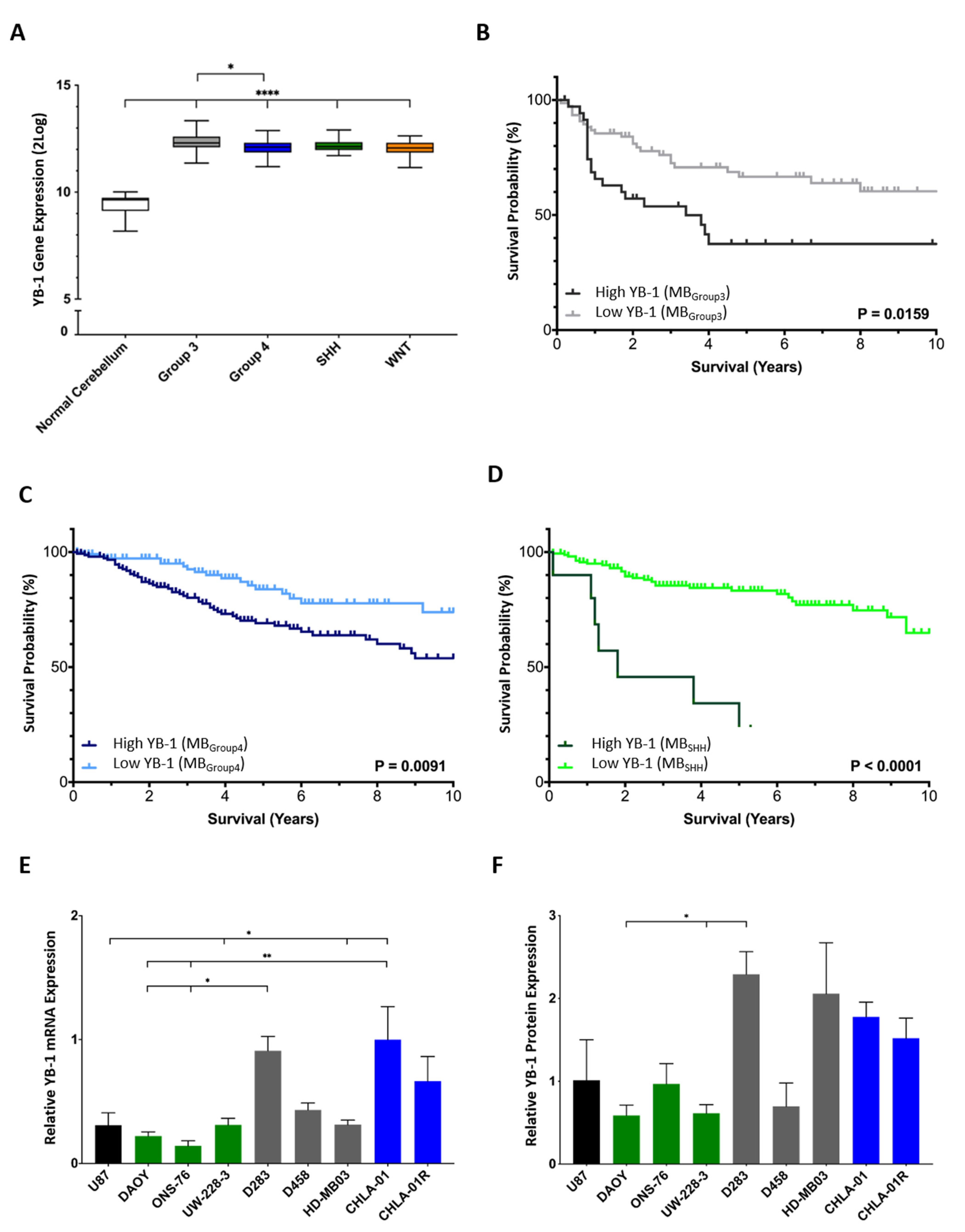
3.1. High YB-1 Expression Correlates with Poor Overall Survival in Medulloblastoma
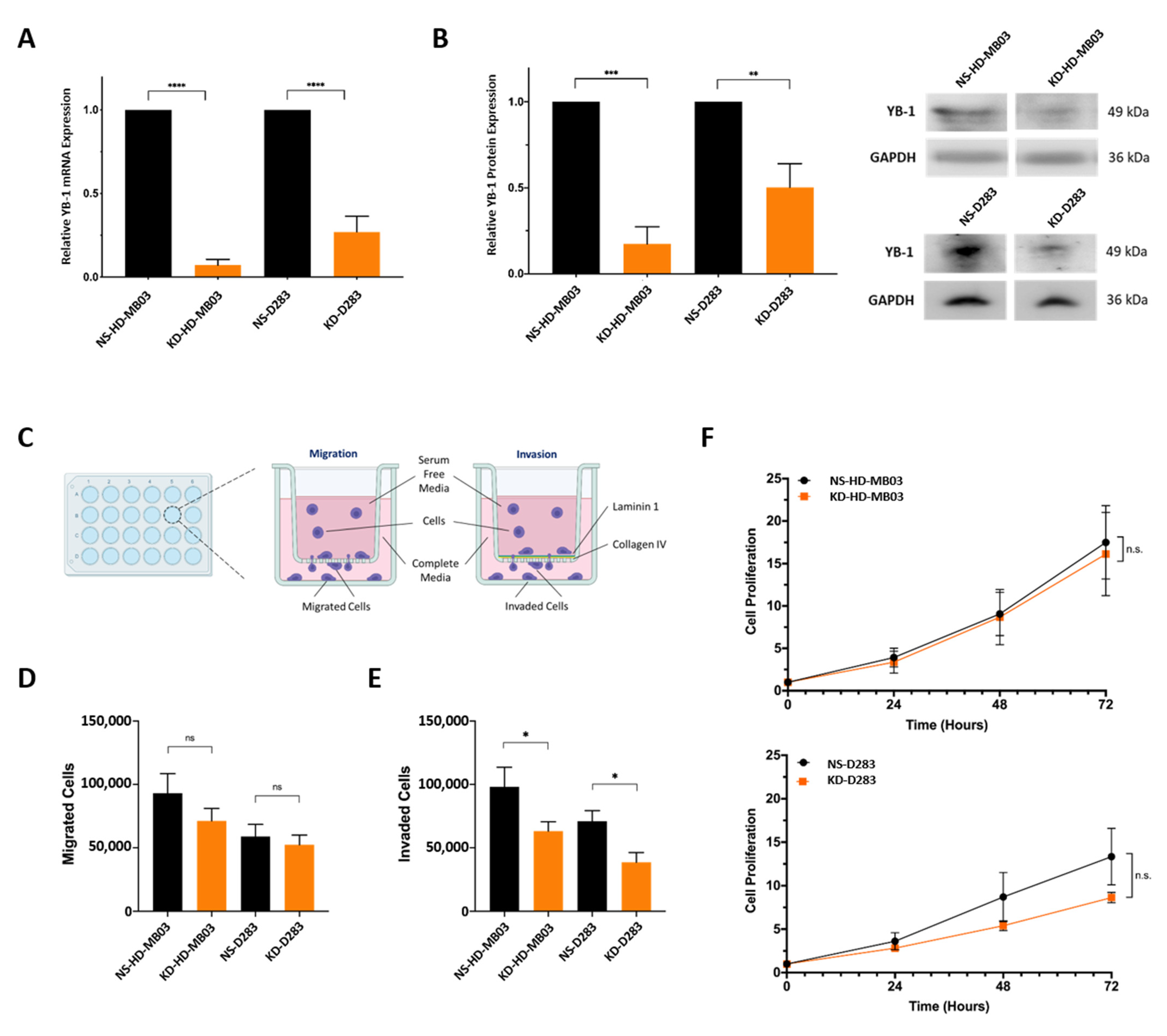
3.2. YB-1 Depletion Impedes the Invasive Capability of Medulloblastoma Cells
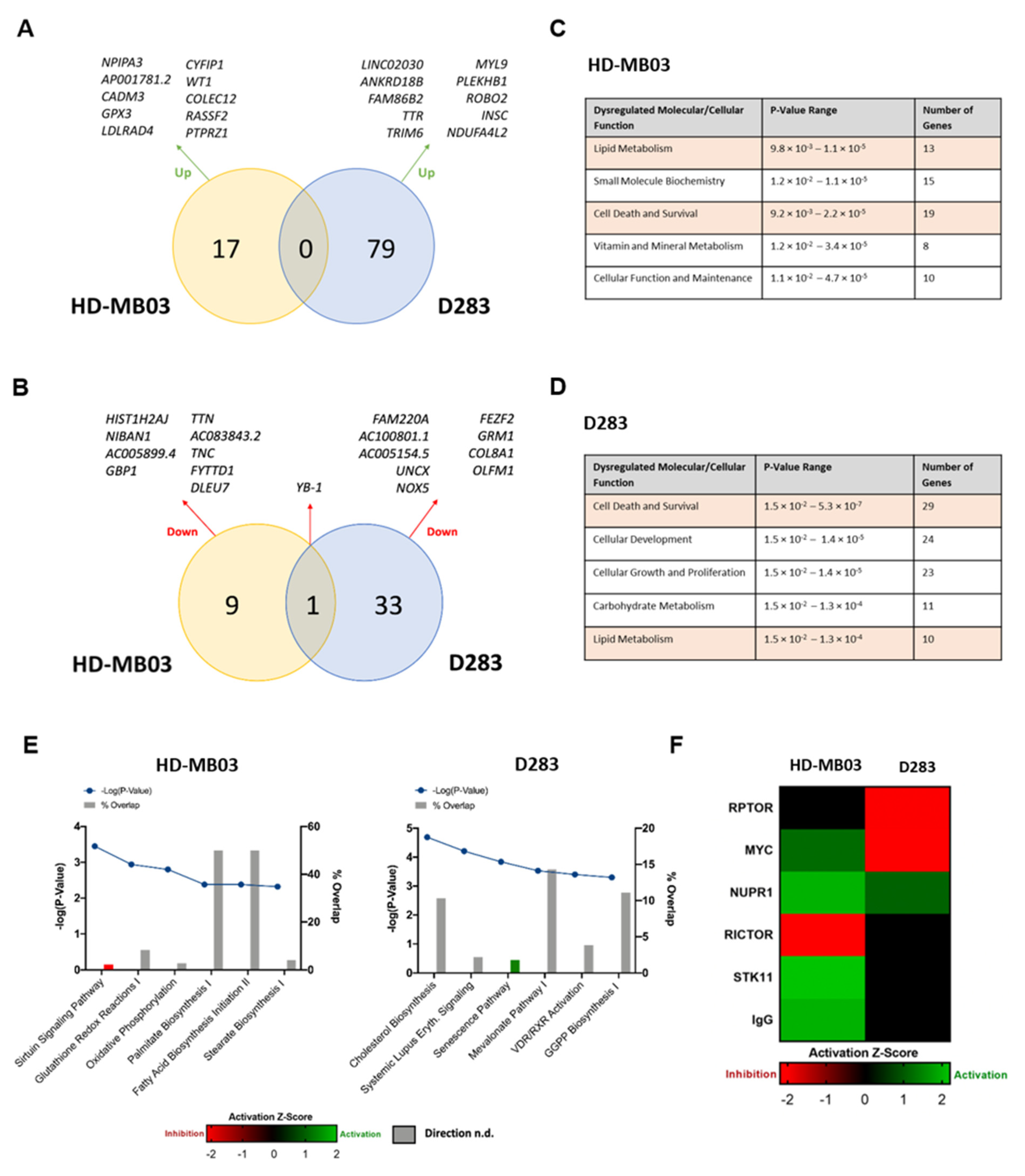
3.3. YB-1 Functions in Numerous Key Cellular Pathways in Medulloblastoma Cells
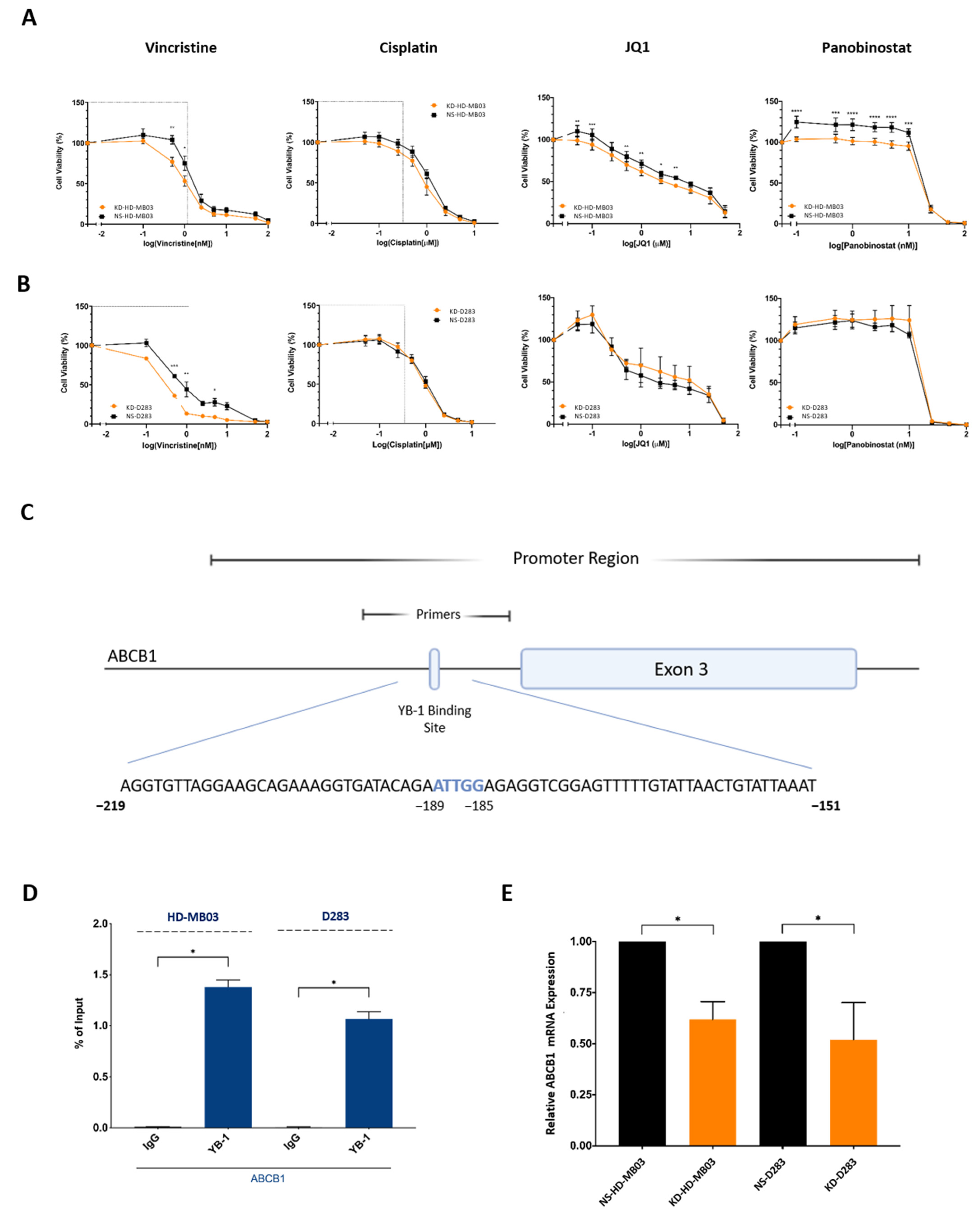
3.4. YB-1 Inhibition Is Associated with Increased Cellular Sensitivity to Vincristine, Partly through Reduced Expression of ABCB1
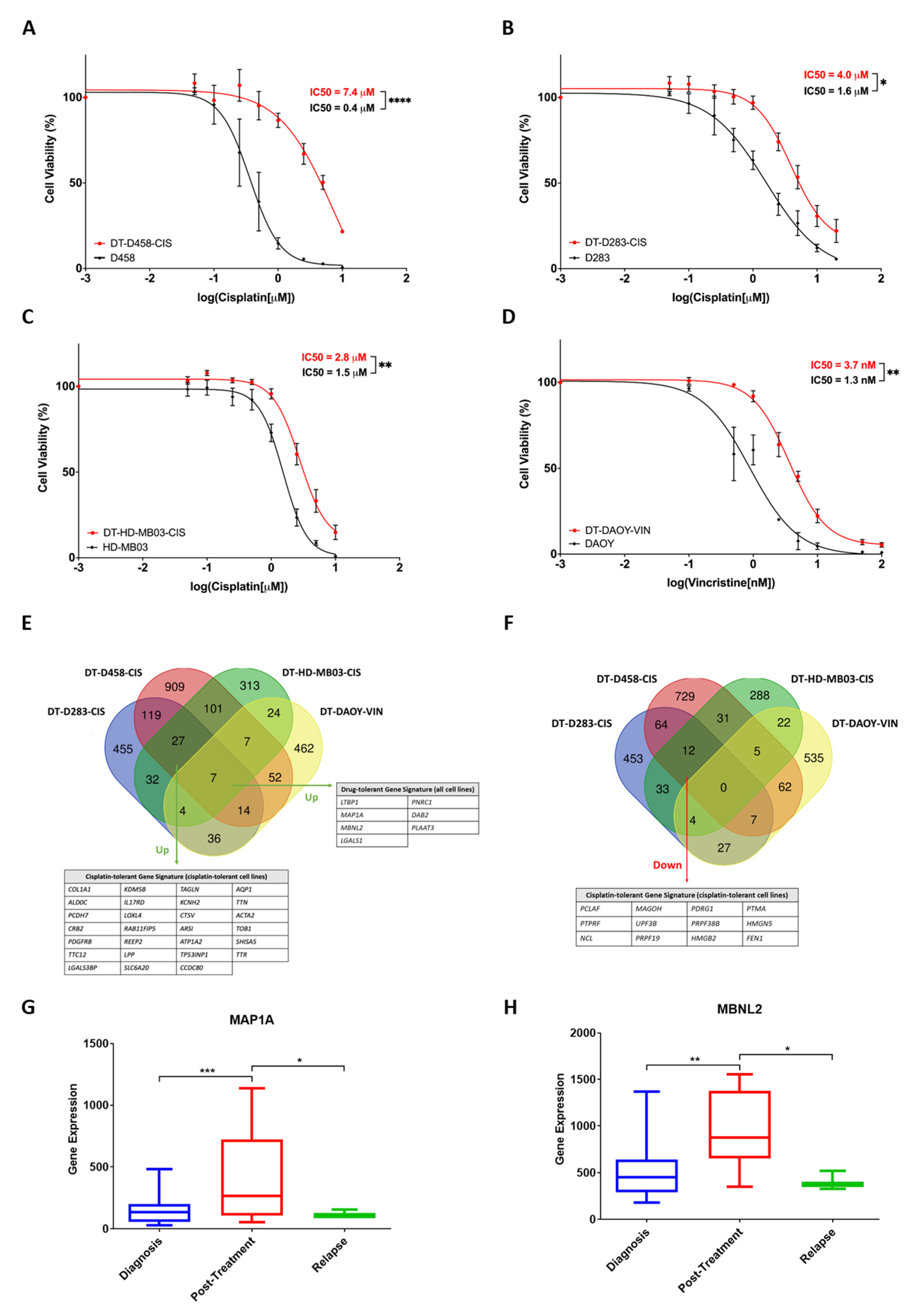
3.5. Drug-Tolerant Medulloblastoma Cell Lines Exhibit a Common Gene Signature Associated with Chemoresistance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharma, T.; Schwalbe, E.; Williamson, D.; Sill, M.; Hovestadt, V.; Mynarek, M.; Rutkowski, S.; Robinson, G.; Gajjar, A.; Cavalli, F.; et al. Second-generation molecular subgrouping of medulloblastoma: An international meta-analysis of Group 3 and Group 4 subtypes. Acta Neuropathol. 2019, 138, 309–326. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.; Northcott, P.; Korshunov, A.; Remke, M.; Cho, Y.-J.; Clifford, S.; Eberhart, C.; Parsons, W.; Rutkowski, S.; Gajjar, A.; et al. Molecular subgroups of medulloblastoma: The current consensus. Acta Neuropathol. 2012, 123, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.M.; Richardson, S.; Schwalbe, E.C.; Hicks, D.; Lindsey, J.C.; Crosier, S.; Rafiee, G.; Grabovska, Y.; Wharton, S.B.; Jacques, T.S.; et al. Time, pattern, and outcome of medulloblastoma relapse and their association with tumour biology at diagnosis and therapy: A multicentre cohort study. Lancet Child Adolesc. Health 2020, 4, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, K.C.; Faria, C.C.; Skowron, P.; Luck, A.; Garzia, L.; Wu, X.; Agnihotri, S.; Smith, C.A.; Taylor, M.D.; Mack, S.C.; et al. A functional genomics approach to identify pathways of drug resistance in medulloblastoma. Acta Neuropathol. Commun. 2018, 6, 146. [Google Scholar] [CrossRef]
- Eliseeva, I.A.; Kim, E.R.; Guryanov, S.G.; Ovchinnikov, L.P.; Lyabin, D.N. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry 2011, 76, 1402–1433. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Geng, H.; Qi, C.; Liu, Y.; Yue, D. Overexpression of YB1 and EZH2 are associated with cancer metastasis and poor prognosis in renal cell carcinomas. Tumor Biol. 2015, 36, 7159–7166. [Google Scholar] [CrossRef]
- Wang, X.; Guo, X.B.; Shen, X.C.; Zhou, H.; Wan, D.W.; Xue, X.F.; Han, Y.; Yuan, B.; Zhou, J.; Zhao, H.; et al. Prognostic role of YB-1 expression in breast cancer: A meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 1780–1791. [Google Scholar]
- Fujiwara-Okada, Y.; Matsumoto, Y.; Fukushi, J.; Setsu, N.; Matsuura, S.; Kamura, S.; Fujiwara, T.; Iida, K.; Hatano, M.; Nabeshima, A.; et al. Y-box binding protein-1 regulates cell proliferation and is associated with clinical outcomes of osteosarcoma. Br. J. Cancer 2013, 108, 836–847. [Google Scholar] [CrossRef]
- Kolk, A.; Jubitz, N.; Mengele, K.; Mantwill, K.; Bissinger, O.; Schmitt, M.; Kremer, M.; Holm, P.S. Expression of Y-box-binding protein YB-1 allows stratification into long- and short-term survivors of head and neck cancer patients. Br. J. Cancer 2011, 105, 1864–1873. [Google Scholar] [CrossRef]
- Giménez-Bonafé, P.; Fedoruk, M.N.; Whitmore, T.G.; Akbari, M.; Ralph, J.L.; Ettinger, S.; Gleave, M.E.; Nelson, C.C. YB-1 is upregulated during prostate cancer tumor progression and increases P-glycoprotein activity. Prostate 2004, 59, 337–349. [Google Scholar] [CrossRef]
- Zheng, J.; Dong, W.; Zhang, J.; Li, G.; Gong, H. YB-1, a new biomarker of glioma progression, is associated with the prognosis of glioma patients. Acta Biochim. Biophys. Sin. 2016, 48, 318–325. [Google Scholar] [CrossRef]
- Shibahara, K.; Sugio, K.; Osaki, T.; Uchiumi, T.; Maehara, Y.; Kohno, K.; Yasumoto, K.; Sugimachi, K.; Kuwano, K. Nuclear Expression of the Y-Box Binding Protein, YB-1, as a Novel Marker of Disease Progression in Non-Small Cell Lung Cancer. Clin. Cancer Res. 2001, 7, 3151–3155. [Google Scholar]
- Taylor, L.; Kerr, I.D.; Coyle, B. Y-Box Binding Protein-1: A Neglected Target in Pediatric Brain Tumors? Mol. Cancer Res. 2021, 19, 375–387. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Cavalli, F.M.G.; Remke, M.; Rampasek, L.; Peacock, J.; Shih, D.J.H.; Luu, B.; Garzia, L.; Torchia, J.; Nor, C.; Morrissy, A.S.; et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2017, 31, 737–754.e6. [Google Scholar] [CrossRef]
- Northcott, P.; Buchhalter, I.; Morrissy, A.S.; Hovestadt, V.; Weischenfeldt, J.; Ehrenberger, T.; Gröbner, S.; Segura-Wang, M.; Zichner, T.; Rudneva, V.A.; et al. The whole-genome landscape of medulloblastoma subtypes. Nature 2017, 547, 311–317. [Google Scholar] [CrossRef]
- Wang, X.; Dubuc, A.M.; Ramaswamy, V.; Mack, S.; Gendoo, D.M.; Remke, M.; Wu, X.; Garzia, L.; Luu, B.; Cavalli, F.; et al. Medulloblastoma subgroups remain stable across primary and metastatic compartments. Acta Neuropathol. 2015, 129, 449–457. [Google Scholar] [CrossRef]
- Weishaupt, H.; Johansson, P.; Sundström, A.; Lubovac-Pilav, Z.; Olsson, B.; Nelander, S.; Swartling, F.J. Batch-normalization of cerebellar and medulloblastoma gene expression datasets utilizing empirically defined negative control genes. Bioinformatics 2019, 35, 3357–3364. [Google Scholar] [CrossRef]
- Friedman, H.S.; Burger, P.C.; Bigner, S.H.; Trojanowski, J.Q.; Wikstrand, C.J.; Halperin, E.C.; Bigner, D.D. Establishment and characterization of the human medulloblastoma cell line and transplantable xenograft D283 Med. J. Neuropathol. Exp. Neurol. 1985, 44, 592–605. [Google Scholar] [CrossRef]
- Bigner, S.H.; Friedman, H.S.; Vogelstein, B.; Oakes, W.J.; Bigner, D.D. Amplification of the c-myc gene in human medulloblastoma cell lines and xenografts. Cancer Res. 1990, 50, 2347–2350. [Google Scholar]
- Milde, T.; Lodrini, M.; Savelyeva, L.; Korshunov, A.; Kool, M.; Brueckner, L.M.; Antunes, A.S.; Oehme, I.; Pekrun, A.; Pfister, S.M.; et al. HD-MB03 is a novel Group 3 medulloblastoma model demonstrating sensitivity to histone deacetylase inhibitor treatment. J. Neurooncol. 2012, 110, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Margol, A.S.; Shukla, A.; Ren, X.; Finlay, J.L.; Krieger, M.D.; Gilles, F.H.; Couch, F.J.; Aziz, M.; Fung, E.T.; et al. Disseminated Medulloblastoma in a Child with Germline BRCA2 6174delT Mutation and without Fanconi Anemia. Front. Oncol. 2015, 5, 191. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Shimizu, K.; Tamura, K.; Okamoto, Y.; Matsui, Y.; Moriuchi, S.; Park, K.; Mabuchi, E.; Yamamoto, K.; Hayakawa, T. [Establishment and biological characterization of human medulloblastoma cell lines]. No To Shinkei 1989, 41, 695–702. [Google Scholar] [PubMed]
- Jacobsen, P.F.; Jenkyn, D.J.; Papadimitriou, J.M. Establishment of a human medulloblastoma cell line and its heterotransplantation into nude mice. J. Neuropathol. Exp. Neurol. 1985, 44, 472–485. [Google Scholar] [CrossRef] [PubMed]
- Keles, G.E.; Berger, M.S.; Srinivasan, J.; Kolstoe, D.D.; Bobola, M.S.; Silber, J.R. Establishment and characterization of four human medulloblastoma-derived cell lines. Oncol. Res. 1995, 7, 493–503. [Google Scholar]
- Jackson, D.V.; Sethi, V.S.; Spurr, C.L.; McWhorter, J.M. Pharmacokinetics of vincristine in the cerebrospinal fluid of humans. Cancer Res. 1981, 41, 1466–1468. [Google Scholar]
- Balayssac, D.; Cayre, A.; Authier, N.; Bourdu, S.; Penault-Llorca, F.; Gillet, J.P.; Maublant, J.; Eschalier, A.; Coudore, F. Patterns of P-glycoprotein activity in the nervous system during vincristine-induced neuropathy in rats. J. Peripher. Nerv. Syst. 2005, 10, 301–310. [Google Scholar] [CrossRef]
- Beretta, G.L.; Benedetti, V.; Cossa, G.; Assaraf, Y.G.; Bram, E.; Gatti, L.; Corna, E.; Carenini, N.; Colangelo, D.; Howell, S.B.; et al. Increased levels and defective glycosylation of MRPs in ovarian carcinoma cells resistant to oxaliplatin. Biochem. Pharmacol. 2010, 79, 1108–1117. [Google Scholar] [CrossRef]
- Dey, A.; Robitaille, M.; Remke, M.; Maier, C.; Malhotra, A.; Gregorieff, A.; Wrana, J.L.; Taylor, M.D.; Angers, S.; Kenney, A.M. YB-1 is elevated in medulloblastoma and drives proliferation in Sonic hedgehog-dependent cerebellar granule neuron progenitor cells and medulloblastoma cells. Oncogene 2016, 35, 4256–4268. [Google Scholar] [CrossRef]
- Bosch-Presegué, L.; Vaquero, A. The dual role of sirtuins in cancer. Genes Cancer 2011, 2, 648–662. [Google Scholar] [CrossRef]
- Sinha, S.; Sharma, S.; Vora, J.; Shrivastava, N. Emerging role of sirtuins in breast cancer metastasis and multidrug resistance: Implication for novel therapeutic strategies targeting sirtuins. Pharmacol. Res. 2020, 158, 104880. [Google Scholar] [CrossRef]
- Tiberi, L.; Bonnefont, J.; van den Ameele, J.; Le Bon, S.D.; Herpoel, A.; Bilheu, A.; Baron, B.W.; Vanderhaeghen, P. A BCL6/BCOR/SIRT1 complex triggers neurogenesis and suppresses medulloblastoma by repressing Sonic Hedgehog signaling. Cancer Cell 2014, 26, 797–812. [Google Scholar] [CrossRef]
- Ma, J.X.; Li, H.; Chen, X.M.; Yang, X.H.; Wang, Q.; Wu, M.L.; Kong, Q.Y.; Li, Z.X.; Liu, J. Expression patterns and potential roles of SIRT1 in human medulloblastoma cells in vivo and in vitro. Neuropathology 2013, 33, 7–16. [Google Scholar] [CrossRef]
- Hill, R.M.; Kuijper, S.; Lindsey, J.C.; Petrie, K.; Schwalbe, E.C.; Barker, K.; Boult, J.K.; Williamson, D.; Ahmad, Z.; Hallsworth, A.; et al. Combined MYC and P53 defects emerge at medulloblastoma relapse and define rapidly progressive, therapeutically targetable disease. Cancer Cell 2015, 27, 72–84. [Google Scholar] [CrossRef]
- Tao, R.; Murad, N.; Xu, Z.; Zhang, P.; Okonechnikov, K.; Kool, M.; Rivero-Hinojosa, S.; Lazarski, C.; Zheng, P.; Liu, Y.; et al. Drives Group 3 Medulloblastoma through Transformation of Sox2. Cancer Res. 2019, 79, 1967–1980. [Google Scholar] [CrossRef]
- Fujii, T.; Seki, N.; Namoto-Matsubayashi, R.; Takahashi, H.; Inoue, Y.; Toh, U.; Kage, M.; Shirouzu, K. YB-1 prevents apoptosis via the mTOR/STAT3 pathway in HER-2-overexpressing breast cancer cells. Future Oncol. 2009, 5, 153–156. [Google Scholar] [CrossRef]
- Gong, H.; Gao, S.; Yu, C.; Li, M.; Liu, P.; Zhang, G.; Song, J.; Zheng, J. Effect and mechanism of YB-1 knockdown on glioma cell growth, migration, and apoptosis. Acta Biochim. Biophys. Sin. 2020, 52, 168–179. [Google Scholar] [CrossRef]
- Lee, C.; Dhillon, J.; Wang, M.Y.; Gao, Y.; Hu, K.; Park, E.; Astanehe, A.; Hung, M.C.; Eirew, P.; Eaves, C.J.; et al. Targeting YB-1 in HER-2 overexpressing breast cancer cells induces apoptosis via the mTOR/STAT3 pathway and suppresses tumor growth in mice. Cancer Res. 2008, 68, 8661–8666. [Google Scholar] [CrossRef]
- Kloetgen, A.; Duggimpudi, S.; Schuschel, K.; Hezaveh, K.; Picard, D.; Schaal, H.; Remke, M.; Klusmann, J.H.; Borkhardt, A.; McHardy, A.C.; et al. YBX1 Indirectly Targets Heterochromatin-Repressed Inflammatory Response-Related Apoptosis Genes through Regulating CBX5 mRNA. Int. J. Mol. Sci. 2020, 21, 4453. [Google Scholar] [CrossRef]
- Gao, Y.; Fotovati, A.; Lee, C.; Wang, M.; Cote, G.; Guns, E.; Toyota, B.; Faury, D.; Jabado, N.; Dunn, S.E. Inhibition of Y-box binding protein-1 slows the growth of glioblastoma multiforme and sensitizes to temozolomide independent O6-methylguanine-DNA methyltransferase. Mol. Cancer Ther. 2009, 8, 3276–3284. [Google Scholar] [CrossRef]
- Lim, J.P.; Shyamasundar, S.; Gunaratne, J.; Scully, O.J.; Matsumoto, K.; Bay, B.H. YBX1 gene silencing inhibits migratory and invasive potential via CORO1C in breast cancer in vitro. BMC Cancer 2017, 17, 201. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Li, X.; Wang, F.; Guo, Y.; Huang, Y.; Zhu, H.; Wang, Y.; Lu, Y.; Wang, Z. YB-1 expression promotes pancreatic cancer metastasis that is inhibited by microRNA-216a. Exp. Cell Res. 2017, 359, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Lasham, A.; Samuel, W.; Cao, H.; Patel, R.; Mehta, R.; Stern, J.L.; Reid, G.; Woolley, A.G.; Miller, L.D.; Black, M.A.; et al. YB-1, the E2F pathway, and regulation of tumor cell growth. J. Natl. Cancer Inst. 2012, 104, 133–146. [Google Scholar] [CrossRef]
- Bommert, K.S.; Effenberger, M.; Leich, E.; Küspert, M.; Murphy, D.; Langer, C.; Moll, R.; Janz, S.; Mottok, A.; Weissbach, S.; et al. The feed-forward loop between YB-1 and MYC is essential for multiple myeloma cell survival. Leukemia 2013, 27, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Marneth, A.E.; Alexe, G.; Walker, S.R.; Gandler, H.I.; Ye, D.Q.; Labella, K.; Mathur, R.; Toniolo, P.A.; Tillgren, M.; et al. The kinases IKBKE and TBK1 regulate MYC-dependent survival pathways through YB-1 in AML and are targets for therapy. Blood Adv. 2018, 2, 3428–3442. [Google Scholar] [CrossRef] [PubMed]
- Hann, S.R. MYC cofactors: Molecular switches controlling diverse biological outcomes. Cold Spring Harb. Perspect. Med. 2014, 4, a014399. [Google Scholar] [CrossRef]
- Agrawal, P.; Yu, K.; Salomon, A.R.; Sedivy, J.M. Proteomic profiling of Myc-associated proteins. Cell Cycle 2010, 9, 4908–4921. [Google Scholar] [CrossRef]
- Othman, R.T.; Kimishi, I.; Bradshaw, T.D.; Storer, L.C.D.; Korshunov, A.; Pfister, S.M.; Grundy, R.G.; Kerr, I.D.; Coyle, B. Overcoming multiple drug resistance mechanisms in medulloblastoma. Acta Neuropathol. Commun. 2014, 2, 57. [Google Scholar] [CrossRef]
- Lasham, A.; Mehta, S.Y.; Fitzgerald, S.J.; Woolley, A.G.; Hearn, J.I.; Hurley, D.G.; Ruza, I.; Algie, M.; Shelling, A.N.; Braithwaite, A.W.; et al. A novel EGR-1 dependent mechanism for YB-1 modulation of paclitaxel response in a triple negative breast cancer cell line. Int. J. Cancer 2016, 139, 1157–1170. [Google Scholar] [CrossRef]
- Dolfini, D.; Mantovani, R. Targeting the Y/CCAAT box in cancer: YB-1 (YBX1) or NF-Y? Cell Death Differ. 2013, 20, 676–685. [Google Scholar] [CrossRef]
- Ghatak, S.; Hascall, V.C.; Markwald, R.R.; Misra, S. FOLFOX Therapy Induces Feedback Upregulation of CD44v6 through YB-1 to Maintain Stemness in Colon Initiating Cells. Int. J. Mol. Sci. 2021, 22, 753. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Xu, W.; Luo, W.; Zhou, L.; Yong, W.; Chen, F.; Wu, C.; Chen, Q.; Han, X. Upregulation of mdr1 gene is related to activation of the MAPK/ERK signal transduction pathway and YB-1 nuclear translocation in B-cell lymphoma. Exp. Hematol. 2011, 39, 558–569. [Google Scholar] [CrossRef] [PubMed]
- van Helvoort, A.; Smith, A.J.; Sprong, H.; Fritzsche, I.; Schinkel, A.H.; Borst, P.; van Meer, G. MDR1 P-glycoprotein is a lipid translocase of broad specificity, while MDR3 P-glycoprotein specifically translocates phosphatidylcholine. Cell 1996, 87, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Morita, S.Y.; Matsuo, M.; Ueda, K. Mechanism of multidrug recognition by MDR1/ABCB1. Cancer Sci. 2007, 98, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Garrigues, A.; Escargueil, A.E.; Orlowski, S. The multidrug transporter, P-glycoprotein, actively mediates cholesterol redistribution in the cell membrane. Proc. Natl. Acad. Sci. USA 2002, 99, 10347–10352. [Google Scholar] [CrossRef]
- Barreto-Ojeda, E.; Corradi, V.; Gu, R.X.; Tieleman, D.P. Coarse-grained molecular dynamics simulations reveal lipid access pathways in P-glycoprotein. J. Gen. Physiol. 2018, 150, 417–429. [Google Scholar] [CrossRef]
- Nasir, A.; Cardall, A.; Othman, R.T.; Nicolaou, N.; Lourdusamy, A.; Linke, F.; Onion, D.; Ryzhova, M.; Cameron, H.; Valente, C.; et al. ABCB1 inhibition provides a novel therapeutic target to block TWIST1-induced migration in medulloblastoma. Neurooncol. Adv. 2021, 3, vdab030. [Google Scholar] [CrossRef]
- Cheng, C.; Geng, F.; Cheng, X.; Guo, D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun. 2018, 38, 27. [Google Scholar] [CrossRef]
- Jeffords, E.; Freeman, S.; Cole, B.; Root, K.; Chekouo, T.; Melvin, R.G.; Bemis, L.; Simmons, G.E. Y-box binding protein 1 acts as a negative regulator of stearoyl CoA desaturase 1 in clear cell renal cell carcinoma. Oncol. Lett. 2020, 20, 165. [Google Scholar] [CrossRef]
- Bhatia, B.; Hsieh, M.; Kenney, A.M.; Nahlé, Z. Mitogenic Sonic hedgehog signaling drives E2F1-dependent lipogenesis in progenitor cells and medulloblastoma. Oncogene 2011, 30, 410–422. [Google Scholar] [CrossRef]
- Huang, D.; Liu, J.; Eldridge, R.C.; Gaul, D.A.; Paine, M.R.L.; Uppal, K.; MacDonald, T.J.; Fernández, F.M. Lipidome signatures of metastasis in a transgenic mouse model of sonic hedgehog medulloblastoma. Anal. Bioanal. Chem. 2020, 412, 7017–7027. [Google Scholar] [CrossRef]
- Susanto, E.; Marin Navarro, A.; Zhou, L.; Sundström, A.; van Bree, N.; Stantic, M.; Moslem, M.; Tailor, J.; Rietdijk, J.; Zubillaga, V.; et al. Modeling SHH-driven medulloblastoma with patient iPS cell-derived neural stem cells. Proc. Natl. Acad. Sci. USA 2020, 117, 20127–20138. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taylor, L.; Wade, P.K.; Johnson, J.E.C.; Aldighieri, M.; Morlando, S.; Di Leva, G.; Kerr, I.D.; Coyle, B. Drug Resistance in Medulloblastoma Is Driven by YB-1, ABCB1 and a Seven-Gene Drug Signature. Cancers 2023, 15, 1086. https://doi.org/10.3390/cancers15041086
Taylor L, Wade PK, Johnson JEC, Aldighieri M, Morlando S, Di Leva G, Kerr ID, Coyle B. Drug Resistance in Medulloblastoma Is Driven by YB-1, ABCB1 and a Seven-Gene Drug Signature. Cancers. 2023; 15(4):1086. https://doi.org/10.3390/cancers15041086
Chicago/Turabian StyleTaylor, Louisa, Philippa K. Wade, James E. C. Johnson, Macha Aldighieri, Sonia Morlando, Gianpiero Di Leva, Ian D. Kerr, and Beth Coyle. 2023. "Drug Resistance in Medulloblastoma Is Driven by YB-1, ABCB1 and a Seven-Gene Drug Signature" Cancers 15, no. 4: 1086. https://doi.org/10.3390/cancers15041086
APA StyleTaylor, L., Wade, P. K., Johnson, J. E. C., Aldighieri, M., Morlando, S., Di Leva, G., Kerr, I. D., & Coyle, B. (2023). Drug Resistance in Medulloblastoma Is Driven by YB-1, ABCB1 and a Seven-Gene Drug Signature. Cancers, 15(4), 1086. https://doi.org/10.3390/cancers15041086






