RNA-Binding Proteins in Bladder Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. RNA Binding Motif 3 (RBM3)
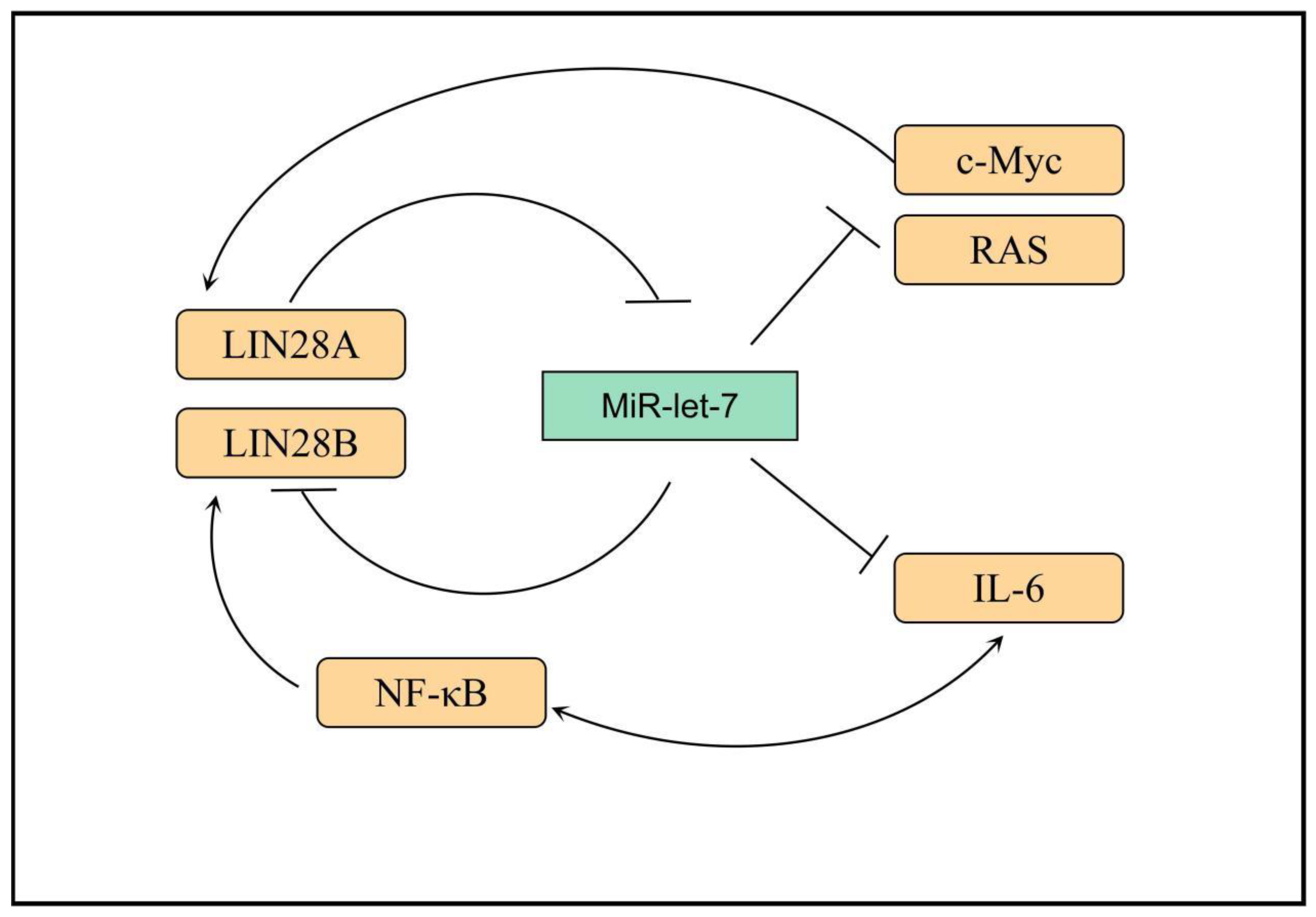
3. LIN28
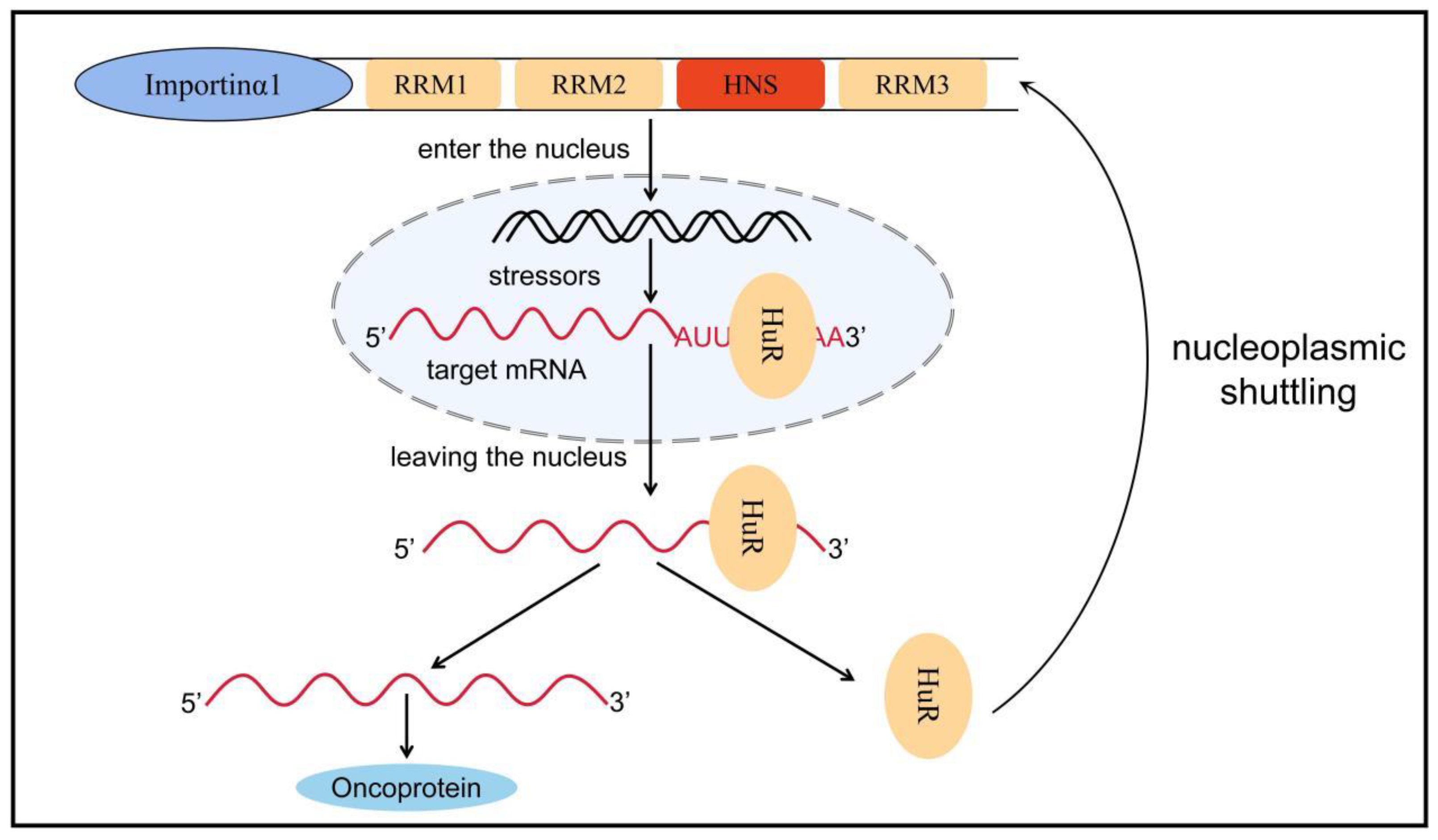
4. HuR
5. Heterogeneous Nuclear RNPs (hnRNPs)
| hnRNP- | Molecular Weight (kDa) | RBD | Binding Sequence | Functions | References |
|---|---|---|---|---|---|
| A1 | 34 | 2 of RRM, RGG box, Gly-enrich domain | UAGGGA/U | Splicing Translational regulation Influence of mRNA stability | [111,116,118] |
| A2/B1 | 36/38 | 2 of RRM, RGG box, Gly-enrich domain | UUAGGG | Splicing Influence of mRNA stability | [124,125,129] |
| C1/C2 | 41/43 | RRM | Poly U | Splicing Translational regulation Transcriptional regulation | [111,113] |
| D | 44~48 | 2 of RRM | AU-rich domain | Translational regulation Telomere maintenance | [113] |
| E | 39 | 3 of KH | Poly C | Splicing Translational regulation Transcriptional regulation Influence of mRNA stability | [119] |
| F | 53 | 3 of quasi -RRM 2 of Gly-enrich domain | UUAGG | Splicing Telomere maintenance | [131,132] |
| K | 62 | 3 of KH | Poly C | Splicing Translational regulation Transcriptional regulation Influence of mRNA stability | [117,133,134,135] |
| L | 68 | 4 of RRM, Gly-enrich domain | CA-repeat sequence | Splicing Influence of mRNA stability | [127,136] |
| M | 77 | 3 of RRM | Poly G/C | Splicing | [121] |
| R | 31 | 3 of RRM, RGG box | UCUAUC | Translational regulation Transcriptional regulation | [113] |
| U | 120 | RGG, Gly-enrich domain | GGACUGCR-RUCGC | Splicing Transcriptional regulation | [115] |
6. Others
6.1. IGF2BP3
6.2. NCL
6.3. QKI
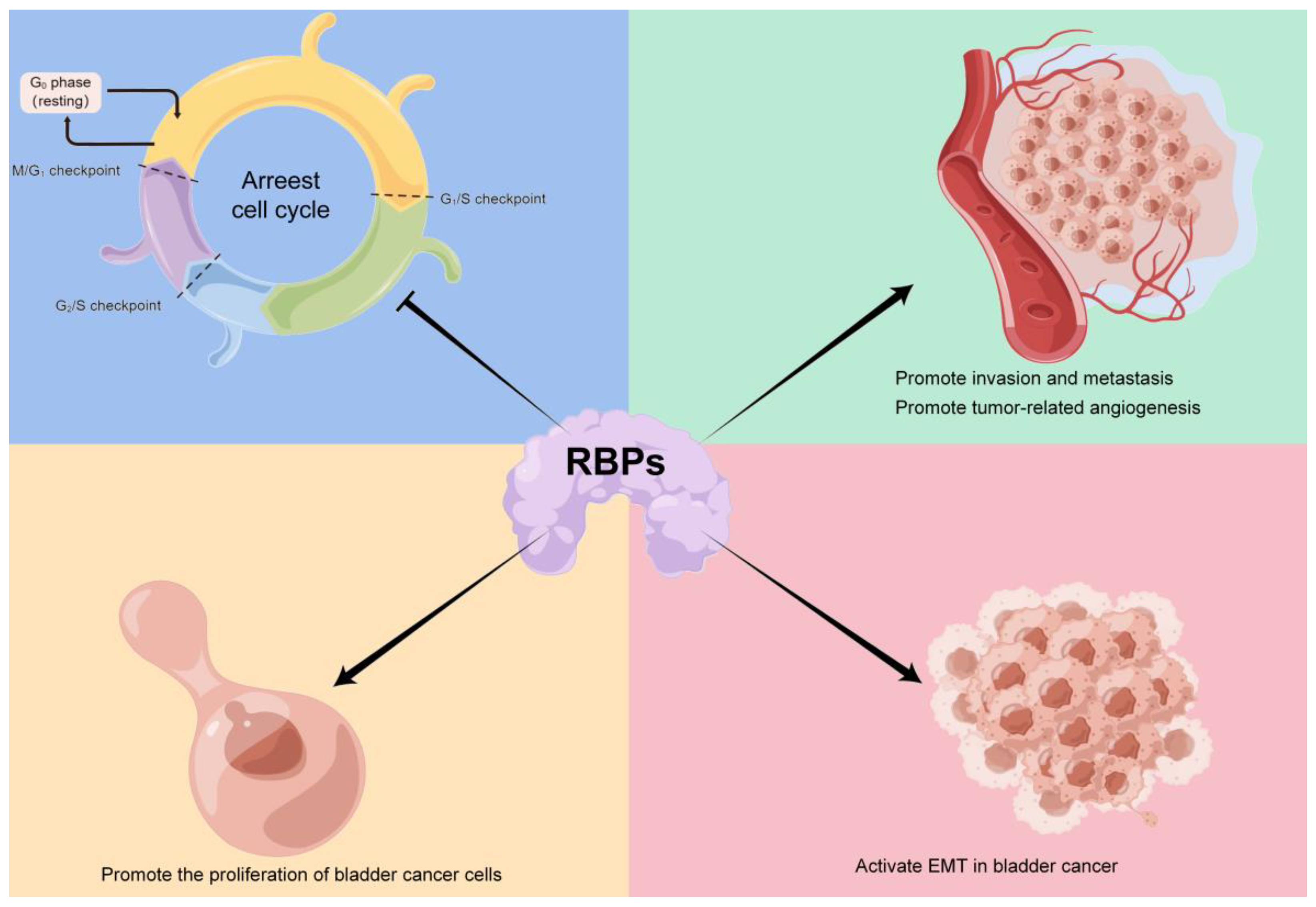
7. Discussion
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.; Miller, K.; Wagle, N.; Jemal, A. Cancer statistics, 2023. CA A Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.; Miller, K.; Fuchs, H.; Jemal, A. Cancer statistics, 2022. CA A Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Xia, C.; Dong, X.; Li, H.; Cao, M.; Sun, D.; He, S.; Yang, F.; Yan, X.; Zhang, S.; Li, N.; et al. Cancer statistics in China and United States, 2022: Profiles, trends, and determinants. Chin. Med. J. 2022, 135, 584–590. [Google Scholar] [CrossRef]
- Ritch, C.; Velasquez, M.; Kwon, D.; Becerra, M.; Soodana-Prakash, N.; Atluri, V.; Almengo, K.; Alameddine, M.; Kineish, O.; Kava, B.; et al. Use and Validation of the AUA/SUO Risk Grouping for Nonmuscle Invasive Bladder Cancer in a Contemporary Cohort. J. Urol. 2020, 203, 505–511. [Google Scholar] [CrossRef]
- Alfred Witjes, J.; Lebret, T.; Compérat, E.; Cowan, N.; De Santis, M.; Bruins, H.; Hernández, V.; Espinós, E.; Dunn, J.; Rouanne, M.; et al. Updated 2016 EAU Guidelines on Muscle-invasive and Metastatic Bladder Cancer. Eur. Urol. 2017, 71, 462–475. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, B.; Luo, Y.; Lin, Q.; Liu, S.; Zhang, X.; Zhou, H.; Yang, J.; Qu, L. Comprehensive Genomic Characterization of RNA-Binding Proteins across Human Cancers. Cell Rep. 2018, 22, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.; Valcárcel, J. Alternative pre-mRNA splicing: The logic of combinatorial control. Trends Biochem. Sci. 2000, 25, 381–388. [Google Scholar] [CrossRef]
- Michlewski, G.; Sanford, J.; Cáceres, J. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol. Cell 2008, 30, 179–189. [Google Scholar] [CrossRef]
- Fabian, M.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef]
- Kim, H.; Kuwano, Y.; Srikantan, S.; Lee, E.; Martindale, J.; Gorospe, M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009, 23, 1743–1748. [Google Scholar] [CrossRef] [Green Version]
- Sureban, S.; Murmu, N.; Rodriguez, P.; May, R.; Maheshwari, R.; Dieckgraefe, B.; Houchen, C.; Anant, S. Functional antagonism between RNA binding proteins HuR and CUGBP2 determines the fate of COX-2 mRNA translation. Gastroenterology 2007, 132, 1055–1065. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, K.; Noubissi, F.; Katti, P.; Hahn, C.; Davey, S.; Lundsmith, E.; Klein-Szanto, A.; Rhim, A.; Spiegelman, V.; Rustgi, A. IMP1 promotes tumor growth, dissemination and a tumor-initiating cell phenotype in colorectal cancer cell xenografts. Carcinogenesis 2013, 34, 2647–2654. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Hämmerle, M.; Pazaitis, N.; Bley, N.; Fiskin, E.; Uckelmann, H.; Heim, A.; Groβ, M.; Hofmann, N.; Geffers, R.; et al. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) is an important protumorigenic factor in hepatocellular carcinoma. Hepatology 2014, 59, 1900–1911. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Huang, Z.; Liu, X.; Huang, W.; Chen, S.; Zhou, Y.; Li, D.; Singer, R.; Gu, W. IMP1 suppresses breast tumor growth and metastasis through the regulation of its target mRNAs. Oncotarget 2016, 7, 15690–15702. [Google Scholar] [CrossRef]
- Hamilton, K.; Chatterji, P.; Lundsmith, E.; Andres, S.; Giroux, V.; Hicks, P.; Noubissi, F.; Spiegelman, V.; Rustgi, A. Loss of Stromal IMP1 Promotes a Tumorigenic Microenvironment in the Colon. Mol. Cancer Res. MCR 2015, 13, 1478–1486. [Google Scholar] [CrossRef]
- Zhou, R.; Lu, X.; Zhang, C.; Yin, D. RNA binding motif protein 3: A potential biomarker in cancer and therapeutic target in neuroprotection. Oncotarget 2017, 8, 22235–22250. [Google Scholar] [CrossRef]
- Gonatopoulos-Pournatzis, T.; Cowling, V. Cap-binding complex (CBC). Biochem. J. 2014, 457, 231–242. [Google Scholar] [CrossRef]
- Chappell, S.; Mauro, V. The internal ribosome entry site (IRES) contained within the RNA-binding motif protein 3 (Rbm3) mRNA is composed of functionally distinct elements. J. Biol. Chem. 2003, 278, 33793–33800. [Google Scholar] [CrossRef]
- Zhan, X.; Qian, B.; Cao, F.; Wu, W.; Yang, L.; Guan, Q.; Gu, X.; Wang, P.; Okusolubo, T.; Dunn, S.; et al. An Arabidopsis PWI and RRM motif-containing protein is critical for pre-mRNA splicing and ABA responses. Nat. Commun. 2015, 6, 8139. [Google Scholar] [CrossRef]
- Gu, L.; Chen, Y.; Li, X.; Mei, Y.; Zhou, J.; Ma, J.; Zhang, M.; Hou, T.; He, D.; Zeng, J. Integrated Analysis and Identification of Critical RNA-Binding Proteins in Bladder Cancer. Cancers 2022, 14, 3739. [Google Scholar] [CrossRef]
- Guo, C.; Shao, T.; Jiang, X.; Wei, D.; Wang, Z.; Li, M.; Bao, G. Comprehensive analysis of the functions and prognostic significance of RNA-binding proteins in bladder urothelial carcinoma. Am. J. Transl. Res. 2020, 12, 7160–7173. [Google Scholar] [PubMed]
- Kishore, S.; Luber, S.; Zavolan, M. Deciphering the role of RNA-binding proteins in the post-transcriptional control of gene expression. Brief. Funct. Genomics 2010, 9, 391–404. [Google Scholar] [CrossRef] [PubMed]
- Phadtare, S.; Alsina, J.; Inouye, M. Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 1999, 2, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Bührer, C.; Wellmann, S. Cold-inducible proteins CIRP and RBM3, a unique couple with activities far beyond the cold. Cell. Mol. Life Sci. CMLS 2016, 73, 3839–3859. [Google Scholar] [CrossRef] [PubMed]
- Godin, K.; Varani, G. How arginine-rich domains coordinate mRNA maturation events. RNA Biol. 2007, 4, 69–75. [Google Scholar] [CrossRef]
- Smart, F.; Aschrafi, A.; Atkins, A.; Owens, G.; Pilotte, J.; Cunningham, B.; Vanderklish, P. Two isoforms of the cold-inducible mRNA-binding protein RBM3 localize to dendrites and promote translation. J. Neurochem. 2007, 101, 1367–1379. [Google Scholar] [CrossRef]
- Sureban, S.; Ramalingam, S.; Natarajan, G.; May, R.; Subramaniam, D.; Bishnupuri, K.; Morrison, A.; Dieckgraefe, B.; Brackett, D.; Postier, R.; et al. Translation regulatory factor RBM3 is a proto-oncogene that prevents mitotic catastrophe. Oncogene 2008, 27, 4544–4556. [Google Scholar] [CrossRef]
- Dresios, J.; Aschrafi, A.; Owens, G.; Vanderklish, P.; Edelman, G.; Mauro, V. Cold stress-induced protein Rbm3 binds 60S ribosomal subunits, alters microRNA levels, and enhances global protein synthesis. Proc. Natl. Acad. Sci. USA 2005, 102, 1865–1870. [Google Scholar] [CrossRef]
- Pilotte, J.; Dupont-Versteegden, E.; Vanderklish, P. Widespread regulation of miRNA biogenesis at the Dicer step by the cold-inducible RNA-binding protein, RBM3. PLoS ONE 2011, 6, e28446. [Google Scholar] [CrossRef]
- Wong, J.; Au, A.; Gao, D.; Pinello, N.; Kwok, C.; Thoeng, A.; Lau, K.; Gordon, J.; Schmitz, U.; Feng, Y.; et al. RBM3 regulates temperature sensitive miR-142-5p and miR-143 (thermomiRs), which target immune genes and control fever. Nucleic Acids Res. 2016, 44, 2888–2897. [Google Scholar] [CrossRef] [Green Version]
- Venugopal, A.; Subramaniam, D.; Balmaceda, J.; Roy, B.; Dixon, D.; Umar, S.; Weir, S.; Anant, S. RNA binding protein RBM3 increases β-catenin signaling to increase stem cell characteristics in colorectal cancer cells. Mol. Carcinog. 2016, 55, 1503–1516. [Google Scholar] [CrossRef]
- Matsuda, A.; Ogawa, M.; Yanai, H.; Naka, D.; Goto, A.; Ao, T.; Tanno, Y.; Takeda, K.; Watanabe, Y.; Honda, K.; et al. Generation of mice deficient in RNA-binding motif protein 3 (RBM3) and characterization of its role in innate immune responses and cell growth. Biochem. Biophys. Res. Commun. 2011, 411, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Ehlén, Å.; Nodin, B.; Rexhepaj, E.; Brändstedt, J.; Uhlén, M.; Alvarado-Kristensson, M.; Pontén, F.; Brennan, D.; Jirström, K. RBM3-regulated genes promote DNA integrity and affect clinical outcome in epithelial ovarian cancer. Transl. Oncol. 2011, 4, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.; Chen, M.; Wen, X.; Gao, Y.; Huang, D.; Cao, H.; Peng, Y.; Guo, N.; Ni, J.; Zhang, S. Endoplasmic Reticulum Stress and Tumor Microenvironment in Bladder Cancer: The Missing Link. Front. Cell Dev. Biol. 2021, 9, 683940. [Google Scholar] [CrossRef]
- Lin, J.; Li, H.; Yasumura, D.; Cohen, H.; Zhang, C.; Panning, B.; Shokat, K.; Lavail, M.; Walter, P. IRE1 signaling affects cell fate during the unfolded protein response. Science 2007, 318, 944–949. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zelmer, A.; Kapfhammer, J.; Wellmann, S. Cold-inducible RBM3 inhibits PERK phosphorylation through cooperation with NF90 to protect cells from endoplasmic reticulum stress. FASEB J. 2016, 30, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Rzechorzek, N.; Connick, P.; Patani, R.; Selvaraj, B.; Chandran, S. Hypothermic Preconditioning of Human Cortical Neurons Requires Proteostatic Priming. EBioMedicine 2015, 2, 528–535. [Google Scholar] [CrossRef]
- Poone, G.; Hasseldam, H.; Munkholm, N.; Rasmussen, R.; Grønberg, N.; Johansen, F. The Hypothermic Influence on CHOP and Ero1-α in an Endoplasmic Reticulum Stress Model of Cerebral Ischemia. Brain Sci. 2015, 5, 178–187. [Google Scholar] [CrossRef]
- Boman, K.; Andersson, G.; Wennersten, C.; Nodin, B.; Ahlgren, G.; Jirström, K. Podocalyxin-like and RNA-binding motif protein 3 are prognostic biomarkers in urothelial bladder cancer: A validatory study. Biomark. Res. 2017, 5, 10. [Google Scholar] [CrossRef]
- Boman, K.; Segersten, U.; Ahlgren, G.; Eberhard, J.; Uhlén, M.; Jirström, K.; Malmström, P. Decreased expression of RNA-binding motif protein 3 correlates with tumour progression and poor prognosis in urothelial bladder cancer. BMC Urol. 2013, 13, 17. [Google Scholar] [CrossRef]
- Florianova, L.; Xu, B.; Traboulsi, S.; Elmansi, H.; Tanguay, S.; Aprikian, A.; Kassouf, W.; Brimo, F. Evaluation of RNA-binding motif protein 3 expression in urothelial carcinoma of the bladder: An immunohistochemical study. World J. Surg. Oncol. 2015, 13, 317. [Google Scholar] [CrossRef] [PubMed]
- Bartkova, J.; Rajpert-De Meyts, E.; Skakkebaek, N.; Lukas, J.; Bartek, J. DNA damage response in human testes and testicular germ cell tumours: Biology and implications for therapy. Int. J. Androl. 2007, 30, 282–291, discussion 291. [Google Scholar] [CrossRef] [PubMed]
- Karnevi, E.; Dror, L.; Mardinoglu, A.; Elebro, J.; Heby, M.; Olofsson, S.; Nodin, B.; Eberhard, J.; Gallagher, W.; Uhlén, M.; et al. Translational study reveals a two-faced role of RBM3 in pancreatic cancer and suggests its potential value as a biomarker for improved patient stratification. Oncotarget 2018, 9, 6188–6200. [Google Scholar] [CrossRef] [PubMed]
- Moss, E.; Lee, R.; Ambros, V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 1997, 88, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Ng, S.; Chng, W. LIN28/LIN28B: An emerging oncogenic driver in cancer stem cells. Int. J. Biochem. Cell Biol. 2013, 45, 973–978. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y. A mirror of two faces: Lin28 as a master regulator of both miRNA and mRNA. Wiley Interdiscip. Rev. RNA 2012, 3, 483–494. [Google Scholar] [CrossRef]
- Heo, I.; Joo, C.; Cho, J.; Ha, M.; Han, J.; Kim, V. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell 2008, 32, 276–284. [Google Scholar] [CrossRef]
- Lu, L.; Katsaros, D.; Shaverdashvili, K.; Qian, B.; Wu, Y.; de la Longrais, I.; Preti, M.; Menato, G.; Yu, H. Pluripotent factor lin-28 and its homologue lin-28b in epithelial ovarian cancer and their associations with disease outcomes and expression of let-7a and IGF-II. Eur. J. Cancer 2009, 45, 2212–2218. [Google Scholar] [CrossRef]
- Lei, X.; Xu, J.; Ma, W.; Qiao, C.; Newman, M.; Hammond, S.; Huang, Y. Determinants of mRNA recognition and translation regulation by Lin28. Nucleic Acids Res. 2012, 40, 3574–3584. [Google Scholar] [CrossRef]
- Balzer, E.; Moss, E. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 2007, 4, 16–25. [Google Scholar] [CrossRef] [Green Version]
- Piskounova, E.; Polytarchou, C.; Thornton, J.; LaPierre, R.; Pothoulakis, C.; Hagan, J.; Iliopoulos, D.; Gregory, R. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 2011, 147, 1066–1079. [Google Scholar] [CrossRef]
- Calin, G.; Sevignani, C.; Dumitru, C.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef] [PubMed]
- Chirshev, E.; Oberg, K.; Ioffe, Y.; Unternaehrer, J. Let-7 as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin. Transl. Med. 2019, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xie, C.; Zheng, X.; Nie, X.; Wang, Z.; Liu, H.; Zhao, Y. let-7LIN28//PD-L1 Pathway as a Target for Cancer Immunotherapy. Cancer Immunol. Res. 2019, 7, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V.; Horvitz, H. Heterochronic mutants of the nematode Caenorhabditis elegans. Science 1984, 226, 409–416. [Google Scholar] [CrossRef]
- Balzeau, J.; Menezes, M.; Cao, S.; Hagan, J. The LIN28/let-7 Pathway in Cancer. Front. Genet. 2017, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Lee, K.; Lee, H.; Jeong, C.; Ku, J.; Kim, H.; Kwak, C. Concurrent treatment with simvastatin and NF-κB inhibitor in human castration-resistant prostate cancer cells exerts synergistic anti-cancer effects via control of the NF-κB/LIN28/let-7 miRNA signaling pathway. PLoS ONE 2017, 12, e0184644. [Google Scholar] [CrossRef]
- Büssing, I.; Slack, F.; Grosshans, H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008, 14, 400–409. [Google Scholar] [CrossRef]
- Viswanathan, S.; Powers, J.; Einhorn, W.; Hoshida, Y.; Ng, T.; Toffanin, S.; O’Sullivan, M.; Lu, J.; Phillips, L.; Lockhart, V.; et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 2009, 41, 843–848. [Google Scholar] [CrossRef]
- Rybak, A.; Fuchs, H.; Smirnova, L.; Brandt, C.; Pohl, E.; Nitsch, R.; Wulczyn, F. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008, 10, 987–993. [Google Scholar] [CrossRef]
- Dangi-Garimella, S.; Yun, J.; Eves, E.; Newman, M.; Erkeland, S.; Hammond, S.; Minn, A.; Rosner, M. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009, 28, 347–358. [Google Scholar] [CrossRef]
- Chang, T.; Zeitels, L.; Hwang, H.; Chivukula, R.; Wentzel, E.; Dews, M.; Jung, J.; Gao, P.; Dang, C.; Beer, M.; et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc. Natl. Acad. Sci. USA 2009, 106, 3384–3389. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Hirsch, H.; Struhl, K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 2009, 139, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Shee, K.; Seigne, J.; Karagas, M.; Marsit, C.; Hinds, J.; Schned, A.; Pettus, J.; Armstrong, D.; Miller, T.; Andrew, A. Identification of Let-7f-5p as a novel biomarker of recurrence in non-muscle invasive bladder cancer. Cancer Biomark. Sect. A Dis. Mrk. 2020, 29, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, H.; Lai, C.; Du, X.; Su, Z.; Gao, S. The Lin28/let-7a/c-Myc pathway plays a role in non-muscle invasive bladder cancer. Cell Tissue Res. 2013, 354, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, C.; Ma, Q.; Zhuang, C.; Ye, J.; Zhang, F.; Gui, Y. LncRNA GClnc1 promotes proliferation and invasion of bladder cancer through activation of MYC. FASEB J. 2019, 33, 11045–11059. [Google Scholar] [CrossRef]
- Pan, P.; Chen, T.; Zhang, Y.; Qi, Z.; Qin, J.; Cui, G.; Guo, X. LIN28A inhibits lysosome-associated membrane glycoprotein 1 protein expression in embryonic stem and bladder cancer cells. Mol. Med. Rep. 2018, 18, 399–406. [Google Scholar] [CrossRef]
- Shi, H.; Xie, J.; Wang, K.; Li, W.; Yin, L.; Wang, G.; Wu, Z.; Ni, J.; Mao, W.; Guo, C.; et al. LINC01451 drives epithelial-mesenchymal transition and progression in bladder cancer cells via LIN28/TGF-β/Smad pathway. Cell. Signal. 2021, 81, 109932. [Google Scholar] [CrossRef]
- Park, S.; Shim, J.; Park, H.; Eum, D.; Park, M.; Mi Yi, J.; Choi, S.; Kim, S.; Son, T.; Lu, W.; et al. MacroH2A1 downregulation enhances the stem-like properties of bladder cancer cells by transactivation of Lin28B. Oncogene 2016, 35, 1292–1301. [Google Scholar] [CrossRef]
- Lv, K.; Liu, L.; Wang, L.; Yu, J.; Liu, X.; Cheng, Y.; Dong, M.; Teng, R.; Wu, L.; Fu, P.; et al. Lin28 mediates paclitaxel resistance by modulating p21, Rb and Let-7a miRNA in breast cancer cells. PLoS ONE 2012, 7, e40008. [Google Scholar] [CrossRef]
- Liao, W.; Wang, W.; Chang, W.; Tseng, J. The RNA-binding protein HuR stabilizes cytosolic phospholipase A2α mRNA under interleukin-1β treatment in non-small cell lung cancer A549 Cells. J. Biol. Chem. 2011, 286, 35499–35508. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Cheng, S.; Campbell, C.; Wright, A.; Furneaux, H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 1996, 271, 8144–8151. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, K.; Gorospe, M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip. Rev. RNA 2010, 1, 214–229. [Google Scholar] [CrossRef]
- Zarei, M.; Lal, S.; Parker, S.; Nevler, A.; Vaziri-Gohar, A.; Dukleska, K.; Mambelli-Lisboa, N.; Moffat, C.; Blanco, F.; Chand, S.; et al. Posttranscriptional Upregulation of IDH1 by HuR Establishes a Powerful Survival Phenotype in Pancreatic Cancer Cells. Cancer Res. 2017, 77, 4460–4471. [Google Scholar] [CrossRef] [PubMed]
- von Roretz, C.; Di Marco, S.; Mazroui, R.; Gallouzi, I. Turnover of AU-rich-containing mRNAs during stress: A matter of survival. Wiley Interdiscip. Rev. RNA 2011, 2, 336–347. [Google Scholar] [CrossRef]
- Scheiba, R.; de Opakua, A.; Díaz-Quintana, A.; Cruz-Gallardo, I.; Martínez-Cruz, L.; Martínez-Chantar, M.; Blanco, F.; Díaz-Moreno, I. The C-terminal RNA binding motif of HuR is a multi-functional domain leading to HuR oligomerization and binding to U-rich RNA targets. RNA Biol. 2014, 11, 1250–1261. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, J. HuR stabilizes TFAM mRNA in an ATM/p38-dependent manner in ionizing irradiated cancer cells. Cancer Sci. 2018, 109, 2446–2457. [Google Scholar] [CrossRef]
- Reglero, C.; Lafarga, V.; Rivas, V.; Albitre, Á.; Ramos, P.; Berciano, S.; Tapia, O.; Martínez-Chantar, M.; Mayor, F.; Penela, P. GRK2-Dependent HuR Phosphorylation Regulates HIF1α Activation under Hypoxia or Adrenergic Stress. Cancers 2020, 12, 1216. [Google Scholar] [CrossRef]
- Zarei, M.; Lal, S.; Vaziri-Gohar, A.; O’Hayer, K.; Gunda, V.; Singh, P.; Brody, J.; Winter, J. RNA-Binding Protein HuR Regulates Both Mutant and Wild-Type IDH1 in IDH1-Mutated Cancer. Mol. Cancer Res. MCR 2019, 17, 508–520. [Google Scholar] [CrossRef]
- Deka, K.; Saha, S. Heat stress induced arginylation of HuR promotes alternative polyadenylation of Hsp70.3 by regulating HuR stability and RNA binding. Cell Death Differ. 2021, 28, 730–747. [Google Scholar] [CrossRef]
- Jehung, J.; Kitamura, T.; Yanagawa-Matsuda, A.; Kuroshima, T.; Towfik, A.; Yasuda, M.; Sano, H.; Kitagawa, Y.; Minowa, K.; Shindoh, M.; et al. Adenovirus infection induces HuR relocalization to facilitate virus replication. Biochem. Biophys. Res. Commun. 2018, 495, 1795–1800. [Google Scholar] [CrossRef]
- Cai, J.; Wang, H.; Jiao, X.; Huang, R.; Qin, Q.; Zhang, J.; Chen, H.; Feng, D.; Tian, X.; Wang, H. The RNA-Binding Protein HuR Confers Oxaliplatin Resistance of Colorectal Cancer by Upregulating CDC6. Mol. Cancer Ther. 2019, 18, 1243–1254. [Google Scholar] [CrossRef]
- Liu, Y.; Wei, W.; Wang, Y.; Wan, C.; Bai, Y.; Sun, X.; Ma, J.; Zheng, F. TNF-α/calreticulin dual signaling induced NLRP3 inflammasome activation associated with HuR nucleocytoplasmic shuttling in rheumatoid arthritis. Inflamm. Res. 2019, 68, 597–611. [Google Scholar] [CrossRef]
- Doller, A.; Schlepckow, K.; Schwalbe, H.; Pfeilschifter, J.; Eberhardt, W. Tandem phosphorylation of serines 221 and 318 by protein kinase Cdelta coordinates mRNA binding and nucleocytoplasmic shuttling of HuR. Mol. Cell. Biol. 2010, 30, 1397–1410. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Park, S.; Kilburn, B.; Jelinek, M.; Henschen-Edman, A.; Aswad, D.; Stallcup, M.; Laird-Offringa, I. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase. J. Biol. Chem. 2002, 277, 44623–44630. [Google Scholar] [CrossRef] [PubMed]
- Vancura, A.; Nagar, S.; Kaur, P.; Bu, P.; Bhagwat, M.; Vancurova, I. Reciprocal Regulation of AMPK/SNF1 and Protein Acetylation. Int. J. Mol. Sci. 2018, 19, 3314. [Google Scholar] [CrossRef]
- Kulawik, A.; Engesser, R.; Ehlting, C.; Raue, A.; Albrecht, U.; Hahn, B.; Lehmann, W.; Gaestel, M.; Klingmüller, U.; Häussinger, D.; et al. IL-1β-induced and p38-dependent activation of the mitogen-activated protein kinase-activated protein kinase 2 (MK2) in hepatocytes: Signal transduction with robust and concentration-independent signal amplification. J. Biol. Chem. 2017, 292, 6291–6302. [Google Scholar] [CrossRef]
- García-Mauriño, S.; Rivero-Rodríguez, F.; Velázquez-Cruz, A.; Hernández-Vellisca, M.; Díaz-Quintana, A.; De la Rosa, M.; Díaz-Moreno, I. RNA Binding Protein Regulation and Cross-Talk in the Control of AU-rich mRNA Fate. Front. Mol. Biosci. 2017, 4, 71. [Google Scholar] [CrossRef] [PubMed]
- Donahue, J.; Chang, E.; Xiao, L.; Wang, P.; Rao, J.; Turner, D.; Wang, J.; Battafarano, R. The RNA-binding protein HuR stabilizes survivin mRNA in human oesophageal epithelial cells. Biochem. J. 2011, 437, 89–96. [Google Scholar] [CrossRef]
- Subramaniam, D.; Ramalingam, S.; May, R.; Dieckgraefe, B.; Berg, D.; Pothoulakis, C.; Houchen, C.; Wang, T.; Anant, S. Gastrin-mediated interleukin-8 and cyclooxygenase-2 gene expression: Differential transcriptional and posttranscriptional mechanisms. Gastroenterology 2008, 134, 1070–1082. [Google Scholar] [CrossRef]
- Xie, M.; Yu, T.; Jing, X.; Ma, L.; Fan, Y.; Yang, F.; Ma, P.; Jiang, H.; Wu, X.; Shu, Y.; et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol. Cancer 2020, 19, 112. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Christodoulou-Vafeiadou, E.; Rao, J.; Zou, T.; Xiao, L.; Chung, H.; Yang, H.; Gorospe, M.; Kontoyiannis, D.; Wang, J. RNA-binding protein HuR promotes growth of small intestinal mucosa by activating the Wnt signaling pathway. Mol. Biol. Cell 2014, 25, 3308–3318. [Google Scholar] [CrossRef] [PubMed]
- Miyata, Y.; Watanabe, S.; Sagara, Y.; Mitsunari, K.; Matsuo, T.; Ohba, K.; Sakai, H. High expression of HuR in cytoplasm, but not nuclei, is associated with malignant aggressiveness and prognosis in bladder cancer. PLoS ONE 2013, 8, e59095. [Google Scholar] [CrossRef] [PubMed]
- Fus, Ł.; Pihowicz, P.; Koperski, Ł.; Marczewska, J.; Górnicka, B. High cytoplasmic HuR expression is associated with advanced pT stage, high grade and increased microvessel density in urothelial bladder carcinoma. Ann. Diagn. Pathol. 2018, 33, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, T.; Miyata, Y.; Asai, A.; Sagara, Y.; Furusato, B.; Fukuoka, J.; Sakai, H. Green Tea Polyphenol Induces Changes in Cancer-Related Factors in an Animal Model of Bladder Cancer. PLoS ONE 2017, 12, e0171091. [Google Scholar] [CrossRef]
- Guo, J.; Lv, J.; Chang, S.; Chen, Z.; Lu, W.; Xu, C.; Liu, M.; Pang, X. Inhibiting cytoplasmic accumulation of HuR synergizes genotoxic agents in urothelial carcinoma of the bladder. Oncotarget 2016, 7, 45249–45262. [Google Scholar] [CrossRef]
- Xiao, M.; Liu, J.; Xiang, L.; Zhao, K.; He, D.; Zeng, Q.; Zhang, Q.; Xie, D.; Deng, M.; Zhu, Y.; et al. MAFG-AS1 promotes tumor progression via regulation of the HuR/PTBP1 axis in bladder urothelial carcinoma. Clin. Transl. Med. 2020, 10, e241. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Zhang, Y.; Yang, L.; Lu, W.; Mai, L.; Guo, H.; Liu, X. Long noncoding RNA HCG22 suppresses proliferation and metastasis of bladder cancer cells by regulation of PTBP1. J. Cell. Physiol. 2020, 235, 1711–1722. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Zhang, C.; Gui, J. RNA-binding protein HuR promotes bladder cancer progression by competitively binding to the long noncoding HOTAIR with miR-1. OncoTargets Ther. 2017, 10, 2609–2619. [Google Scholar] [CrossRef]
- Hua, X.; Huang, M.; Deng, X.; Xu, J.; Luo, Y.; Xie, Q.; Xu, J.; Tian, Z.; Li, J.; Zhu, J.; et al. The inhibitory effect of compound ChlA-F on human bladder cancer cell invasion can be attributed to its blockage of SOX2 protein. Cell Death Differ. 2020, 27, 632–645. [Google Scholar] [CrossRef]
- Tian, Z.; Luo, Y.; Zhu, J.; Hua, X.; Xu, J.; Huang, C.; Jin, H.; Huang, H.; Huang, C. Transcriptionally elevation of miR-494 by new ChlA-F compound via a HuR/JunB axis inhibits human bladder cancer cell invasion. Biochim. Et Biophys. Acta. Gene Regul. Mech. 2019, 1862, 822–833. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Peng, W.; Furuuchi, N.; Gerhart, J.; Rhodes, K.; Mukherjee, N.; Jimbo, M.; Gonye, G.; Brody, J.; Getts, R.; et al. Delivery of Therapeutics Targeting the mRNA-Binding Protein HuR Using 3DNA Nanocarriers Suppresses Ovarian Tumor Growth. Cancer Res. 2016, 76, 1549–1559. [Google Scholar] [CrossRef] [PubMed]
- Muralidharan, R.; Babu, A.; Amreddy, N.; Basalingappa, K.; Mehta, M.; Chen, A.; Zhao, Y.; Kompella, U.; Munshi, A.; Ramesh, R. Folate receptor-targeted nanoparticle delivery of HuR-RNAi suppresses lung cancer cell proliferation and migration. J. Nanobiotechnol. 2016, 14, 47. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hjelmeland, A.; Nabors, L.; King, P. Anti-cancer effects of the HuR inhibitor, MS-444, in malignant glioma cells. Cancer Biol. Ther. 2019, 20, 979–988. [Google Scholar] [CrossRef]
- Geuens, T.; Bouhy, D.; Timmerman, V. The hnRNP family: Insights into their role in health and disease. Hum. Genet. 2016, 135, 851–867. [Google Scholar] [CrossRef]
- Han, S.; Tang, Y.; Smith, R. Functional diversity of the hnRNPs: Past, present and perspectives. Biochem. J. 2010, 430, 379–392. [Google Scholar] [CrossRef]
- Piñol-Roma, S.; Choi, Y.; Matunis, M.; Dreyfuss, G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 1988, 2, 215–227. [Google Scholar] [CrossRef]
- Pawson, T. Protein modules and signalling networks. Nature 1995, 373, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Kiledjian, M.; Dreyfuss, G. Primary structure and binding activity of the hnRNP U protein: Binding RNA through RGG box. EMBO J. 1992, 11, 2655–2664. [Google Scholar] [CrossRef]
- Cartegni, L.; Maconi, M.; Morandi, E.; Cobianchi, F.; Riva, S.; Biamonti, G. hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J. Mol. Biol. 1996, 259, 337–348. [Google Scholar] [CrossRef]
- Chen, Y.; Zeng, Y.; Xiao, Z.; Chen, S.; Li, Y.; Zou, J.; Zeng, X. Role of heterogeneous nuclear ribonucleoprotein K in tumor development. J. Cell. Biochem. 2019, 120, 14296–14305. [Google Scholar] [CrossRef]
- Beusch, I.; Barraud, P.; Moursy, A.; Cléry, A.; Allain, F. Tandem hnRNP A1 RNA recognition motifs act in concert to repress the splicing of survival motor neuron exon 7. eLife 2017, 6, e25736. [Google Scholar] [CrossRef]
- Jensen, K.; Dredge, B.; Stefani, G.; Zhong, R.; Buckanovich, R.; Okano, H.; Yang, Y.; Darnell, R. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 2000, 25, 359–371. [Google Scholar] [CrossRef]
- Piñol-Roma, S.; Dreyfuss, G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature 1992, 355, 730–732. [Google Scholar] [CrossRef]
- Rebane, A.; Aab, A.; Steitz, J. Transportins 1 and 2 are redundant nuclear import factors for hnRNP A1 and HuR. RNA 2004, 10, 590–599. [Google Scholar] [CrossRef] [PubMed]
- Allemand, E.; Guil, S.; Myers, M.; Moscat, J.; Cáceres, J.; Krainer, A. Regulation of heterogenous nuclear ribonucleoprotein A1 transport by phosphorylation in cells stressed by osmotic shock. Proc. Natl. Acad. Sci. USA 2005, 102, 3605–3610. [Google Scholar] [CrossRef] [PubMed]
- Amano, N.; Matsumoto, K.; Shimizu, Y.; Nakamura, M.; Tsumura, H.; Ishii, D.; Sato, Y.; Iwamura, M. High HNRNPA3 expression is associated with lymph node metastasis and poor prognosis in patients treated with radical cystectomy. Urol. Oncol. 2021, 39, 196.e1–196.e7. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Su, J.; Lin, J.; Chen, C.; Hwang, W.; Huang, H.; Wu, Y. An investigation into the cytotoxic effects of 13-acetoxysarcocrassolide from the soft coral Sarcophyton crassocaule on bladder cancer cells. Mar. Drugs 2011, 9, 2622–2642. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, C.; Zhang, X.; Yang, T.; Zuo, S.; Fu, C.; Zhang, Y.; Yang, C.; Chen, L. Discovery of bladder cancer biomarkers in paired pre- and postoperative urine samples. Transl. Androl. Urol. 2021, 10, 3402–3414. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Hao, L.; Han, X.; Wu, Z.; Pang, K.; Dong, Y.; Qin, J.; Wang, G.; Zhang, X.; Xia, T.; et al. Targeting HNRNPU to overcome cisplatin resistance in bladder cancer. Mol. Cancer 2022, 21, 37. [Google Scholar] [CrossRef]
- Micalizzi, D.; Farabaugh, S.; Ford, H. Epithelial-mesenchymal transition in cancer: Parallels between normal development and tumor progression. J. Mammary Gland Biol. Neoplasia 2010, 15, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.; Wu, H.; Xing, R.; Shu, F.; Lei, B.; Lei, C.; Zhou, X.; Wan, B.; Yang, Y.; Zhong, L.; et al. HnRNP-L mediates bladder cancer progression by inhibiting apoptotic signaling and enhancing MAPK signaling pathways. Oncotarget 2017, 8, 13586–13599. [Google Scholar] [CrossRef]
- Li, F.; Zhao, H.; Su, M.; Xie, W.; Fang, Y.; Du, Y.; Yu, Z.; Hou, L.; Tan, W. HnRNP-F regulates EMT in bladder cancer by mediating the stabilization of Snail1 mRNA by binding to its 3’ UTR. EBioMedicine 2019, 45, 208–219. [Google Scholar] [CrossRef]
- Chen, C.; Luo, Y.; He, W.; Zhao, Y.; Kong, Y.; Liu, H.; Zhong, G.; Li, Y.; Li, J.; Huang, J.; et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J. Clin. Investig. 2020, 130, 404–421. [Google Scholar] [CrossRef]
- Zeng, J.; Xu, H.; Huang, C.; Sun, Y.; Xiao, H.; Yu, G.; Zhou, H.; Zhang, Y.; Yao, W.; Xiao, W.; et al. CD46 splice variant enhances translation of specific mRNAs linked to an aggressive tumor cell phenotype in bladder cancer. Mol. Ther. Nucleic Acids 2021, 24, 140–153. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Su, M.; Zhao, H.; Xie, W.; Cao, S.; Xu, Y.; Chen, W.; Wang, L.; Hou, L.; Tan, W. HnRNP-F promotes cell proliferation by regulating TPX2 in bladder cancer. Am. J. Transl. Res. 2019, 11, 7035–7048. [Google Scholar] [PubMed]
- Li, F.; Xie, W.; Fang, Y.; Xie, K.; Liu, W.; Hou, L.; Tan, W. HnRNP-F promotes the proliferation of bladder cancer cells mediated by PI3K/AKT/FOXO1. J. Cancer 2021, 12, 281–291. [Google Scholar] [CrossRef]
- Chen, X.; Gu, P.; Xie, R.; Han, J.; Liu, H.; Wang, B.; Xie, W.; Xie, W.; Zhong, G.; Chen, C.; et al. Heterogeneous nuclear ribonucleoprotein K is associated with poor prognosis and regulates proliferation and apoptosis in bladder cancer. J. Cell. Mol. Med. 2017, 21, 1266–1279. [Google Scholar] [CrossRef]
- Aboushousha, T.; Hammam, O.; Helal, N.; El Dahshan, S. Impact of Cyclin D1 and Heterogeneous Nuclear Ribonucleoprotein-K (HnRNP-K) on Urinary Bladder Carcinogenesis. Asian Pac. J. Cancer Prev. APJCP 2018, 19, 513–519. [Google Scholar] [CrossRef]
- Zhao, F.; Qin, S.; Huang, L.; Ding, L.; Shi, X.; Zhong, G. Long noncoding RNA AK023096 interacts with hnRNP-K and contributes to the maintenance of self-renewal in bladder cancer stem-like cells. Exp. Cell Res. 2021, 409, 112909. [Google Scholar] [CrossRef]
- Li, J.; Chen, Y.; Tiwari, M.; Bansal, V.; Sen, G. Regulation of integrin and extracellular matrix genes by HNRNPL is necessary for epidermal renewal. PLoS Biol. 2021, 19, e3001378. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Wang, N.; Li, Z.; Chen, R.; Duan, J.; Peng, Y.; Wu, Z.; Zhang, Z.; Jiang, L.; Zheng, X.; et al. FXR1 can bind with the CFIm25/CFIm68 complex and promote the progression of urothelial carcinoma of the bladder by stabilizing TRAF1 mRNA. Cell Death Dis. 2022, 13, 170. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Mi, Q.; Yan, S.; Xie, Y.; Cui, Z.; Zhang, S.; Wang, Y.; Gao, H.; Wang, Y.; Li, J.; et al. Characterization of circSCL38A1 as a novel oncogene in bladder cancer via targeting ILF3/TGF-β2 signaling axis. Cell Death Dis. 2023, 14, 59. [Google Scholar] [CrossRef]
- Mancarella, C.; Scotlandi, K. IGF2BP3 From Physiology to Cancer: Novel Discoveries, Unsolved Issues, and Future Perspectives. Front. Cell Dev. Biol. 2019, 7, 363. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, H.; Guo, X.; Zhu, Z.; Cai, H.; Kong, X. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in cancer. J. Hematol. Oncol. 2018, 11, 88. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.; Mesquita, A.; Liu, C.; Yuan, C.; et al. Publisher Correction: Recognition of RNA N-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2020, 22, 1288. [Google Scholar] [CrossRef]
- Schneider, T.; Hung, L.; Aziz, M.; Wilmen, A.; Thaum, S.; Wagner, J.; Janowski, R.; Müller, S.; Schreiner, S.; Friedhoff, P.; et al. Combinatorial recognition of clustered RNA elements by the multidomain RNA-binding protein IMP3. Nat. Commun. 2019, 10, 2266. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, X.; Liu, M.; Zeng, Z.; Lin, C.; Xu, W.; Ma, W.; Wang, J.; Xiang, Q.; Johnston, R.; et al. The Oncogenic Functions of Insulin-like Growth Factor 2 mRNA-Binding Protein 3 in Human Carcinomas. Curr. Pharm. Des. 2020, 26, 3939–3954. [Google Scholar] [CrossRef]
- Lee, D.; Xylinas, E.; Rieken, M.; Khani, F.; Klatte, T.; Wood, C.; Karam, J.; Weizer, A.; Raman, J.; Remzi, M.; et al. Insulin-like growth factor messenger RNA-binding protein 3 expression helps prognostication in patients with upper tract urothelial carcinoma. Eur. Urol. 2014, 66, 379–385. [Google Scholar] [CrossRef]
- Huang, W.; Li, Y.; Zhang, C.; Zha, H.; Zhou, X.; Fu, B.; Guo, J.; Wang, G. IGF2BP3 facilitates cell proliferation and tumorigenesis via modulation of JAK/STAT signalling pathway in human bladder cancer. J. Cell. Mol. Med. 2020, 24, 13949–13960. [Google Scholar] [CrossRef]
- Jia, W.; Yao, Z.; Zhao, J.; Guan, Q.; Gao, L. New perspectives of physiological and pathological functions of nucleolin (NCL). Life Sci. 2017, 186, 1–10. [Google Scholar] [CrossRef]
- Berger, C.; Gaume, X.; Bouvet, P. The roles of nucleolin subcellular localization in cancer. Biochimie 2015, 113, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, M.; McBride, O.; Fleming, P.; Pollard, H.; Burns, A. Genomic organization and chromosomal localization of the human nucleolin gene. J. Biol. Chem. 1990, 265, 14922–14931. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhao, G.; Zhang, S.; Nigim, F.; Zhou, G.; Yu, Z.; Song, Y.; Chen, Y.; Li, Y. AS1411-Induced Growth Inhibition of Glioma Cells by Up-Regulation of p53 and Down-Regulation of Bcl-2 and Akt1 via Nucleolin. PLoS ONE 2016, 11, e0167094. [Google Scholar] [CrossRef] [PubMed]
- Ginisty, H.; Sicard, H.; Roger, B.; Bouvet, P. Structure and functions of nucleolin. J. Cell Sci. 1999, 112, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Xu, X. Roles of nucleolin. Focus on cancer and anti-cancer therapy. Saudi Med. J. 2016, 37, 1312–1318. [Google Scholar] [CrossRef]
- Ugrinova, I.; Monier, K.; Ivaldi, C.; Thiry, M.; Storck, S.; Mongelard, F.; Bouvet, P. Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol. Biol. 2007, 8, 66. [Google Scholar] [CrossRef]
- Xu, J.; Lu, S.; Xu, X.; Hu, S.; Li, B.; Qi, R.; Chen, L.; Chang, J. Knocking Down Nucleolin Expression Enhances the Radiosensitivity of Non-Small Cell Lung Cancer by Influencing DNA-PKcs Activity. Asian Pac. J. Cancer Prev. APJCP 2015, 16, 3301–3306. [Google Scholar] [CrossRef]
- Benedetti, E.; Antonosante, A.; d’Angelo, M.; Cristiano, L.; Galzio, R.; Destouches, D.; Florio, T.; Dhez, A.; Astarita, C.; Cinque, B.; et al. Nucleolin antagonist triggers autophagic cell death in human glioblastoma primary cells and decreased in vivo tumor growth in orthotopic brain tumor model. Oncotarget 2015, 6, 42091–42104. [Google Scholar] [CrossRef]
- Subramanian, N.; Srimany, A.; Kanwar, J.; Kanwar, R.; Akilandeswari, B.; Rishi, P.; Khetan, V.; Vasudevan, M.; Pradeep, T.; Krishnakumar, S. Nucleolin-aptamer therapy in retinoblastoma: Molecular changes and mass spectrometry-based imaging. Mol. Ther. Nucleic Acids 2016, 5, e358. [Google Scholar] [CrossRef] [Green Version]
- Shang, Y.; Kakinuma, S.; Nishimura, M.; Kobayashi, Y.; Nagata, K.; Shimada, Y. Interleukin-9 receptor gene is transcriptionally regulated by nucleolin in T-cell lymphoma cells. Mol. Carcinog. 2012, 51, 619–627. [Google Scholar] [CrossRef]
- D’Avino, C.; Palmieri, D.; Braddom, A.; Zanesi, N.; James, C.; Cole, S.; Salvatore, F.; Croce, C.; De Lorenzo, C. A novel fully human anti-NCL immunoRNase for triple-negative breast cancer therapy. Oncotarget 2016, 7, 87016–87030. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, D.; Richmond, T.; Piovan, C.; Sheetz, T.; Zanesi, N.; Troise, F.; James, C.; Wernicke, D.; Nyei, F.; Gordon, T.; et al. Human anti-nucleolin recombinant immunoagent for cancer therapy. Proc. Natl. Acad. Sci. USA 2015, 112, 9418–9423. [Google Scholar] [CrossRef] [PubMed]
- Gilles, M.; Maione, F.; Cossutta, M.; Carpentier, G.; Caruana, L.; Di Maria, S.; Houppe, C.; Destouches, D.; Shchors, K.; Prochasson, C.; et al. Nucleolin Targeting Impairs the Progression of Pancreatic Cancer and Promotes the Normalization of Tumor Vasculature. Cancer Res. 2016, 76, 7181–7193. [Google Scholar] [CrossRef]
- Fogal, V.; Sugahara, K.; Ruoslahti, E.; Christian, S. Cell surface nucleolin antagonist causes endothelial cell apoptosis and normalization of tumor vasculature. Angiogenesis 2009, 12, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Song, N.; Liu, C.; He, T.; Zhuo, W.; He, X.; Chen, Y.; Song, X.; Fu, Y.; Luo, Y. Heat shock cognate 70 regulates the translocation and angiogenic function of nucleolin. Arterioscler. Thromb. Vasc. Biol. 2012, 32, e126–e134. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.; Yao, Y.; Liu, Y.; Yan, M.; Huang, Y.; Chen, Y. Nucleolin identified by comparative mass-spectra analysis is a potential marker for invasive progression of hepatocellular carcinoma. Mol. Med. Rep. 2014, 10, 1489–1494. [Google Scholar] [CrossRef]
- Li, Y.; Tang, Y.; Ye, L.; Liu, B.; Liu, K.; Chen, J.; Xue, Q. Establishment of a hepatocellular carcinoma cell line with unique metastatic characteristics through in vivo selection and screening for metastasis-related genes through cDNA microarray. J. Cancer Res. Clin. Oncol. 2003, 129, 43–51. [Google Scholar] [CrossRef]
- Choi, W.; Yang, Y.; Xu, Y.; An, J. Targeting chemokine receptor CXCR4 for treatment of HIV-1 infection, tumor progression, and metastasis. Curr. Top. Med. Chem. 2014, 14, 1574–1589. [Google Scholar] [CrossRef]
- Niu, H.; Yang, X.; Xu, Z.; Du, T.; Wang, R. Cell surface nucleolin interacts with CXCR4 receptor via the 212 c-terminal portion. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2015, 36, 1099–1104. [Google Scholar] [CrossRef]
- Yu, Y.; Jin, H.; Xu, J.; Gu, J.; Li, X.; Xie, Q.; Huang, H.; Li, J.; Tian, Z.; Jiang, G.; et al. XIAP overexpression promotes bladder cancer invasion in vitro and lung metastasis in vivo via enhancing nucleolin-mediated Rho-GDIβ mRNA stability. Int. J. Cancer 2018, 142, 2040–2055. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Zhang, N.; Shen, L.; Lu, Y.; Chang, Y.; Lin, Z.; Sun, N.; Zhang, Y.; Xu, J.; Huang, H.; et al. Lnc00892 competes with c-Jun to block NCL transcription, reducing the stability of RhoA/RhoC mRNA and impairing bladder cancer invasion. Oncogene 2021, 40, 6579–6589. [Google Scholar] [CrossRef]
- Jin, H.; Yu, Y.; Hu, Y.; Lu, C.; Li, J.; Gu, J.; Zhang, L.; Huang, H.; Zhang, D.; Wu, X.; et al. Divergent behaviors and underlying mechanisms of cell migration and invasion in non-metastatic T24 and its metastatic derivative T24T bladder cancer cell lines. Oncotarget 2015, 6, 522–536. [Google Scholar] [CrossRef] [PubMed]
- Daizumoto, K.; Yoshimaru, T.; Matsushita, Y.; Fukawa, T.; Uehara, H.; Ono, M.; Komatsu, M.; Kanayama, H.; Katagiri, T. A DDX31/Mutant-p53/EGFR Axis Promotes Multistep Progression of Muscle-Invasive Bladder Cancer. Cancer Res. 2018, 78, 2233–2247. [Google Scholar] [CrossRef]
- Lubet, R.; Lu, Y.; Bode, A.; You, M.; Verney, Z.; Steele, V.; Townsend, R.; Juliana, M.; Grubbs, C. Efficacy of the EGFr inhibitor Iressa on development of chemically-induced urinary bladder cancers: Dose dependency and modulation of biomarkers. Oncol. Rep. 2011, 25, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
- Vernet, C.; Artzt, K. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. TIG 1997, 13, 479–484. [Google Scholar] [CrossRef]
- Artzt, K.; Wu, J. STAR trek: An introduction to STAR family proteins and review of quaking (QKI). Adv. Exp. Med. Biol. 2010, 693, 1–24. [Google Scholar]
- Teplova, M.; Hafner, M.; Teplov, D.; Essig, K.; Tuschl, T.; Patel, D. Structure-function studies of STAR family Quaking proteins bound to their in vivo RNA target sites. Genes Dev. 2013, 27, 928–940. [Google Scholar] [CrossRef]
- Darbelli, L.; Richard, S. Emerging functions of the Quaking RNA-binding proteins and link to human diseases. Wiley Interdiscip. Rev. RNA 2016, 7, 399–412. [Google Scholar] [CrossRef]
- Farnsworth, B.; Peuckert, C.; Zimmermann, B.; Jazin, E.; Kettunen, P.; Emilsson, L. Gene Expression of Quaking in Sporadic Alzheimer’s Disease Patients is Both Upregulated and Related to Expression Levels of Genes Involved in Amyloid Plaque and Neurofibrillary Tangle Formation. J. Alzheimer’s Dis. 2016, 53, 209–219. [Google Scholar] [CrossRef]
- Galarneau, A.; Richard, S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat. Struct. Mol. Biol. 2005, 12, 691–698. [Google Scholar] [CrossRef]
- Ryder, S.; Frater, L.; Abramovitz, D.; Goodwin, E.; Williamson, J. RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nat. Struct. Mol. Biol. 2004, 11, 20–28. [Google Scholar] [CrossRef]
- Zhang, R.; Yang, J.; Peng, L.; Zheng, L.; Xie, P.; Wang, M.; Cao, Y.; Zhang, Z.; Zhou, F.; Qian, C.; et al. RNA-binding protein QKI-5 inhibits the proliferation of clear cell renal cell carcinoma via post-transcriptional stabilization of RASA1 mRNA. Cell Cycle 2016, 15, 3094–3104. [Google Scholar] [CrossRef]
- Novikov, L.; Park, J.; Chen, H.; Klerman, H.; Jalloh, A.; Gamble, M. QKI-mediated alternative splicing of the histone variant MacroH2A1 regulates cancer cell proliferation. Mol. Cell. Biol. 2011, 31, 4244–4255. [Google Scholar] [CrossRef]
- Bian, Y.; Wang, L.; Lu, H.; Yang, G.; Zhang, Z.; Fu, H.; Lu, X.; Wei, M.; Sun, J.; Zhao, Q.; et al. Downregulation of tumor suppressor QKI in gastric cancer and its implication in cancer prognosis. Biochem. Biophys. Res. Commun. 2012, 422, 187–193. [Google Scholar] [CrossRef]
- Yang, G.; Fu, H.; Zhang, J.; Lu, X.; Yu, F.; Jin, L.; Bai, L.; Huang, B.; Shen, L.; Feng, Y.; et al. RNA-binding protein quaking, a critical regulator of colon epithelial differentiation and a suppressor of colon cancer. Gastroenterology 2010, 138, 231–240.e5. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, G.; Wei, M.; Lu, X.; Fu, H.; Feng, F.; Wang, S.; Lu, W.; Wu, N.; Lu, Z.; et al. The tumor suppressing effects of QKI-5 in prostate cancer: A novel diagnostic and prognostic protein. Cancer Biol. Ther. 2014, 15, 108–118. [Google Scholar] [CrossRef]
- Shi, F.; Deng, Z.; Zhou, Z.; Jiang, C.; Zhao, R.; Sun, F.; Cui, D.; Bei, X.; Yang, B.; Sun, Q.; et al. QKI-6 inhibits bladder cancer malignant behaviours through down-regulating E2F3 and NF-κB signalling. J. Cell. Mol. Med. 2019, 23, 6578–6594. [Google Scholar] [CrossRef]
- Wei, X.; Wang, B.; Wang, Q.; Yang, X.; Yang, Y.; Fang, Z.; Yi, C.; Shi, L.; Fan, X.; Tao, J.; et al. MiR-362-5p, Which Is Regulated by Long Non-Coding RNA MBNL1-AS1, Promotes the Cell Proliferation and Tumor Growth of Bladder Cancer by Targeting QKI. Front. Pharmacol. 2020, 11, 164. [Google Scholar] [CrossRef]
- LeBleu, V.; Kalluri, R. A peek into cancer-associated fibroblasts: Origins, functions and translational impact. Dis. Model. Mech. 2018, 11, dmm029447. [Google Scholar] [CrossRef]
- Quail, D.; Joyce, J. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Kharaishvili, G.; Simkova, D.; Bouchalova, K.; Gachechiladze, M.; Narsia, N.; Bouchal, J. The role of cancer-associated fibroblasts, solid stress and other microenvironmental factors in tumor progression and therapy resistance. Cancer Cell Int. 2014, 14, 41. [Google Scholar] [CrossRef]
- Zhou, Z.; Cui, D.; Sun, M.; Huang, J.; Deng, Z.; Han, B.; Sun, X.; Xia, S.; Sun, F.; Shi, F. CAFs-derived MFAP5 promotes bladder cancer malignant behavior through NOTCH2/HEY1 signaling. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 7970–7988. [Google Scholar] [CrossRef]
- Bley, N.; Hmedat, A.; Müller, S.; Rolnik, R.; Rausch, A.; Lederer, M.; Hüttelmaier, S. Musashi-1-A Stemness RBP for Cancer Therapy? Biology 2021, 10, 407. [Google Scholar] [CrossRef]
- Sun, J.; Sheng, W.; Ma, Y.; Dong, M. Potential Role of Musashi-2 RNA-Binding Protein in Cancer EMT. OncoTargets Ther. 2021, 14, 1969–1980. [Google Scholar] [CrossRef]
- Qi, X.; Zhang, F.; Wu, H.; Liu, J.; Zong, B.; Xu, C.; Jiang, J. Wilms’ tumor 1 (WT1) expression and prognosis in solid cancer patients: A systematic review and meta-analysis. Sci. Rep. 2015, 5, 8924. [Google Scholar] [CrossRef]
- Gupta, G.; Bebawy, M.; Pinto, T.; Chellappan, D.; Mishra, A.; Dua, K. Role of the Tristetraprolin (Zinc Finger Protein 36 Homolog) Gene in Cancer. Crit. Rev. Eukaryot. Gene Expr. 2018, 28, 217–221. [Google Scholar] [CrossRef]
- Saini, Y.; Chen, J.; Patial, S. The Tristetraprolin Family of RNA-Binding Proteins in Cancer: Progress and Future Prospects. Cancers 2020, 12, 1539. [Google Scholar] [CrossRef]
- Schuschel, K.; Helwig, M.; Hüttelmaier, S.; Heckl, D.; Klusmann, J.; Hoell, J. RNA-Binding Proteins in Acute Leukemias. Int. J. Mol. Sci. 2020, 21, 3409. [Google Scholar] [CrossRef]
- Wang, E.; Lu, S.; Pastore, A.; Chen, X.; Imig, J.; Chun-Wei Lee, S.; Hockemeyer, K.; Ghebrechristos, Y.; Yoshimi, A.; Inoue, D.; et al. Targeting an RNA-Binding Protein Network in Acute Myeloid Leukemia. Cancer Cell 2019, 35, 369–384.e7. [Google Scholar] [CrossRef]
- Yamauchi, T.; Masuda, T.; Canver, M.; Seiler, M.; Semba, Y.; Shboul, M.; Al-Raqad, M.; Maeda, M.; Schoonenberg, V.; Cole, M.; et al. Genome-wide CRISPR-Cas9 Screen Identifies Leukemia-Specific Dependence on a Pre-mRNA Metabolic Pathway Regulated by DCPS. Cancer Cell 2018, 33, 386–400.e5. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Kang, D.; Turnbull, G.; Dong, Y. Delivery of CRISPR-Cas9 system for screening and editing RNA binding proteins in cancer. Adv. Drug Deliv. Rev. 2022, 180, 114042. [Google Scholar] [CrossRef]
- Wang, F.; Song, W.; Zhao, H.; Ma, Y.; Li, Y.; Zhai, D.; Pi, J.; Si, Y.; Xu, J.; Dong, L.; et al. The RNA-binding protein QKI5 regulates primary miR-124-1 processing via a distal RNA motif during erythropoiesis. Cell Res. 2017, 27, 416–439. [Google Scholar] [CrossRef]
- Chen, A.; Paik, J.; Zhang, H.; Shukla, S.; Mortensen, R.; Hu, J.; Ying, H.; Hu, B.; Hurt, J.; Farny, N.; et al. STAR RNA-binding protein Quaking suppresses cancer via stabilization of specific miRNA. Genes Dev. 2012, 26, 1459–1472. [Google Scholar] [CrossRef]
- He, B.; Gao, S.; Huang, L.; Huang, Y.; Zhang, Q.; Zhou, M.; Shi, H.; Song, Q.; Shan, Y. MicroRNA-155 promotes the proliferation and invasion abilities of colon cancer cells by targeting quaking. Mol. Med. Rep. 2015, 11, 2355–2359. [Google Scholar] [CrossRef]
- Danan-Gotthold, M.; Golan-Gerstl, R.; Eisenberg, E.; Meir, K.; Karni, R.; Levanon, E. Identification of recurrent regulated alternative splicing events across human solid tumors. Nucleic Acids Res. 2015, 43, 5130–5144. [Google Scholar] [CrossRef]
- Xi, Z.; Wang, P.; Xue, Y.; Shang, C.; Liu, X.; Ma, J.; Li, Z.; Li, Z.; Bao, M.; Liu, Y. Overexpression of miR-29a reduces the oncogenic properties of glioblastoma stem cells by downregulating Quaking gene isoform 6. Oncotarget 2017, 8, 24949–24963. [Google Scholar] [CrossRef]
- Fu, X.; Feng, Y. QKI-5 suppresses cyclin D1 expression and proliferation of oral squamous cell carcinoma cells via MAPK signalling pathway. Int. J. Oral Maxillofac. Surg. 2015, 44, 562–567. [Google Scholar] [CrossRef]
- Zhou, X.; Li, X.; Sun, C.; Shi, C.; Hua, D.; Yu, L.; Wen, Y.; Hua, F.; Wang, Q.; Zhou, Q.; et al. Quaking-5 suppresses aggressiveness of lung cancer cells through inhibiting β-catenin signaling pathway. Oncotarget 2017, 8, 82174–82184. [Google Scholar] [CrossRef]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.; Strein, C.; Davey, N.; Humphreys, D.; Preiss, T.; Steinmetz, L.; et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 149, 1393–1406. [Google Scholar] [CrossRef]
- Wang, T.; Xiao, G.; Chu, Y.; Zhang, M.; Corey, D.; Xie, Y. Design and bioinformatics analysis of genome-wide CLIP experiments. Nucleic Acids Res. 2015, 43, 5263–5274. [Google Scholar] [CrossRef] [Green Version]
- King, C.; Cuatrecasas, M.; Castells, A.; Sepulveda, A.; Lee, J.; Rustgi, A. LIN28B promotes colon cancer progression and metastasis. Cancer Res. 2011, 71, 4260–4268. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Jiménez, C.; Ludeña, M.; Izquierdo, J. T-cell intracellular antigens function as tumor suppressor genes. Cell Death Dis. 2015, 6, e1669. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, J.; Domingo-Fernández, R.; Ebus, M.; Lindner, S.; Koster, J.; Drabek, K.; Mestdagh, P.; van Sluis, P.; Valentijn, L.; van Nes, J.; et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat. Genet. 2012, 44, 1199–1206. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Hayama, S.; Yamabuki, T.; Ishikawa, N.; Miyamoto, M.; Ito, T.; Tsuchiya, E.; Kondo, S.; Nakamura, Y.; Daigo, Y. Increased expression of insulin-like growth factor-II messenger RNA-binding protein 1 is associated with tumor progression in patients with lung cancer. Clin. Cancer Res. 2007, 13, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Lan, L.; Wilson, D.; Marquez, R.; Tsao, W.; Gao, P.; Roy, A.; Turner, B.; McDonald, P.; Tunge, J.; et al. Identification and validation of novel small molecule disruptors of HuR-mRNA interaction. ACS Chem. Biol. 2015, 10, 1476–1484. [Google Scholar] [CrossRef] [PubMed]
- Roos, M.; Pradère, U.; Ngondo, R.; Behera, A.; Allegrini, S.; Civenni, G.; Zagalak, J.; Marchand, J.; Menzi, M.; Towbin, H.; et al. A Small-Molecule Inhibitor of Lin28. ACS Chem. Biol. 2016, 11, 2773–2781. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Y.; Cao, H.; Huang, D.; Zheng, L.; Nie, Z.; Zhang, S. RNA-Binding Proteins in Bladder Cancer. Cancers 2023, 15, 1150. https://doi.org/10.3390/cancers15041150
Gao Y, Cao H, Huang D, Zheng L, Nie Z, Zhang S. RNA-Binding Proteins in Bladder Cancer. Cancers. 2023; 15(4):1150. https://doi.org/10.3390/cancers15041150
Chicago/Turabian StyleGao, Yuanhui, Hui Cao, Denggao Huang, Linlin Zheng, Zhenyu Nie, and Shufang Zhang. 2023. "RNA-Binding Proteins in Bladder Cancer" Cancers 15, no. 4: 1150. https://doi.org/10.3390/cancers15041150
APA StyleGao, Y., Cao, H., Huang, D., Zheng, L., Nie, Z., & Zhang, S. (2023). RNA-Binding Proteins in Bladder Cancer. Cancers, 15(4), 1150. https://doi.org/10.3390/cancers15041150





