NSC243928 Treatment Induces Anti-Tumor Immune Response in Mouse Mammary Tumor Models
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells
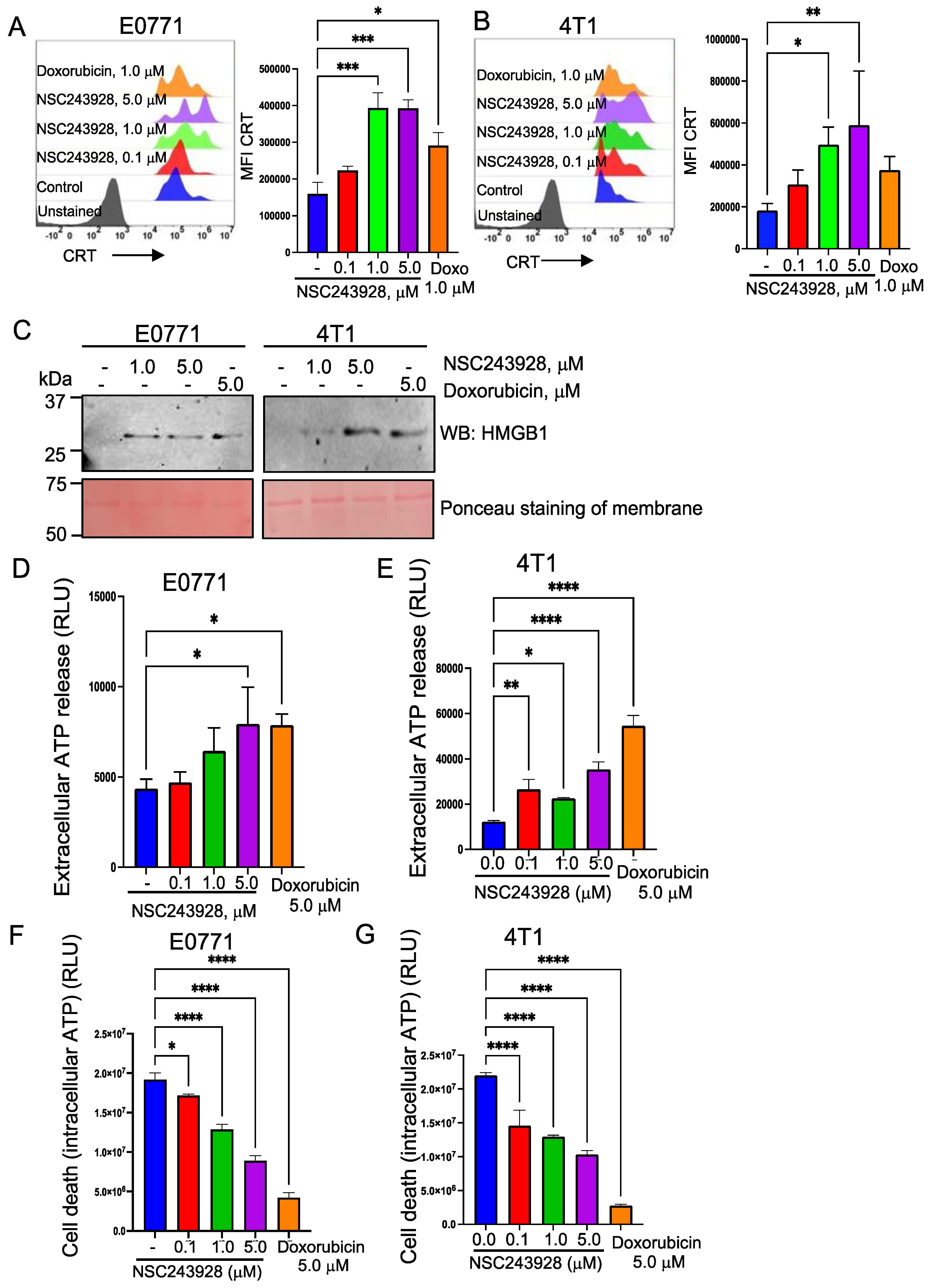
2.2. Calreticulin (CRT) Cell Surface Expression
2.3. HMGB1 Release Assay
2.4. ATP Assay for Extracellular ATP Release and Cell Viability
2.5. Isograft Mouse Model, Tumor Measurements, and Treatment
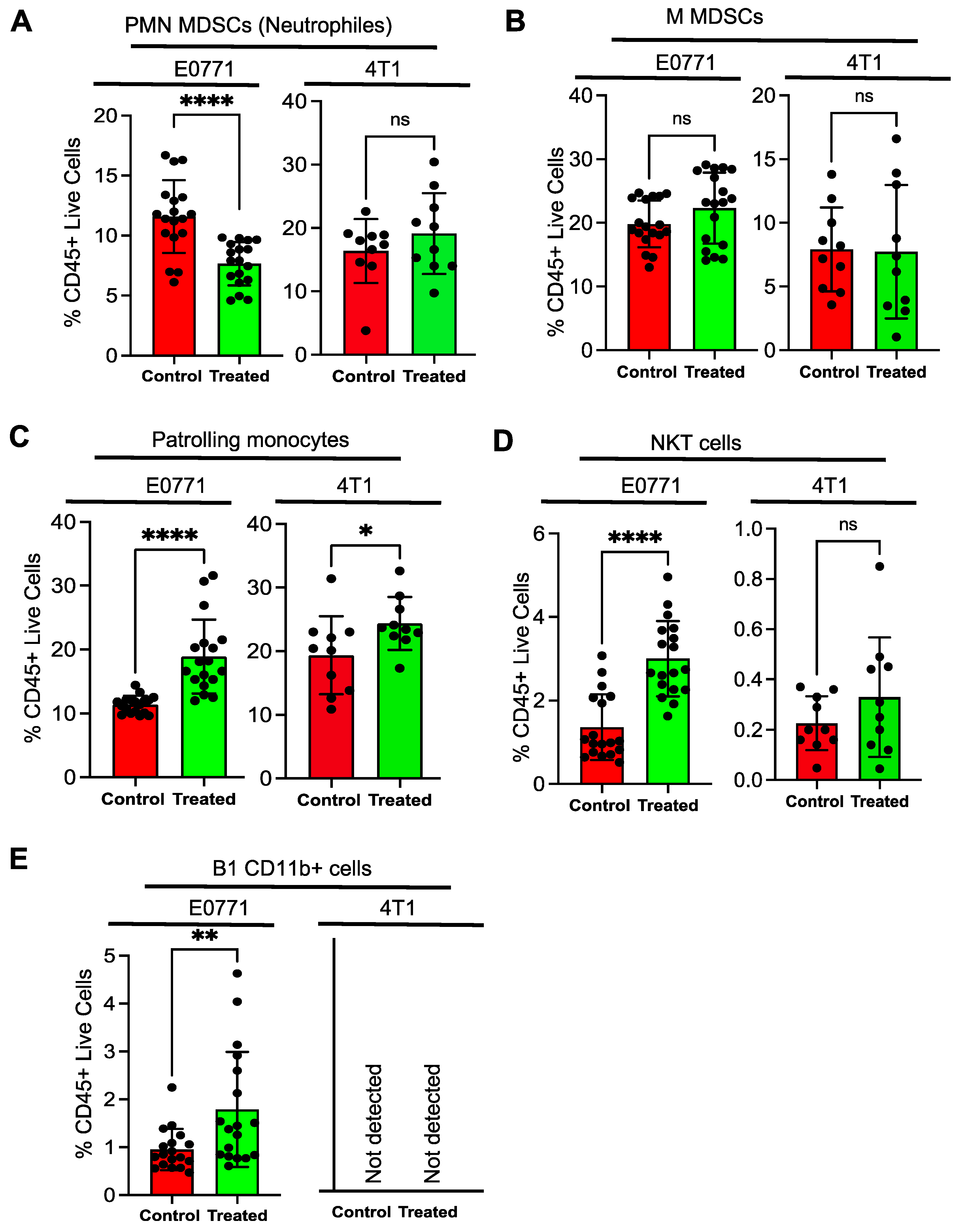
2.6. Blood Collection and MDSC Analysis
2.7. Immunophenotyping of the Isografts
2.8. Total RNA Sequencing and Data Analysis
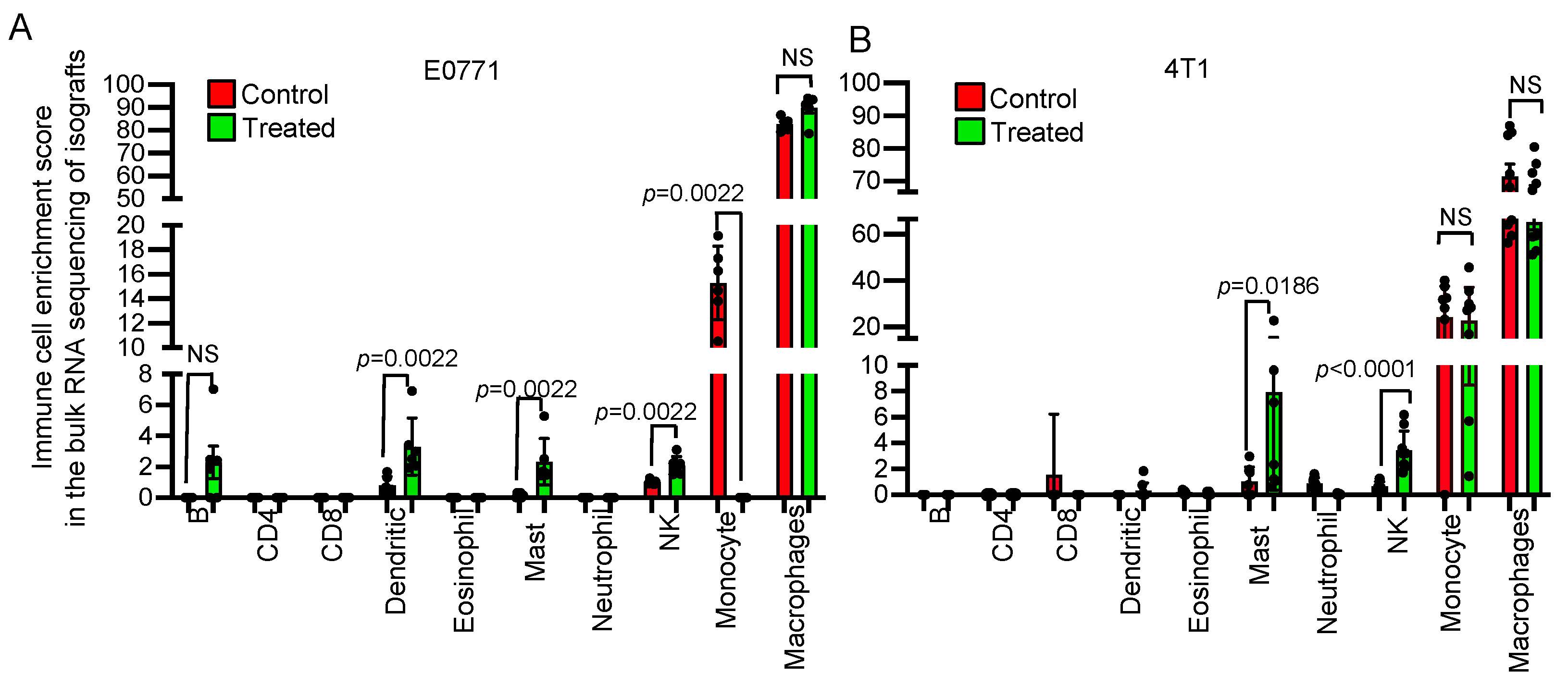
2.9. Immune Cell Compositions of 4T1 and E0771 Isografts
3. Results
3.1. NSC243928 Induces Immunogenic Cell Death in the E0771 and 4T1 Cell Lines
3.2. NSC243928 Reduces the Tumor Growth In Vivo and Induces a Systemic Anti-Tumor Immune Response
3.3. Transcriptome Analysis Revealed That NSC243928 Induces an Immune Responsive Tumor Microenvironment
3.4. Tumor Infiltrating Lymphocytes Show a Distinct Pattern upon NSC243928 Treatment in the E0771 and 4T1 Models
3.5. Immune Cells from NSC243928 Treated E0771 Tumor Isografts Generate a Better Response in Cytokine Production Ex Vivo Compared to the 4T1 Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Luo, L.; McGarvey, P.; Madhavan, S.; Kumar, R.; Gusev, Y.; Upadhyay, G. Distinct lymphocyte antigens 6 (Ly6) family members Ly6D, Ly6E, Ly6K and Ly6H drive tumorigenesis and clinical outcome. Oncotarget 2016, 7, 11165–11193. [Google Scholar] [CrossRef] [PubMed]
- Benti, S.; Tiwari, P.B.; Goodlett, D.W.; Daneshian, L.; Kern, G.B.; Smith, M.D.; Uren, A.; Chruszcz, M.; Shimizu, L.S.; Upadhyay, G. Small Molecule Binds with Lymphocyte Antigen 6K to Induce Cancer Cell Death. Cancers 2020, 12, 509. [Google Scholar] [CrossRef]
- Ferguson, L.R.; Denny, W.A. Potential antitumor agents. 30. Mutagenic activity of some 9-anilinoacridines: Relationships between structure, mutagenic potential, and antileukemic activity. J. Med. Chem. 1979, 22, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Mezencev, R.; Wang, L.; McDonald, J.F. Identification of inhibitors of ovarian cancer stem-like cells by high-throughput screening. J. Ovarian. Res. 2012, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- AlHossiny, M.; Luo, L.; Frazier, W.R.; Steiner, N.; Gusev, Y.; Kallakury, B.; Glasgow, E.; Creswell, K.; Madhavan, S.; Kumar, R.; et al. Ly6E/K Signaling to TGFbeta Promotes Breast Cancer Progression, Immune Escape, and Drug Resistance. Cancer Res. 2016, 76, 3376–3386. [Google Scholar] [CrossRef]
- Schrörs, B.; Boegel, S.; Albrecht, C.; Bukur, T.; Bukur, V.; Holtsträter, C.; Ritzel, C.; Manninen, K.; Tadmor, A.D.; Vormehr, M.; et al. Multi-Omics Characterization of the 4T1 Murine Mammary Gland Tumor Model. Front. Oncol. 2020, 10, 1195. [Google Scholar] [CrossRef]
- Le Naour, A.; Koffi, Y.; Diab, M.; Le Guennec, D.; Rougé, S.; Aldekwer, S.; Goncalves-Mendes, N.; Talvas, J.; Farges, M.-C.; Caldefie-Chezet, F.; et al. EO771, the first luminal B mammary cancer cell line from C57BL/6 mice. Cancer Cell Int. 2020, 20, 328. [Google Scholar] [CrossRef]
- Faustino-Rocha, A.; Oliveira, P.A.; Pinho-Oliveira, J.; Teixeira-Guedes, C.; Soares-Maia, R.; da Costa, R.G.; Colaço, B.; Pires, M.J.; Colaço, J.; Ferreira, R.; et al. Estimation of rat mammary tumor volume using caliper and ultrasonography measurements. Lab. Anim. 2013, 42, 217–224. [Google Scholar] [CrossRef]
- Hather, G.; Liu, R.; Bandi, S.; Mettetal, J.; Manfredi, M.; Shyu, W.C.; Donelan, J.; Chakravarty, A. Growth rate analysis and efficient experimental design for tumor xenograft studies. Cancer Inf. 2014, 13 (Suppl. S4), 65–72. [Google Scholar] [CrossRef]
- Cassetta, L.; Baekkevold, E.S.; Brandau, S.; Bujko, A.; Cassatella, M.A.; Dorhoi, A.; Krieg, C.; Lin, A.; Lore, K.; Marini, O.; et al. Deciphering myeloid-derived suppressor cells: Isolation and markers in humans, mice and non-human primates. Cancer Immunol. Immunother. 2019, 68, 687–697. [Google Scholar] [CrossRef]
- Wang, K.; Singh, D.; Zeng, Z.; Coleman, S.J.; Huang, Y.; Savich, G.L.; He, X.; Mieczkowski, P.; Grimm, S.A.; Perou, C.M.; et al. MapSplice: Accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010, 38, e178. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Ewels, P.A.; Peltzer, A.; Fillinger, S.; Patel, H.; Alneberg, J.; Wilm, A.; Garcia, M.U.; Di Tommaso, P.; Nahnsen, S. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 2020, 38, 276–278. [Google Scholar] [CrossRef]
- Chen, Z.; Quan, L.; Huang, A.; Zhao, Q.; Yuan, Y.; Yuan, X.; Shen, Q.; Shang, J.; Ben, Y.; Qin, F.X.-F.; et al. seq-ImmuCC: Cell-Centric View of Tissue Transcriptome Measuring Cellular Compositions of Immune Microenvironment From Mouse RNA-Seq Data. Front. Immunol. 2018, 9, 1286. [Google Scholar] [CrossRef]
- Lin, T.; Chen, T.; Liu, J.; Tu, X.M. Extending the Mann-Whitney-Wilcoxon rank sum test to survey data for comparing mean ranks. Stat. Med. 2021, 40, 1705–1717. [Google Scholar] [CrossRef]
- Wang, Y.J.; Fletcher, R.; Yu, J.; Zhang, L. Immunogenic effects of chemotherapy-induced tumor cell death. Genes Dis. 2018, 5, 194–203. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, G.; Chen, Y.; Wang, H.; Hua, Y.; Cai, Z. Immunogenic cell death in cancer therapy: Present and emerging inducers. J. Cell. Mol. Med. 2019, 23, 4854–4865. [Google Scholar] [CrossRef]
- Fucikova, J.; Kepp, O.; Kasikova, L.; Petroni, G.; Yamazaki, T.; Liu, P.; Zhao, L.; Spisek, R.; Kroemer, G.; Galluzzi, L. Detection of immunogenic cell death and its relevance for cancer therapy. Cell Death Dis. 2020, 11, 1013. [Google Scholar] [CrossRef]
- Ghaffari, A.; Peterson, N.; Khalaj, K.; Vitkin, N.; Robinson, A.; Francis, J.A.; Koti, M. STING agonist therapy in combination with PD-1 immune checkpoint blockade enhances response to carboplatin chemotherapy in high-grade serous ovarian cancer. Br. J. Cancer 2018, 119, 440–449. [Google Scholar] [CrossRef]
- Yang, S.; Wang, J.; Brand, D.D.; Zheng, S.G. Role of TNF-TNF Receptor 2 Signal in Regulatory T Cells and Its Therapeutic Implications. Front. Immunol 2018, 9, 784. [Google Scholar] [CrossRef]
- Wu, Y.C.; Kipling, D.; Leong, H.S.; Martin, V.; Ademokun, A.A.; Dunn-Walters, D.K. High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood 2010, 116, 1070–1078. [Google Scholar] [CrossRef] [PubMed]
- Timmer, J.R.; Mak, T.W.; Manova, K.; Anderson, K.V.; Niswander, L. Tissue morphogenesis and vascular stability require the Frem2 protein, product of the mouse myelencephalic blebs gene. Proc. Natl. Acad. Sci. USA 2005, 102, 11746–11750. [Google Scholar] [CrossRef]
- Hess, D.A.; Strelau, K.M.; Karki, A.; Jiang, M.; Azevedo-Pouly, A.C.; Lee, A.H.; Deering, T.G.; Hoang, C.Q.; MacDonald, R.J.; Konieczny, S.F. MIST1 Links Secretion and Stress as both Target and Regulator of the Unfolded Protein Response. Mol. Cell. Biol. 2016, 36, 2931–2944. [Google Scholar] [CrossRef]
- Lou, W.; Ding, B.; Wang, S.; Fu, P. Overexpression of GPX3, a potential biomarker for diagnosis and prognosis of breast cancer, inhibits progression of breast cancer cells in vitro. Cancer Cell. Int. 2020, 20, 378. [Google Scholar] [CrossRef]
- Chen, C.L.; Hsu, S.C.; Ann, D.K.; Yen, Y.; Kung, H.J. Arginine Signaling and Cancer Metabolism. Cancers 2021, 13, 3541. [Google Scholar] [CrossRef]
- Sever, R.; Brugge, J.S. Signal transduction in cancer. Cold Spring Harb. Perspect. Med. 2015, 5, a006098. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.Y.S.; Reimer, D.; Zehrer, A.; Lu, M.; Mielenz, D.; Körner, H. Expression of Membrane-Bound CC Chemokine Ligand 20 on Follicular T Helper Cells in T-B-Cell Conjugates. Front. Immunol. 2017, 8, 1871. [Google Scholar] [CrossRef]
- Todd, J.R.; Ryall, K.A.; Vyse, S.; Wong, J.P.; Natrajan, R.C.; Yuan, Y.; Tan, A.C.; Huang, P.H. Systematic analysis of tumour cell-extracellular matrix adhesion identifies independent prognostic factors in breast cancer. Oncotarget 2016, 7, 62939–62953. [Google Scholar] [CrossRef] [PubMed]
- Nishikimi, A.; Meller, N.; Uekawa, N.; Isobe, K.; Schwartz, M.A.; Maruyama, M. Zizimin2: A novel, DOCK180-related Cdc42 guanine nucleotide exchange factor expressed predominantly in lymphocytes. FEBS Lett. 2005, 579, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Sinkala, M.; Mulder, N.; Patrick Martin, D. Metabolic gene alterations impact the clinical aggressiveness and drug responses of 32 human cancers. Commun. Biol. 2019, 2, 414. [Google Scholar] [CrossRef] [PubMed]
- Giudicelli, V.; Chaume, D.; Lefranc, M.P. IMGT/GENE-DB: A comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005, 33, D256–D261. [Google Scholar] [CrossRef] [PubMed]
- Djurec, M.; Graña, O.; Lee, A.; Troulé, K.; Espinet, E.; Cabras, L.; Navas, C.; Blasco, M.T.; Martín-Díaz, L.; Burdiel, M.; et al. Saa3 is a key mediator of the protumorigenic properties of cancer-associated fibroblasts in pancreatic tumors. Proc. Natl. Acad. Sci. USA 2018, 115, E1147–E1156. [Google Scholar] [CrossRef]
- Wang, J.; Manni, M.; Bärenwaldt, A.; Wieboldt, R.; Kirchhammer, N.; Ivanek, R.; Stanczak, M.; Zippelius, A.; König, D.; Rodrigues Manutano, N.; et al. Siglec Receptors Modulate Dendritic Cell Activation and Antigen Presentation to T Cells in Cancer. Front. Cell Dev. Biol 2022, 10, 828916. [Google Scholar] [CrossRef] [PubMed]
- Baechler, S.A.; Factor, V.M.; Dalla Rosa, I.; Ravji, A.; Becker, D.; Khiati, S.; Miller Jenkins, L.M.; Lang, M.; Sourbier, C.; Michaels, S.A.; et al. The mitochondrial type IB topoisomerase drives mitochondrial translation and carcinogenesis. Nat. Commun. 2019, 10, 83. [Google Scholar] [CrossRef]
- Xu, L.; Yang, Z.; Zhao, Q.; Feng, H.; Kuang, J.; Liu, Z.; Chen, L.; Zhan, L.; Yan, J.; Cai, W.; et al. Effect of FOXP2 transcription factor on immune infiltration of thyroid cancer and its potential clinical value. Front. Immunol. 2022, 13, 982812. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef]
- Veglia, F.; Hashimoto, A.; Dweep, H.; Sanseviero, E.; De Leo, A.; Tcyganov, E.; Kossenkov, A.; Mulligan, C.; Nam, B.; Masters, G.; et al. Analysis of classical neutrophils and polymorphonuclear myeloid-derived suppressor cells in cancer patients and tumor-bearing mice. J. Exp. Med. 2021, 218, e20201803. [Google Scholar] [CrossRef]
- Yang, P.; Liu, L.; Sun, L.; Fang, P.; Snyder, N.; Saredy, J.; Ji, Y.; Shen, W.; Qin, X.; Wu, Q.; et al. Immunological Feature and Transcriptional Signaling of Ly6C Monocyte Subsets From Transcriptome Analysis in Control and Hyperhomocysteinemic Mice. Front. Immunol. 2021, 12, 632333. [Google Scholar] [CrossRef]
- Haro, M.A.; Dyevoich, A.M.; Phipps, J.P.; Haas, K.M. Activation of B-1 Cells Promotes Tumor Cell Killing in the Peritoneal Cavity. Cancer Res. 2019, 79, 159–170. [Google Scholar] [CrossRef]
- Ramesh, P.; Shivde, R.; Jaishankar, D.; Saleiro, D.; Le Poole, I.C. A Palette of Cytokines to Measure Anti-Tumor Efficacy of T Cell-Based Therapeutics. Cancers 2021, 13, 821. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Miao, W.; He, D.; Wang, S.; Lou, J.; Jiang, Y.; Wang, S. Recent Progress on Immunotherapy for Breast Cancer: Tumor Microenvironment, Nanotechnology and More. Front. Bioeng Biotechnol. 2021, 9, 680315. [Google Scholar] [CrossRef]
- Crosby, E.J.; Wei, J.; Yang, X.Y.; Lei, G.; Wang, T.; Liu, C.X.; Agarwal, P.; Korman, A.J.; Morse, M.A.; Gouin, K.; et al. Complimentary mechanisms of dual checkpoint blockade expand unique T-cell repertoires and activate adaptive anti-tumor immunity in triple-negative breast tumors. Oncoimmunology 2018, 7, e1421891. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Skora, A.D.; Li, Z.; Liu, Q.; Tam, A.J.; Blosser, R.L.; Diaz, L.A., Jr.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B.; et al. Eradication of metastatic mouse cancers resistant to immune checkpoint blockade by suppression of myeloid-derived cells. Proc. Natl. Acad. Sci. USA 2014, 111, 11774–11779. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.M.; Rahim, M.M.A.; Sayed, C.; Mahmoud, A.B.; Makrigiannis, A.P. Immunosurveillance and Immunoediting of Breast Cancer via Class I MHC Receptors. Cancer Immunol. Res. 2017, 5, 1016–1028. [Google Scholar] [CrossRef]
- Hoover, R.; Gullickson, G.; Kornbluth, J. Natural killer lytic-associated molecule plays a role in controlling tumor dissemination and metastasis. Front. Immunol. 2012, 3, 393. [Google Scholar] [CrossRef]
- Gebremeskel, S.; Clattenburg, D.R.; Slauenwhite, D.; Lobert, L.; Johnston, B. Natural killer T cell activation overcomes immunosuppression to enhance clearance of postsurgical breast cancer metastasis in mice. Oncoimmunology 2015, 4, e995562. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, K.A.; Graham, L.V.; Pallero, M.A.; Murphy-Ullrich, J.E. Calreticulin regulates transforming growth factor-beta-stimulated extracellular matrix production. J. Biol Chem. 2013, 288, 14584–14598. [Google Scholar] [CrossRef] [PubMed]
- Kramer, E.D.; Abrams, S.I. Granulocytic Myeloid-Derived Suppressor Cells as Negative Regulators of Anticancer Immunity. Front. Immunol. 2020, 11, 1963. [Google Scholar] [CrossRef]
- Yang, Z.; Guo, J.; Weng, L.; Tang, W.; Jin, S.; Ma, W. Myeloid-derived suppressor cells—New and exciting players in lung cancer. J. Hematol. Oncol. 2020, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, P.B.; Eggert, T.; Zhu, Y.P.; Marcovecchio, P.; Meyer, M.A.; Wu, R.; Hedrick, C.C. Patrolling Monocytes Control NK Cell Expression of Activating and Stimulatory Receptors to Curtail Lung Metastases. J. Immunol. 2020, 204, 192. [Google Scholar] [CrossRef] [PubMed]
- Hanna, R.N.; Cekic, C.; Sag, D.; Tacke, R.; Thomas, G.D.; Nowyhed, H.; Herrley, E.; Rasquinha, N.; McArdle, S.; Wu, R.; et al. Patrolling monocytes control tumor metastasis to the lung. Science 2015, 350, 985–990. [Google Scholar] [CrossRef]
- Robinson, A.; Han, C.Z.; Glass, C.K.; Pollard, J.W. Monocyte Regulation in Homeostasis and Malignancy. Trends Immunol. 2021, 42, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Kmieciak, M.; Basu, D.; Payne, K.K.; Toor, A.; Yacoub, A.; Wang, X.Y.; Smith, L.; Bear, H.D.; Manjili, M.H. Activated NKT cells and NK cells render T cells resistant to myeloid-derived suppressor cells and result in an effective adoptive cellular therapy against breast cancer in the FVBN202 transgenic mouse. J. Immunol. 2011, 187, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, Q.; Qin, L.; Zhao, S.; Wang, J.; Chen, X. Transition of tumor-associated macrophages from MHC class II(hi) to MHC class II(low) mediates tumor progression in mice. BMC Immunol. 2011, 12, 43. [Google Scholar] [CrossRef]
- DeNardo, D.G.; Ruffell, B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 2019, 19, 369–382. [Google Scholar] [CrossRef]



| Condition | Upregulated Genes in Bold, Downregulated Genes in Italic | Function |
|---|---|---|
| Gene expression changes in NSC243928 treated E0771 isografts | Tnfrsf21, Tspan9, Hacl1 | Immune activation [22] |
| Zfp953, Bnip5 | B-cell pathway associated genes [23] | |
| Frem2, Celsr3, Mamdc2, Tm4sf1, Tnc, Gjb5 | Extracellular matrix regulation [24] | |
| MIST1/Bhlha15 | Unfolded protein response [25] | |
| Gpx3, Aox1 | Oxidation response [26] | |
| Arg1, Irx30s, Rars2 | Arginine signaling [27] | |
| Runx2, Wnt2b, Sox5, Irf5, Stap2, Pacsin1, Fnbp1l, Wrn, Nr2c1, Hspa1b, Bmp15 | Oncogenic pathways [28] | |
| Ccl20, Cxcr5 | T-cell regulatory responses [29] | |
| F12 | Extracellular matrix [30] | |
| Dock10 | GTPase signaling [31] | |
| Gene expression changes in NSC243928 treated 4T1 isografts | Acacb, Cbarp, Has2 | Metabolism [32] |
| Ighv13-2, Cxcr1, Usp31, Cxcl2 | Immune activation [33] | |
| Saa3, Defa39 | Extracellular matrix regulation [34] | |
| Cidn7, Nog, Heg1, Plekhh1, Bicd1, Cox6a1, Flt4 | Oncogenic signaling [28] | |
| Cd80, Siglece | Immune suppression [35] | |
| Top1mt | Mitochondrial metabolism [36] | |
| Common changes in both models | Ccr8, Itgad, Cd3d, Foxp2 | Immune suppression [37] |
| Id4, Spast, Zgrf1, Ctsd, Grk2, Terc | Oncogenic signaling [28] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Selvanesan, B.C.; de Mingo Pulido, A.; Varghese, S.; Rohila, D.; Hupalo, D.; Gusev, Y.; Contente, S.; Wilkerson, M.D.; Dalgard, C.L.; Upadhyay, G. NSC243928 Treatment Induces Anti-Tumor Immune Response in Mouse Mammary Tumor Models. Cancers 2023, 15, 1468. https://doi.org/10.3390/cancers15051468
Selvanesan BC, de Mingo Pulido A, Varghese S, Rohila D, Hupalo D, Gusev Y, Contente S, Wilkerson MD, Dalgard CL, Upadhyay G. NSC243928 Treatment Induces Anti-Tumor Immune Response in Mouse Mammary Tumor Models. Cancers. 2023; 15(5):1468. https://doi.org/10.3390/cancers15051468
Chicago/Turabian StyleSelvanesan, Benson Chellakkan, Alvaro de Mingo Pulido, Sheelu Varghese, Deepak Rohila, Daniel Hupalo, Yuriy Gusev, Sara Contente, Matthew D. Wilkerson, Clifton L. Dalgard, and Geeta Upadhyay. 2023. "NSC243928 Treatment Induces Anti-Tumor Immune Response in Mouse Mammary Tumor Models" Cancers 15, no. 5: 1468. https://doi.org/10.3390/cancers15051468
APA StyleSelvanesan, B. C., de Mingo Pulido, A., Varghese, S., Rohila, D., Hupalo, D., Gusev, Y., Contente, S., Wilkerson, M. D., Dalgard, C. L., & Upadhyay, G. (2023). NSC243928 Treatment Induces Anti-Tumor Immune Response in Mouse Mammary Tumor Models. Cancers, 15(5), 1468. https://doi.org/10.3390/cancers15051468






