Quantitative Evaluation of Stem-like Markers of Human Glioblastoma Using Single-Cell RNA Sequencing Datasets
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Preprocessing
2.3. Clustering and Enrichment Analysis
2.4. Calculation of Abundance and Percentage-Rank
2.5. Data Visualization
3. Results
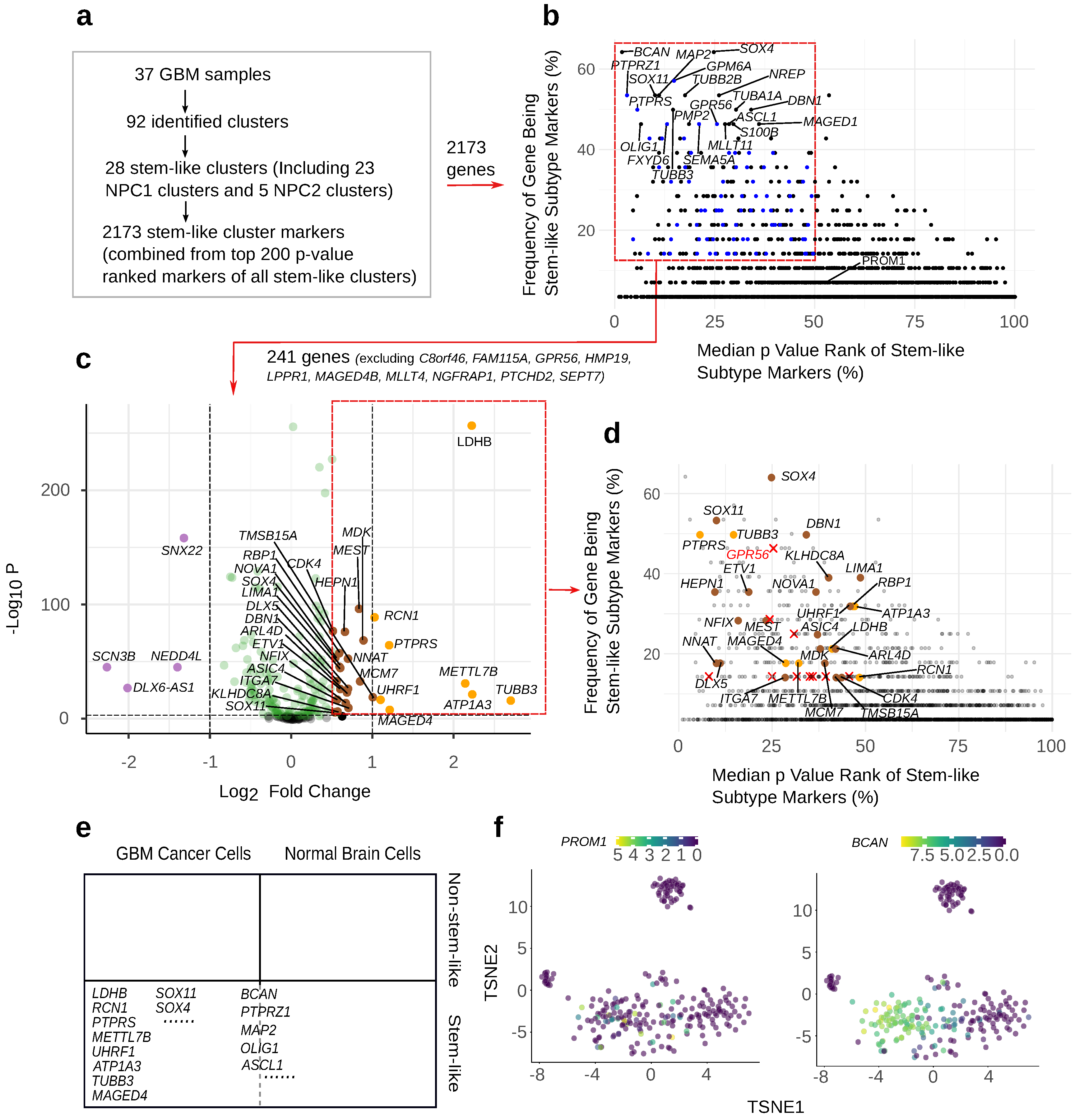
3.1. GBM Stem-like Cluster Identification
3.2. PROM1 Is the Marker Gene for Eight out of 28 Total Stem-like Clusters with Moderate Significance
3.3. Proposing Multiple Standards for Choosing the Optimal GBM Stem-like Markers According to the Application
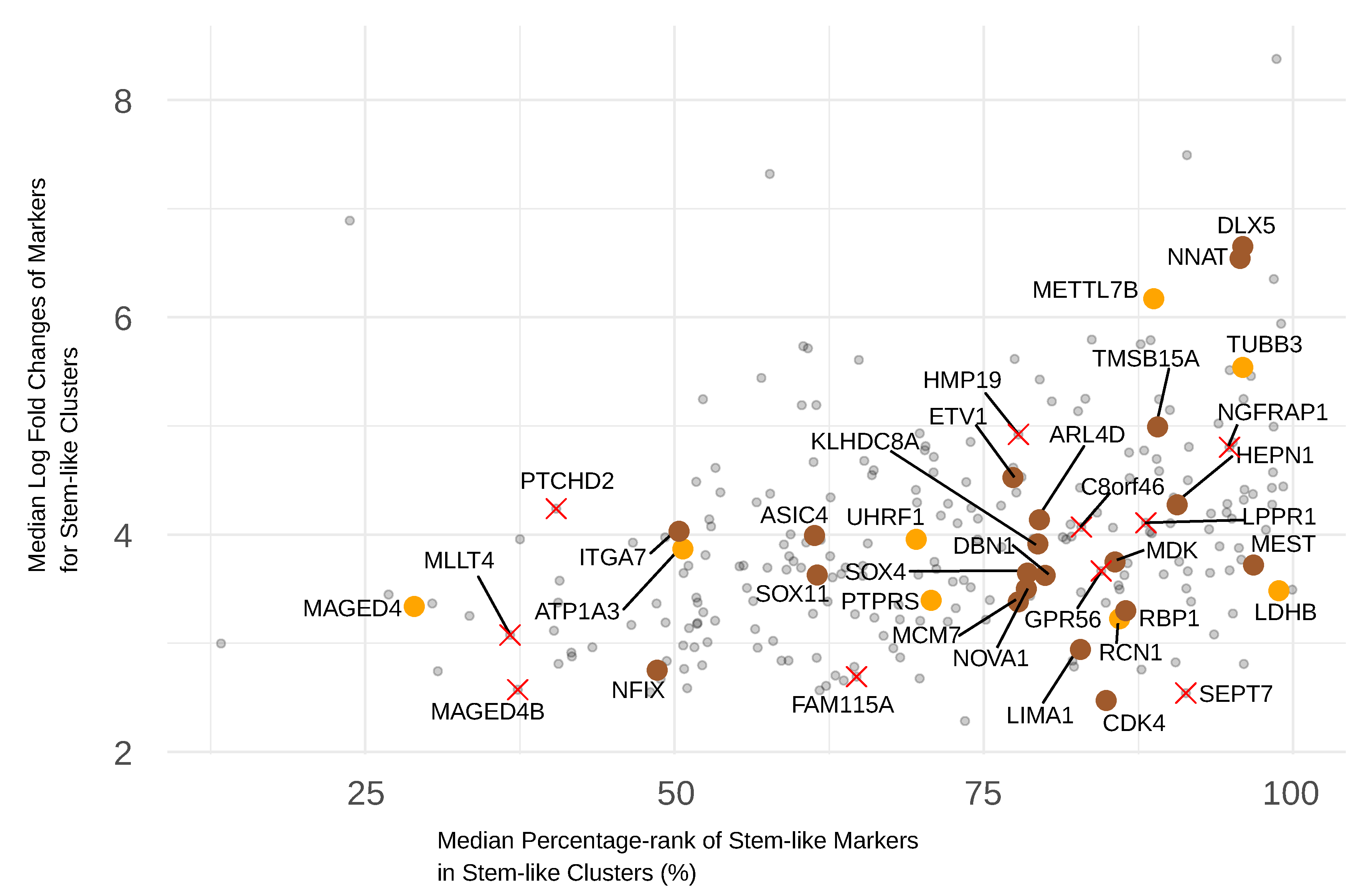
3.4. Selecting Frequent and Significant GBM Stem-like Markers
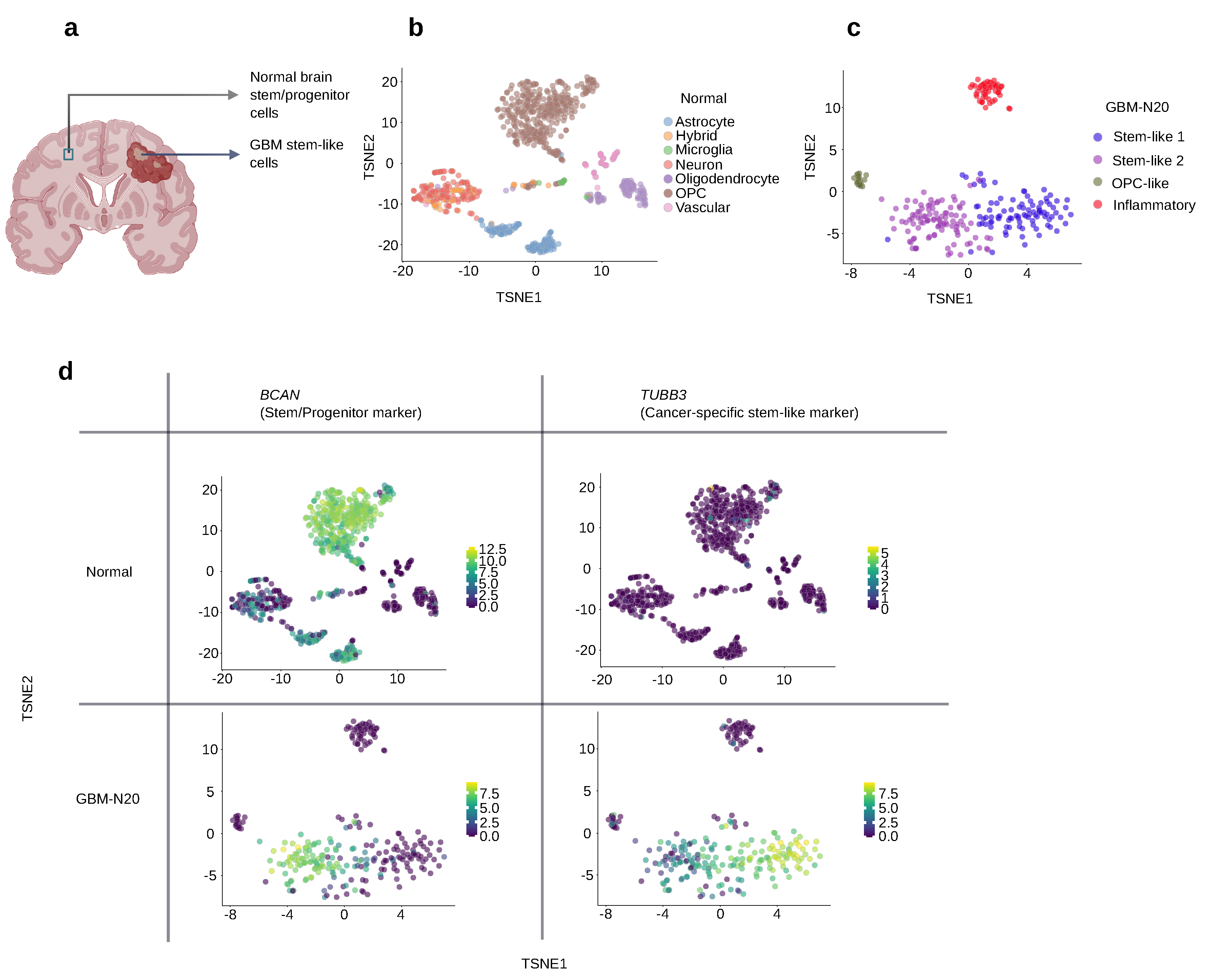
3.5. Selecting Stem-like Markers Overexpressed by GSCs Relative to Normal Cells
3.6. Selection of GBM Stem-like Markers Based on Their Expression Level
3.7. The Location of a Marker Protein Should Be Considered to Achieve Successful Targeting
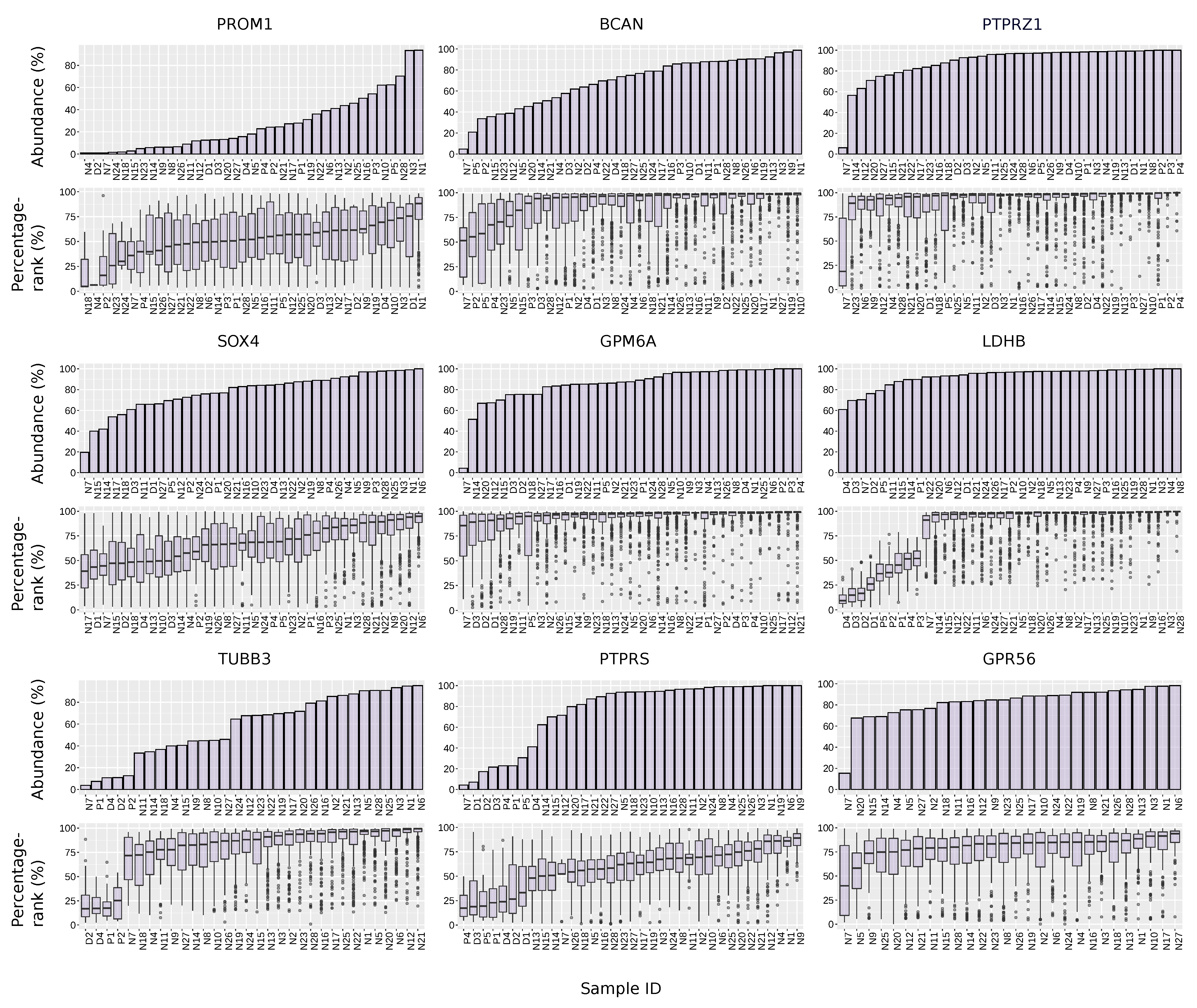
3.8. The Abundance and Expression Level of Selected GSCs Markers across the Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gately, L.; McLachlan, S.A.; Philip, J.; Ruben, J.; Dowling, A. Long-term survivors of glioblastoma: A closer look. J. Neuro-Oncol. 2018, 136, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Delgado-López, P.; Corrales-García, E. Survival in glioblastoma: A review on the impact of treatment modalities. Clin. Transl. Oncol. 2016, 18, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; Van Den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Oronsky, B.; Reid, T.R.; Oronsky, A.; Sandhu, N.; Knox, S.J. A review of newly diagnosed glioblastoma. Front. Oncol. 2021, 10, 574012. [Google Scholar] [CrossRef]
- Fabian, D.; Guillermo Prieto Eibl, M.d.P.; Alnahhas, I.; Sebastian, N.; Giglio, P.; Puduvalli, V.; Gonzalez, J.; Palmer, J.D. Treatment of glioblastoma (GBM) with the addition of tumor-treating fields (TTF): A review. Cancers 2019, 11, 174. [Google Scholar] [CrossRef]
- Prager, B.C.; Bhargava, S.; Mahadev, V.; Hubert, C.G.; Rich, J.N. Glioblastoma stem cells: Driving resilience through chaos. Trends Cancer 2020, 6, 223–235. [Google Scholar] [CrossRef]
- Gimple, R.C.; Bhargava, S.; Dixit, D.; Rich, J.N. Glioblastoma stem cells: Lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 2019, 33, 591–609. [Google Scholar] [CrossRef]
- Knudsen, A.M.; Halle, B.; Cédile, O.; Burton, M.; Baun, C.; Thisgaard, H.; Anand, A.; Hubert, C.; Thomassen, M.; Michaelsen, S.R.; et al. Surgical resection of glioblastomas induces pleiotrophin-mediated self-renewal of glioblastoma stem cells in recurrent tumors. Neuro-Oncology 2022, 24, 1074–1087. [Google Scholar] [CrossRef]
- Eramo, A.; Ricci-Vitiani, L.; Zeuner, A.; Pallini, R.; Lotti, F.; Sette, G.; Pilozzi, E.; Larocca, L.M.; Peschle, C.; De Maria, R. Chemotherapy resistance of glioblastoma stem cells. Cell Death Differ. 2006, 13, 1238–1241. [Google Scholar] [CrossRef]
- Mattei, V.; Santilli, F.; Martellucci, S.; Delle Monache, S.; Fabrizi, J.; Colapietro, A.; Angelucci, A.; Festuccia, C. The importance of tumor stem cells in glioblastoma resistance to therapy. Int. J. Mol. Sci. 2021, 22, 3863. [Google Scholar] [CrossRef]
- Neftel, C.; Laffy, J.; Filbin, M.G.; Hara, T.; Shore, M.E.; Rahme, G.J.; Richman, A.R.; Silverbush, D.; Shaw, M.L.; Hebert, C.M.; et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 2019, 178, 835–849. [Google Scholar] [CrossRef]
- Brescia, P.; Ortensi, B.; Fornasari, L.; Levi, D.; Broggi, G.; Pelicci, G. CD133 is essential for glioblastoma stem cell maintenance. Stem Cells 2013, 31, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Aghajani, M.; Mansoori, B.; Mohammadi, A.; Asadzadeh, Z.; Baradaran, B. New emerging roles of CD133 in cancer stem cell: Signaling pathway and miRNA regulation. J. Cell. Physiol. 2019, 234, 21642–21661. [Google Scholar] [CrossRef] [PubMed]
- Gangemi, R.M.R.; Griffero, F.; Marubbi, D.; Perera, M.; Capra, M.C.; Malatesta, P.; Ravetti, G.L.; Zona, G.L.; Daga, A.; Corte, G. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells 2009, 27, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Mesci, P.; Bernatchez, J.A.; Gimple, R.C.; Wang, X.; Schafer, S.T.; Wettersten, H.I.; Beck, S.; Clark, A.E.; Wu, Q.; et al. Zika virus targets glioblastoma stem cells through a SOX2-integrin αvβ5 axis. Cell Stem Cell 2020, 26, 187–204. [Google Scholar] [CrossRef] [PubMed]
- Soni, P.; Qayoom, S.; Husain, N.; Kumar, P.; Chandra, A.; Ojha, B.K.; Gupta, R.K. CD24 and nanog expression in stem cells in glioblastoma: Correlation with response to chemoradiation and overall survival. Asian Pac. J. Cancer Prev. APJCP 2017, 18, 2215. [Google Scholar]
- Lukenda, A.; Dotlic, S.; Vukojevic, N.; Saric, B.; Vranic, S.; Zarkovic, K. Expression and prognostic value of putative cancer stem cell markers CD117 and CD15 in choroidal and ciliary body melanoma. J. Clin. Pathol. 2016, 69, 234–239. [Google Scholar] [CrossRef]
- Yin, A.H.; Miraglia, S.; Zanjani, E.D.; Almeida-Porada, G.; Ogawa, M.; Leary, A.G.; Olweus, J.; Kearney, J.; Buck, D.W. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood, J. Am. Soc. Hematol. 1997, 90, 5002–5012. [Google Scholar]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar]
- Vora, P.; Venugopal, C.; Salim, S.K.; Tatari, N.; Bakhshinyan, D.; Singh, M.; Seyfrid, M.; Upreti, D.; Rentas, S.; Wong, N.; et al. The rational development of CD133-targeting immunotherapies for glioblastoma. Cell Stem Cell 2020, 26, 832–844. [Google Scholar] [CrossRef]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef]
- Irollo, E.; Pirozzi, G. CD133: To be or not to be, is this the real question? Am. J. Transl. Res. 2013, 5, 563. [Google Scholar]
- Glumac, P.M.; LeBeau, A.M. The role of CD133 in cancer: A concise review. Clin. Transl. Med. 2018, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef] [PubMed]
- Darmanis, S.; Sloan, S.A.; Croote, D.; Mignardi, M.; Chernikova, S.; Samghababi, P.; Zhang, Y.; Neff, N.; Kowarsky, M.; Caneda, C.; et al. Single-cell RNA-seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 2017, 21, 1399–1410. [Google Scholar] [CrossRef] [PubMed]
- Picelli, S.; Faridani, O.R.; Björklund, Å.K.; Winberg, G.; Sagasser, S.; Sandberg, R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014, 9, 171–181. [Google Scholar] [CrossRef]
- Darmanis, S.; Sloan, S.A.; Zhang, Y.; Enge, M.; Caneda, C.; Shuer, L.M.; Hayden Gephart, M.G.; Barres, B.A.; Quake, S.R. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. USA 2015, 112, 7285–7290. [Google Scholar] [CrossRef]
- Andrews, S. Babraham Bioinformatics-FastQC a Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 1 January 2020).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Campbell, K.R.; Lun, A.T.; Wills, Q.F. Scater: Pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 2017, 33, 1179–1186. [Google Scholar] [CrossRef]
- Amezquita, R.A.; Lun, A.T.; Becht, E.; Carey, V.J.; Carpp, L.N.; Geistlinger, L.; Marini, F.; Rue-Albrecht, K.; Risso, D.; Soneson, C.; et al. Orchestrating single-cell analysis with Bioconductor. Nat. Methods 2020, 17, 137–145. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. InterJournal Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Wolf, F.A.; Angerer, P.; Theis, F.J. SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 2018, 19, 15. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g: Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling; R Package Version; 2019; Volume 1, Available online: https://bioconductor.org/packages/release/bioc/html/EnhancedVolcano.html (accessed on 1 January 2020).
- Couturier, C.P.; Ayyadhury, S.; Le, P.U.; Nadaf, J.; Monlong, J.; Riva, G.; Allache, R.; Baig, S.; Yan, X.; Bourgey, M.; et al. Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat. Commun. 2020, 11, 3406. [Google Scholar] [CrossRef]
- Pang, B.; Xu, J.; Hu, J.; Guo, F.; Wan, L.; Cheng, M.; Pang, L. Single-cell RNA-seq reveals the invasive trajectory and molecular cascades underlying glioblastoma progression. Mol. Oncol. 2019, 13, 2588–2603. [Google Scholar] [CrossRef]
- Kiraga, Ł.; Kucharzewska, P.; Paisey, S.; Cheda, Ł.; Domańska, A.; Rogulski, Z.; Rygiel, T.P.; Boffi, A.; Krol, M. Nuclear imaging for immune cell tracking in vivo–Comparison of various cell labeling methods and their application. Coord. Chem. Rev. 2021, 445, 214008. [Google Scholar] [CrossRef]
- Raman, V.; Van Dessel, N.; Hall, C.L.; Wetherby, V.E.; Whitney, S.A.; Kolewe, E.L.; Bloom, S.M.; Sharma, A.; Hardy, J.A.; Bollen, M.; et al. Intracellular delivery of protein drugs with an autonomously lysing bacterial system reduces tumor growth and metastases. Nat. Commun. 2021, 12, 6116. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Liu, H.C.; Chen, C.Y.; Liu, Y.T.; Chu, C.B.; Liang, D.C.; Shih, L.Y.; Lin, C.J. Cross-generation and cross-laboratory predictions of Affymetrix microarrays by rank-based methods. J. Biomed. Informatics 2008, 41, 570–579. [Google Scholar] [CrossRef]
- Lauria, M. Rank-based transcriptional signatures: A novel approach to diagnostic biomarker definition and analysis. Syst. Biomed. 2013, 1, 228–239. [Google Scholar] [CrossRef]
- Richard, M.; Decamps, C.; Chuffart, F.; Brambilla, E.; Rousseaux, S.; Khochbin, S.; Jost, D. PenDA, a rank-based method for personalized differential analysis: Application to lung cancer. PLoS Comput. Biol. 2020, 16, e1007869. [Google Scholar] [CrossRef] [PubMed]
- Lê Cao, K.A.; Rohart, F.; McHugh, L.; Korn, O.; Wells, C.A. YuGene: A simple approach to scale gene expression data derived from different platforms for integrated analyses. Genomics 2014, 103, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Vargo, A.H.; Gilbert, A.C. A rank-based marker selection method for high throughput scRNA-seq data. BMC Bioinform. 2020, 21, 477. [Google Scholar] [CrossRef] [PubMed]
- Wilfinger, W.W.; Miller, R.; Eghbalnia, H.R.; Mackey, K.; Chomczynski, P. Strategies for detecting and identifying biological signals amidst the variation commonly found in RNA sequencing data. BMC Genom. 2021, 22, 322. [Google Scholar] [CrossRef]
- Tilghman, J.; Wu, H.; Sang, Y.; Shi, X.; Guerrero-Cazares, H.; Quinones-Hinojosa, A.; Eberhart, C.G.; Laterra, J.; Ying, M. HMMR Maintains the Stemness and Tumorigenicity of Glioblastoma Stem-like CellsTargeting HMMR Inhibits Glioblastoma Stem Cells. Cancer Res. 2014, 74, 3168–3179. [Google Scholar] [CrossRef]
- Galatro, T.F.d.A.; Uno, M.; Oba-Shinjo, S.M.; Almeida, A.N.; Teixeira, M.J.; Rosemberg, S.; Marie, S.K.N. Differential expression of ID4 and its association with TP53 mutation, SOX2, SOX4 and OCT-4 expression levels. PLoS ONE 2013, 8, e61605. [Google Scholar] [CrossRef]
- Stevanovic, M.; Kovacevic-Grujicic, N.; Mojsin, M.; Milivojevic, M.; Drakulic, D. SOX transcription factors and glioma stem cells: Choosing between stemness and differentiation. World J. Stem Cells 2021, 13, 1417. [Google Scholar] [CrossRef]
- Tsang, S.M.; Oliemuller, E.; Howard, B.A. Regulatory roles for SOX11 in development, stem cells and cancer. Semin. Cancer Biol. 2020, 67, 3–11. [Google Scholar] [CrossRef]
- Wang, L.; Babikir, H.; Müller, S.; Yagnik, G.; Shamardani, K.; Catalan, F.; Kohanbash, G.; Alvarado, B.; Di Lullo, E.; Kriegstein, A.; et al. The Phenotypes of Proliferating Glioblastoma Cells Reside on a Single Axis of VariationA Draft Single-cell Atlas of Human Glioma. Cancer Discov. 2019, 9, 1708–1719. [Google Scholar] [CrossRef]
- Rheinbay, E.; Suvà, M.L.; Gillespie, S.M.; Wakimoto, H.; Patel, A.P.; Shahid, M.; Oksuz, O.; Rabkin, S.D.; Martuza, R.L.; Rivera, M.N.; et al. An aberrant transcription factor network essential for Wnt signaling and stem cell maintenance in glioblastoma. Cell Rep. 2013, 3, 1567–1579. [Google Scholar] [CrossRef]
- Bhaduri, A.; Di Lullo, E.; Jung, D.; Müller, S.; Crouch, E.E.; Espinosa, C.S.; Ozawa, T.; Alvarado, B.; Spatazza, J.; Cadwell, C.R.; et al. Outer radial glia-like cancer stem cells contribute to heterogeneity of glioblastoma. Cell Stem Cell 2020, 26, 48–63. [Google Scholar] [CrossRef] [PubMed]
- Ernst, A.; Hofmann, S.; Ahmadi, R.; Becker, N.; Korshunov, A.; Engel, F.; Hartmann, C.; Felsberg, J.; Sabel, M.; Peterziel, H.; et al. Genomic and expression profiling of glioblastoma stem cell–like spheroid cultures identifies novel tumor-relevant genes associated with survival. Clin. Cancer Res. 2009, 15, 6541–6550. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ping, Y.F.; Zhou, W.; He, Z.C.; Chen, C.; Bian, B.S.J.; Zhang, L.; Chen, L.; Lan, X.; Zhang, X.C.; et al. Tumour-associated macrophages secrete pleiotrophin to promote PTPRZ1 signalling in glioblastoma stem cells for tumour growth. Nat. Commun. 2017, 8, 15080. [Google Scholar] [CrossRef] [PubMed]
- Günther, H.; Schmidt, N.; Phillips, H.; Kemming, D.; Kharbanda, S.; Soriano, R.; Modrusan, Z.; Meissner, H.; Westphal, M.; Lamszus, K. Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene 2008, 27, 2897–2909. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Cui, P.; Lu, Y.; Zhang, X. Requirement of the transcription factor YB-1 for maintaining the stemness of cancer stem cells and reverting differentiated cancer cells into cancer stem cells. Stem Cell Res. Ther. 2019, 10, 233. [Google Scholar] [CrossRef]
- Verma, R.; Chen, X.; Xin, D.; Luo, Z.; Ogurek, S.; Xin, M.; Rao, R.; Berry, K.; Lu, Q.R. Olig1/2-expressing intermediate lineage progenitors are predisposed to PTEN/p53-loss-induced gliomagenesis and harbor specific therapeutic vulnerabilities. Cancer Res. 2023, CAN-22-1577. [Google Scholar] [CrossRef]
- Ng, K.F.; Chen, T.C.; Stacey, M.; Lin, H.H. Role of ADGRG1/GPR56 in tumor progression. Cells 2021, 10, 3352. [Google Scholar] [CrossRef]
- Shashidhar, S.; Lorente, G.; Nagavarapu, U.; Nelson, A.; Kuo, J.; Cummins, J.; Nikolich, K.; Urfer, R.; Foehr, E.D. GPR56 is a GPCR that is overexpressed in gliomas and functions in tumor cell adhesion. Oncogene 2005, 24, 1673–1682. [Google Scholar] [CrossRef]
- Lacore, M.G.; Delmas, C.; Nicaise, Y.; Kowalski-Chauvel, A.; Cohen-Jonathan-Moyal, E.; Seva, C. The Glycoprotein M6a Is Associated with Invasiveness and Radioresistance of Glioblastoma Stem Cells. Cells 2022, 11, 2128. [Google Scholar] [CrossRef]
- Cheng, J.X.; Liu, B.L.; Zhang, X. How powerful is CD133 as a cancer stem cell marker in brain tumors? Cancer Treat. Rev. 2009, 35, 403–408. [Google Scholar] [CrossRef]
- Yin, X.; Wu, Q.; Hao, Z.; Chen, L. Identification of novel prognostic targets in glioblastoma using bioinformatics analysis. BioMedical Eng. OnLine 2022, 21, 26. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Yin, W.; Zhu, H.; Tan, J.; Guo, Y.; Xin, Z.; Zhou, Q.; Cao, Y.; Wu, Z.; Kuang, Y.; et al. METTL7B is a novel prognostic biomarker of lower-grade glioma based on pan-cancer analysis. Cancer Cell Int. 2021, 21, 383. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, C.; Li, Y.; Wu, S.; Liu, W.; Lin, T.; Li, M.; Weng, Y.; Lin, W.; Qiu, S. Characterization of METTL7B to evaluate TME and predict prognosis by integrative analysis of multi-omics data in glioma. Front. Mol. Biosci. 2021, 8, 727481. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Luo, X.; Ding, Y.; Guo, S. Prognostic Potential of METTL7B in Glioma. Neuroimmunomodulation 2022, 29, 186–201. [Google Scholar] [CrossRef]
- Arora, M.; Kumari, S.; Singh, J.; Chopra, A.; Chauhan, S.S. Downregulation of brain enriched type 2 MAGEs is associated with immune infiltration and poor prognosis in glioma. Front. Oncol. 2020, 10, 573378. [Google Scholar] [CrossRef]
- Zhang, Q.M.; Shen, N.; Xie, S.; Bi, S.Q.; Luo, B.; Lin, Y.D.; Fu, J.; Zhou, S.F.; Luo, G.R.; Xie, X.X.; et al. MAGED4 expression in glioma and upregulation in glioma cell lines with 5-aza-2’-deoxycytidine treatment. Asian Pac. J. Cancer Prev. 2014, 15, 3495–3501. [Google Scholar] [CrossRef]
- Li, C.; Chen, Y.; Bai, P.; Wang, J.; Liu, Z.; Wang, T.; Cai, Q. LDHB may be a significant predictor of poor prognosis in osteosarcoma. Am. J. Transl. Res. 2016, 8, 4831. [Google Scholar]
- Du, Y.; Grandis, J.R. Receptor-type protein tyrosine phosphatases in cancer. Chin. J. Cancer 2015, 34, 61–69. [Google Scholar] [CrossRef]
- Wang, Z.C.; Gao, Q.; Shi, J.Y.; Guo, W.J.; Yang, L.X.; Liu, X.Y.; Liu, L.Z.; Ma, L.J.; Duan, M.; Zhao, Y.J.; et al. Protein tyrosine phosphatase receptor S acts as a metastatic suppressor in hepatocellular carcinoma by control of epithermal growth factor receptor–induced epithelial-mesenchymal transition. Hepatology 2015, 62, 1201–1214. [Google Scholar] [CrossRef]
- Lertpanprom, M.; Silsirivanit, A.; Tippayawat, P.; Proungvitaya, T.; Roytrakul, S.; Proungvitaya, S. High expression of protein tyrosine phosphatase receptor S (PTPRS) is an independent prognostic marker for cholangiocarcinoma. Front. Public Health 2022, 10, 835914. [Google Scholar] [CrossRef]
- Ashraf, W.; Ibrahim, A.; Alhosin, M.; Zaayter, L.; Ouararhni, K.; Papin, C.; Ahmad, T.; Hamiche, A.; Mély, Y.; Bronner, C.; et al. The epigenetic integrator UHRF1: On the road to become a universal biomarker for cancer. Oncotarget 2017, 8, 51946. [Google Scholar] [CrossRef]
- Unoki, M.; Kelly, J.; Neal, D.; Ponder, B.; Nakamura, Y.; Hamamoto, R. UHRF1 is a novel molecular marker for diagnosis and the prognosis of bladder cancer. Br. J. Cancer 2009, 101, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, H.; Tang, J.; Lin, Z.; Jiang, R.; Zhang, X.; Ji, J.; Wang, P.; Sun, B. The aberrant expression of MEG3 regulated by UHRF1 predicts the prognosis of hepatocellular carcinoma. Mol. Carcinog. 2016, 55, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Levallet, G.; Bergot, E.; Antoine, M.; Creveuil, C.; Santos, A.O.; Beau-Faller, M.; De Fraipont, F.; Brambilla, E.; Levallet, J.; Morin, F.; et al. High TUBB3 Expression, an Independent Prognostic Marker in Patients with Early Non–Small Cell Lung Cancer Treated by Preoperative Chemotherapy, Is Regulated by K-Ras Signaling PathwayK-Ras and TUBB3 in Early NSCLC. Mol. Cancer Ther. 2012, 11, 1203–1213. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, J.N.; Santoni-Rugiu, E.; Sørensen, J.B. Use of TUBB3 for patient stratification and prognosis in lung cancer. Lung Cancer Manag. 2015, 4, 97–110. [Google Scholar] [CrossRef]
- Sekino, Y.; Han, X.; Babasaki, T.; Miyamoto, S.; Kitano, H.; Kobayashi, G.; Goto, K.; Inoue, S.; Hayashi, T.; Teishima, J.; et al. TUBB3 is associated with high-grade histology, poor prognosis, p53 expression, and cancer stem cell markers in clear cell renal cell carcinoma. Oncology 2020, 98, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Ding, F.; Gao, J.; Huang, X.; Liu, W.; Wang, Y.; Liu, Q.; Xin, T. Molecular and clinical characterization of a novel prognostic and immunologic biomarker FAM111A in diffuse lower-grade glioma. Front. Oncol. 2020, 10, 573800. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Liu, J.; Shao, J.; Huang, C.; Dai, X.; Shen, Y.; Hou, W.; Shen, Y.; Yu, Y. MAGED4B Promotes Glioma Progression via Inactivation of the TNF-α-induced Apoptotic Pathway by Down-regulating TRIM27 Expression. Neurosci. Bull. 2022, 39, 273–291. [Google Scholar] [CrossRef]
- Barzegar Behrooz, A.; Syahir, A.; Ahmad, S. CD133: Beyond a cancer stem cell biomarker. J. Drug Target. 2019, 27, 257–269. [Google Scholar] [CrossRef]
- Lee, A.; Kessler, J.D.; Read, T.A.; Kaiser, C.; Corbeil, D.; Huttner, W.B.; Johnson, J.E.; Wechsler-Reya, R.J. Isolation of neural stem cells from the postnatal cerebellum. Nat. Neurosci. 2005, 8, 723–729. [Google Scholar] [CrossRef]
- Ellis, P.; Fagan, B.M.; Magness, S.T.; Hutton, S.; Taranova, O.; Hayashi, S.; McMahon, A.; Rao, M.; Pevny, L. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev. Neurosci. 2004, 26, 148–165. [Google Scholar] [CrossRef] [PubMed]
- Pruszak, J.; Ludwig, W.; Blak, A.; Alavian, K.; Isacson, O. CD15, CD24, and CD29 define a surface biomarker code for neural lineage differentiation of stem cells. Stem Cells 2009, 27, 2928–2940. [Google Scholar] [CrossRef] [PubMed]
- Ramalho-Santos, M.; Yoon, S.; Matsuzaki, Y.; Mulligan, R.C.; Melton, D.A. “Stemness”: Transcriptional profiling of embryonic and adult stem cells. Science 2002, 298, 597–600. [Google Scholar] [CrossRef] [PubMed]
- Katsetos, C.D.; Dráberová, E.; Legido, A.; Dumontet, C.; Dráber, P. Tubulin targets in the pathobiology and therapy of glioblastoma multiforme. I. class III β-tubulin. J. Cell. Physiol. 2009, 221, 505–513. [Google Scholar] [CrossRef] [PubMed]

| Markers | Cell Subtype | Properties | Methods Used for Validation | References |
|---|---|---|---|---|
| SOX4 | GSC | Stemness regulator, GSC signature marker, transcription factor (TF) highly expressed in embryonic, neural, or tumor stem cells | Transcriptome profiling, tumorigenesis in vivo | [37,38,48,49,50] |
| SOX11 | GSC | GSC signature marker, stemness regulator | Transcriptome profiling | [37,51] |
| ASCL1 | GSC | GSC signature marker | Transcriptome profiling, tumorigenesis in vivo and in vitro, genetic knock-down | [37,38,52,53] |
| PTPRZ1 | GSC | Tumor initiating GSC marker, invasive GSC marker, overexpressed in stem-like phenotype of GBM spheroid | Tumorigenesis in vivo, genetic knock-down, invasion assays, tumorigenesis in vitro, transcriptome profiling | [54,55,56] |
| BCAN | pGSC | pGSC (proneural) signature marker, overexpressed in stem-like phenotype of GBM spheroid | Tumorigenesis in vitro, transcriptome profiling | [52,55,57] |
| OLIG1 | GSC | Stemness regulator, GSC signature marker | Transcriptome profiling, tumorigenesis in vitro | [38,58,59] |
| GPR56 | GSC | overexpressed in stem-like phenotype of GBM spheroid, neural stem cell marker, cancer stem cell (CSC) marker | Tumorigenesis in vitro, transcriptome profiling | [55,60,61] |
| MAP2 | GSC | overexpressed in stem-like phenotype of GBM spheroid | Tumorigenesis in vitro, transcriptome profiling | [55] |
| GPM6A | GSC | GSC signature marker, invasive GSC marker | Tumorigenesis in vitro | [62] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Døssing, K.B.V.; Sloth, A.B.; He, X.; Rossing, M.; Kjaer, A. Quantitative Evaluation of Stem-like Markers of Human Glioblastoma Using Single-Cell RNA Sequencing Datasets. Cancers 2023, 15, 1557. https://doi.org/10.3390/cancers15051557
He Y, Døssing KBV, Sloth AB, He X, Rossing M, Kjaer A. Quantitative Evaluation of Stem-like Markers of Human Glioblastoma Using Single-Cell RNA Sequencing Datasets. Cancers. 2023; 15(5):1557. https://doi.org/10.3390/cancers15051557
Chicago/Turabian StyleHe, Yue, Kristina B. V. Døssing, Ane Beth Sloth, Xuening He, Maria Rossing, and Andreas Kjaer. 2023. "Quantitative Evaluation of Stem-like Markers of Human Glioblastoma Using Single-Cell RNA Sequencing Datasets" Cancers 15, no. 5: 1557. https://doi.org/10.3390/cancers15051557
APA StyleHe, Y., Døssing, K. B. V., Sloth, A. B., He, X., Rossing, M., & Kjaer, A. (2023). Quantitative Evaluation of Stem-like Markers of Human Glioblastoma Using Single-Cell RNA Sequencing Datasets. Cancers, 15(5), 1557. https://doi.org/10.3390/cancers15051557







