Epigenetic Silencing of LRP2 Is Associated with Dedifferentiation and Poor Survival in Multiple Solid Tumor Types
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue
2.2. Antibodies
2.3. Immunohistochemistry
2.4. Data Availability
2.5. Single-Cell RNA Sequencing Analysis
2.6. LRP2 Survival Analysis
2.7. LRP2 Methylation Analysis
2.8. Differential Gene Expression and Gene Set Enrichment Analysis
2.9. LRP2 Tumor Differentiation Analysis
2.10. Statistical Analysis and Data Visualization
3. Results
3.1. LRP2 Expression across Human Cancers
3.2. Tumor LRP2 Expression Is Restricted to Malignant Cells and Correlates with LRP2 Protein Levels
3.3. Confirmation of LRP2 Protein Expression in Luminal Invasive Breast Carcinoma
3.4. Low LRP2 Expression Is Associated with Poor Survival in Multiple Cancers
3.5. Low LRP2 Expression in Human Cancer Is Associated with First Intron CpG Methylation
3.6. LRP2 Silencing Is Associated with Tumor Dedifferentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Brisebarre, A.; Sifrim, A.; Fioramonti, M.; Revenco, T.; Boumahdi, S.; Van Keymeulen, A.; Brown, D.; Moers, V.; Lemaire, S.; et al. Identification of the tumour transition states occurring during EMT. Nature 2018, 556, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Brabletz, T.; Kalluri, R.; Nieto, M.A.; Weinberg, R.A. EMT in cancer. Nat. Rev. Cancer 2018, 18, 128–134. [Google Scholar] [CrossRef]
- McMahon, H.T.; Boucrot, E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev.. Mol. Cell Biol. 2011, 12, 517–533. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.E.; Padilla, B.E.; Hasdemir, B.; Cottrell, G.S.; Bunnett, N.W. Endosomes: A legitimate platform for the signaling train. Proc. Natl. Acad. Sci. USA 2009, 106, 17615–17622. [Google Scholar] [CrossRef]
- Khan, I.; Steeg, P.S. Endocytosis: A pivotal pathway for regulating metastasis. Br. J. Cancer 2021, 124, 66–75. [Google Scholar] [CrossRef]
- Raman, D.; Sai, J.; Hawkins, O.; Richmond, A. Adaptor protein2 (AP2) orchestrates CXCR2-mediated cell migration. Traffic 2014, 15, 451–469. [Google Scholar] [CrossRef]
- Chen, P.H.; Bendris, N.; Hsiao, Y.J.; Reis, C.R.; Mettlen, M.; Chen, H.Y.; Yu, S.L.; Schmid, S.L. Crosstalk between CLCb/Dyn1-Mediated Adaptive Clathrin-Mediated Endocytosis and Epidermal Growth Factor Receptor Signaling Increases Metastasis. Dev. Cell 2017, 40, 278–288.e275. [Google Scholar] [CrossRef]
- Reis, C.R.; Chen, P.H.; Srinivasan, S.; Aguet, F.; Mettlen, M.; Schmid, S.L. Crosstalk between Akt/GSK3beta signaling and dynamin-1 regulates clathrin-mediated endocytosis. EMBO J. 2015, 34, 2132–2146. [Google Scholar] [CrossRef]
- Christensen, E.I.; Birn, H. Megalin and cubilin: Multifunctional endocytic receptors. Nat. Rev. Mol. Cell Biol. 2002, 3, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Christensen, E.I.; Birn, H.; Storm, T.; Weyer, K.; Nielsen, R. Endocytic receptors in the renal proximal tubule. Physiology 2012, 27, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Perez Bay, A.E.; Schreiner, R.; Benedicto, I.; Paz Marzolo, M.; Banfelder, J.; Weinstein, A.M.; Rodriguez-Boulan, E.J. The fast-recycling receptor Megalin defines the apical recycling pathway of epithelial cells. Nat. Commun. 2016, 7, 11550. [Google Scholar] [CrossRef] [PubMed]
- Klassen, R.B.; Allen, P.L.; Batuman, V.; Crenshaw, K.; Hammond, T.G. Light chains are a ligand for megalin. J. Appl. Physiol. (1985) 2005, 98, 257–263. [Google Scholar] [CrossRef]
- Bryniarski, M.A.; Zhao, B.; Chaves, L.D.; Mikkelsen, J.H.; Yee, B.M.; Yacoub, R.; Shen, S.; Madsen, M.; Morris, M.E. Immunoglobulin G Is a Novel Substrate for the Endocytic Protein Megalin. AAPS J. 2021, 23, 40. [Google Scholar] [CrossRef]
- Nielsen, R.; Christensen, E.I.; Birn, H. Megalin and cubilin in proximal tubule protein reabsorption: From experimental models to human disease. Kidney Int. 2016, 89, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Donnai, D.; Barrow, M. Diaphragmatic hernia, exomphalos, absent corpus callosum, hypertelorism, myopia, and sensorineural deafness: A newly recognized autosomal recessive disorder? Am. J. Med. Genet. 1993, 47, 679–682. [Google Scholar] [CrossRef]
- Kantarci, S.; Al-Gazali, L.; Hill, R.S.; Donnai, D.; Black, G.C.; Bieth, E.; Chassaing, N.; Lacombe, D.; Devriendt, K.; Teebi, A.; et al. Mutations in LRP2, which encodes the multiligand receptor megalin, cause Donnai-Barrow and facio-oculo-acoustico-renal syndromes. Nat. Genet. 2007, 39, 957–959. [Google Scholar] [CrossRef]
- Assemat, E.; Chatelet, F.; Chandellier, J.; Commo, F.; Cases, O.; Verroust, P.; Kozyraki, R. Overlapping expression patterns of the multiligand endocytic receptors cubilin and megalin in the CNS, sensory organs and developing epithelia of the rodent embryo. Gene Expr. Patterns 2005, 6, 69–78. [Google Scholar] [CrossRef]
- Fisher, C.E.; Howie, S.E. The role of megalin (LRP-2/Gp330) during development. Dev. Biol. 2006, 296, 279–297. [Google Scholar] [CrossRef]
- Willnow, T.E.; Hilpert, J.; Armstrong, S.A.; Rohlmann, A.; Hammer, R.E.; Burns, D.K.; Herz, J. Defective forebrain development in mice lacking gp330/megalin. Proc. Natl. Acad. Sci. USA 1996, 93, 8460–8464. [Google Scholar] [CrossRef] [PubMed]
- Christ, A.; Herzog, K.; Willnow, T.E. LRP2, an auxiliary receptor that controls sonic hedgehog signaling in development and disease. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2016, 245, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Spoelgen, R.; Hammes, A.; Anzenberger, U.; Zechner, D.; Andersen, O.M.; Jerchow, B.; Willnow, T.E. LRP2/megalin is required for patterning of the ventral telencephalon. Development 2005, 132, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, I.; Lee, C.; Schuster, E.; Hoeren, J.; Trivigno, V.; Riedel, L.; Gorne, J.; Wallingford, J.B.; Hammes, A.; Feistel, K. Neural tube closure requires the endocytic receptor Lrp2 and its functional interaction with intracellular scaffolds. Development 2021, 148, dev195008. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Poon, C.; Chin, D.; Milkowski, S.; Lu, V.; Hallows, K.R.; Chung, E.J. Design and in vivo characterization of kidney-targeting multimodal micelles for renal drug delivery. Nano Res. 2018, 11, 5584–5595. [Google Scholar] [CrossRef]
- Ordikhani, F.; Kasinath, V.; Uehara, M.; Akbarzadeh, A.; Yilmam, O.A.; Dai, L.; Aksu, H.; Jung, S.; Jiang, L.; Li, X.; et al. Selective Trafficking of Light Chain-Conjugated Nanoparticles to the Kidney and Renal Cell Carcinoma. Nano Today 2020, 35, 100990. [Google Scholar] [CrossRef]
- Chlon, T.M.; Taffany, D.A.; Welsh, J.; Rowling, M.J. Retinoids modulate expression of the endocytic partners megalin, cubilin, and disabled-2 and uptake of vitamin D-binding protein in human mammary cells. J. Nutr. 2008, 138, 1323–1328. [Google Scholar] [CrossRef]
- Rowling, M.J.; Kemmis, C.M.; Taffany, D.A.; Welsh, J. Megalin-mediated endocytosis of vitamin D binding protein correlates with 25-hydroxycholecalciferol actions in human mammary cells. J. Nutr. 2006, 136, 2754–2759. [Google Scholar] [CrossRef]
- Andersen, R.K.; Hammer, K.; Hager, H.; Christensen, J.N.; Ludvigsen, M.; Honore, B.; Thomsen, M.B.; Madsen, M. Melanoma tumors frequently acquire LRP2/megalin expression, which modulates melanoma cell proliferation and survival rates. Pigment. Cell Melanoma Res. 2015, 28, 267–280. [Google Scholar] [CrossRef]
- Jensen, L.L.; Andersen, R.K.; Hager, H.; Madsen, M. Lack of megalin expression in adult human terminal ileum suggests megalin-independent cubilin/amnionless activity during vitamin B12 absorption. Physiol. Rep. 2014, 2, e12086. [Google Scholar] [CrossRef]
- Storm, T.; Christensen, E.I.; Christensen, J.N.; Kjaergaard, T.; Uldbjerg, N.; Larsen, A.; Honore, B.; Madsen, M. Megalin Is Predominantly Observed in Vesicular Structures in First and Third Trimester Cytotrophoblasts of the Human Placenta. J. Histochem. Cytochem. 2016, 64, 769–784. [Google Scholar] [CrossRef] [PubMed]
- Storm, T.; Tranebjaerg, L.; Frykholm, C.; Birn, H.; Verroust, P.J.; Neveus, T.; Sundelin, B.; Hertz, J.M.; Holmstrom, G.; Ericson, K.; et al. Renal phenotypic investigations of megalin-deficient patients: Novel insights into tubular proteinuria and albumin filtration. Nephrol. Dial. Transplant. 2013, 28, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Christensen, E.I.; Nielsen, S.; Moestrup, S.K.; Borre, C.; Maunsbach, A.B.; de Heer, E.; Ronco, P.; Hammond, T.G.; Verroust, P. Segmental distribution of the endocytosis receptor gp330 in renal proximal tubules. Eur. J. Cell. Biol. 1995, 66, 349–364. [Google Scholar] [PubMed]
- Vivian, J.; Rao, A.A.; Nothaft, F.A.; Ketchum, C.; Armstrong, J.; Novak, A.; Pfeil, J.; Narkizian, J.; Deran, A.D.; Musselman-Brown, A.; et al. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 2017, 35, 314–316. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell. 2018, 173, 400–416.e411. [Google Scholar] [CrossRef]
- Bi, K.; He, M.X.; Bakouny, Z.; Kanodia, A.; Napolitano, S.; Wu, J.; Grimaldi, G.; Braun, D.A.; Cuoco, M.S.; Mayorga, A.; et al. Tumor and immune reprogramming during immunotherapy in advanced renal cell carcinoma. Cancer Cell 2021, 39, 649–661.e645. [Google Scholar] [CrossRef]
- Wu, S.Z.; Al-Eryani, G.; Roden, D.L.; Junankar, S.; Harvey, K.; Andersson, A.; Thennavan, A.; Wang, C.; Torpy, J.R.; Bartonicek, N.; et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 2021, 53, 1334–1347. [Google Scholar] [CrossRef]
- Lindgren, C.M.; Adams, D.W.; Kimball, B.; Boekweg, H.; Tayler, S.; Pugh, S.L.; Payne, S.H. Simplified and Unified Access to Cancer Proteogenomic Data. J. Proteome Res. 2021, 20, 1902–1910. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Bott, M.; Brevet, M.; Taylor, B.S.; Shimizu, S.; Ito, T.; Wang, L.; Creaney, J.; Lake, R.A.; Zakowski, M.F.; Reva, B.; et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat. Genet. 2011, 43, 668–672. [Google Scholar] [CrossRef] [PubMed]
- Gordon, G.J.; Rockwell, G.N.; Jensen, R.V.; Rheinwald, J.G.; Glickman, J.N.; Aronson, J.P.; Pottorf, B.J.; Nitz, M.D.; Richards, W.G.; Sugarbaker, D.J.; et al. Identification of novel candidate oncogenes and tumor suppressors in malignant pleural mesothelioma using large-scale transcriptional profiling. Am. J. Pathol. 2005, 166, 1827–1840. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell. 2021, 184, 3573–3587.e3529. [Google Scholar] [CrossRef]
- Gyorffy, B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput. Struct. Biotechnol. J. 2021, 19, 4101–4109. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014, 159, 676–690. [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Kassambara, A. ggpubr: ‘ggplot2’ Based Publication Ready Plots. 2020. Available online: https://cran.r-project.org/web/packages/ggpubr/index.html (accessed on 10 January 2023).
- Zheng, G.; Bachinsky, D.R.; Stamenkovic, I.; Strickland, D.K.; Brown, D.; Andres, G.; McCluskey, R.T. Organ distribution in rats of two members of the low-density lipoprotein receptor gene family, gp330 and LRP/alpha 2MR, and the receptor-associated protein (RAP). J. Histochem. Cytochem. 1994, 42, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.T.; Wang, L.; Pasinetti, G.M.; Finch, C.E.; Zlokovic, B.V. Glycoprotein 330/megalin (LRP-2) has low prevalence as mRNA and protein in brain microvessels and choroid plexus. Exp. Neurol. 1999, 157, 194–201. [Google Scholar] [CrossRef]
- Gajera, C.R.; Emich, H.; Lioubinski, O.; Christ, A.; Beckervordersandforth-Bonk, R.; Yoshikawa, K.; Bachmann, S.; Christensen, E.I.; Gotz, M.; Kempermann, G.; et al. LRP2 in ependymal cells regulates BMP signaling in the adult neurogenic niche. J. Cell Sci. 2010, 123, 1922–1930. [Google Scholar] [CrossRef] [PubMed]
- Wicher, G.; Larsson, M.; Fex Svenningsen, A.; Gyllencreutz, E.; Rask, L.; Aldskogius, H. Low density lipoprotein receptor-related protein-2/megalin is expressed in oligodendrocytes in the mouse spinal cord white matter. J. Neurosci. Res. 2006, 83, 864–873. [Google Scholar] [CrossRef]
- Wicher, G.; Larsson, M.; Rask, L.; Aldskogius, H. Low-density lipoprotein receptor-related protein (LRP)-2/megalin is transiently expressed in a subpopulation of neural progenitors in the embryonic mouse spinal cord. J. Comp. Neurol. 2005, 492, 123–131. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, F.; Chandrashekar, D.S.; Varambally, S.; Creighton, C.J. Proteogenomic characterization of 2002 human cancers reveals pan-cancer molecular subtypes and associated pathways. Nat. Commun. 2022, 13, 2669. [Google Scholar] [CrossRef]
- Krug, K.; Jaehnig, E.J.; Satpathy, S.; Blumenberg, L.; Karpova, A.; Anurag, M.; Miles, G.; Mertins, P.; Geffen, Y.; Tang, L.C.; et al. Proteogenomic Landscape of Breast Cancer Tumorigenesis and Targeted Therapy. Cell. 2020, 183, 1436–1456.e1431. [Google Scholar] [CrossRef]
- Clark, D.J.; Dhanasekaran, S.M.; Petralia, F.; Pan, J.; Song, X.; Hu, Y.; da Veiga Leprevost, F.; Reva, B.; Lih, T.M.; Chang, H.Y.; et al. Integrated Proteogenomic Characterization of Clear Cell Renal Cell Carcinoma. Cell. 2019, 179, 964–983.e931. [Google Scholar] [CrossRef]
- Zhao, H.; Ljungberg, B.; Grankvist, K.; Rasmuson, T.; Tibshirani, R.; Brooks, J.D. Gene expression profiling predicts survival in conventional renal cell carcinoma. PLoS Med. 2006, 3, e13. [Google Scholar] [CrossRef]
- Bibikova, M.; Barnes, B.; Tsan, C.; Ho, V.; Klotzle, B.; Le, J.M.; Delano, D.; Zhang, L.; Schroth, G.P.; Gunderson, K.L.; et al. High density DNA methylation array with single CpG site resolution. Genomics 2011, 98, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadi, D.; Esteve-Codina, A.; Piferrer, F. Consistent inverse correlation between DNA methylation of the first intron and gene expression across tissues and species. Epigenet. Chromatin 2018, 11, 37. [Google Scholar] [CrossRef] [PubMed]
- Brenet, F.; Moh, M.; Funk, P.; Feierstein, E.; Viale, A.J.; Socci, N.D.; Scandura, J.M. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS ONE 2011, 6, e14524. [Google Scholar] [CrossRef]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]
- Prat, A.; Parker, J.S.; Karginova, O.; Fan, C.; Livasy, C.; Herschkowitz, J.I.; He, X.; Perou, C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010, 12, R68. [Google Scholar] [CrossRef] [PubMed]
- Hmeljak, J.; Sanchez-Vega, F.; Hoadley, K.A.; Shih, J.; Stewart, C.; Heiman, D.; Tarpey, P.; Danilova, L.; Drill, E.; Gibb, E.A.; et al. Integrative Molecular Characterization of Malignant Pleural Mesothelioma. Cancer Discov. 2018, 8, 1548–1565. [Google Scholar] [CrossRef]
- Park, S.G.; Hannenhalli, S.; Choi, S.S. Conservation in first introns is positively associated with the number of exons within genes and the presence of regulatory epigenetic signals. BMC Genom. 2014, 15, 526. [Google Scholar] [CrossRef] [PubMed]
- Mahadevappa, R.; Nielsen, R.; Christensen, E.I.; Birn, H. Megalin in acute kidney injury: Foe and friend. Am. J. Physiol. Ren. Physiol. 2014, 306, F147–F154. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, A.; Theilig, F.; Schweda, F.; Hocherl, K. Acute endotoxemia in mice induces downregulation of megalin and cubilin in the kidney. Kidney Int. 2012, 82, 53–59. [Google Scholar] [CrossRef]
- Kim, H.J.; Moradi, H.; Yuan, J.; Norris, K.; Vaziri, N.D. Renal mass reduction results in accumulation of lipids and dysregulation of lipid regulatory proteins in the remnant kidney. Am. J. Physiol. Ren. Physiol. 2009, 296, F1297–F1306. [Google Scholar] [CrossRef]
- Yao, S.; Kwan, M.L.; Ergas, I.J.; Roh, J.M.; Cheng, T.D.; Hong, C.C.; McCann, S.E.; Tang, L.; Davis, W.; Liu, S.; et al. Association of Serum Level of Vitamin D at Diagnosis With Breast Cancer Survival: A Case-Cohort Analysis in the Pathways Study. JAMA Oncol. 2017, 3, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Rose, A.A.; Elser, C.; Ennis, M.; Goodwin, P.J. Blood levels of vitamin D and early stage breast cancer prognosis: A systematic review and meta-analysis. Breast Cancer Res. Treat. 2013, 141, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Huss, L.; Butt, S.T.; Borgquist, S.; Elebro, K.; Sandsveden, M.; Rosendahl, A.; Manjer, J. Vitamin D receptor expression in invasive breast tumors and breast cancer survival. Breast Cancer Res. 2019, 21, 84. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.; Kumbrink, J.; Mayr, D.; Seiler, A.; Hagemann, F.; Degenhardt, T.; Sagebiel, S.; Würstlein, R.; Kates, R.; Harbeck, N.; et al. Molecular Prognostic Factors for Distant Metastases in Premenopausal Patients with HR+/HER2- Early Breast Cancer. J. Pers. Med. 2021, 11, 835. [Google Scholar] [CrossRef] [PubMed]


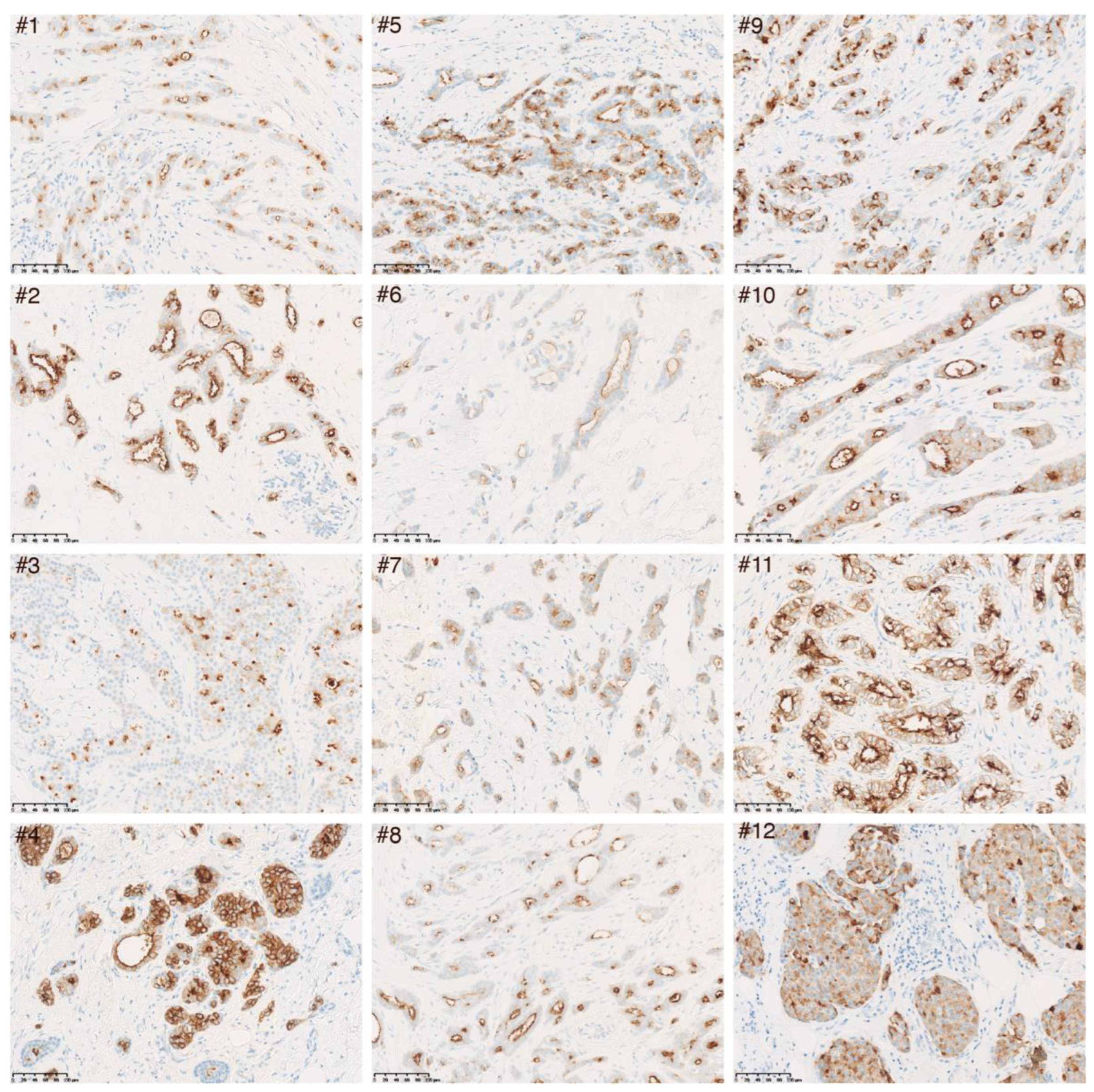



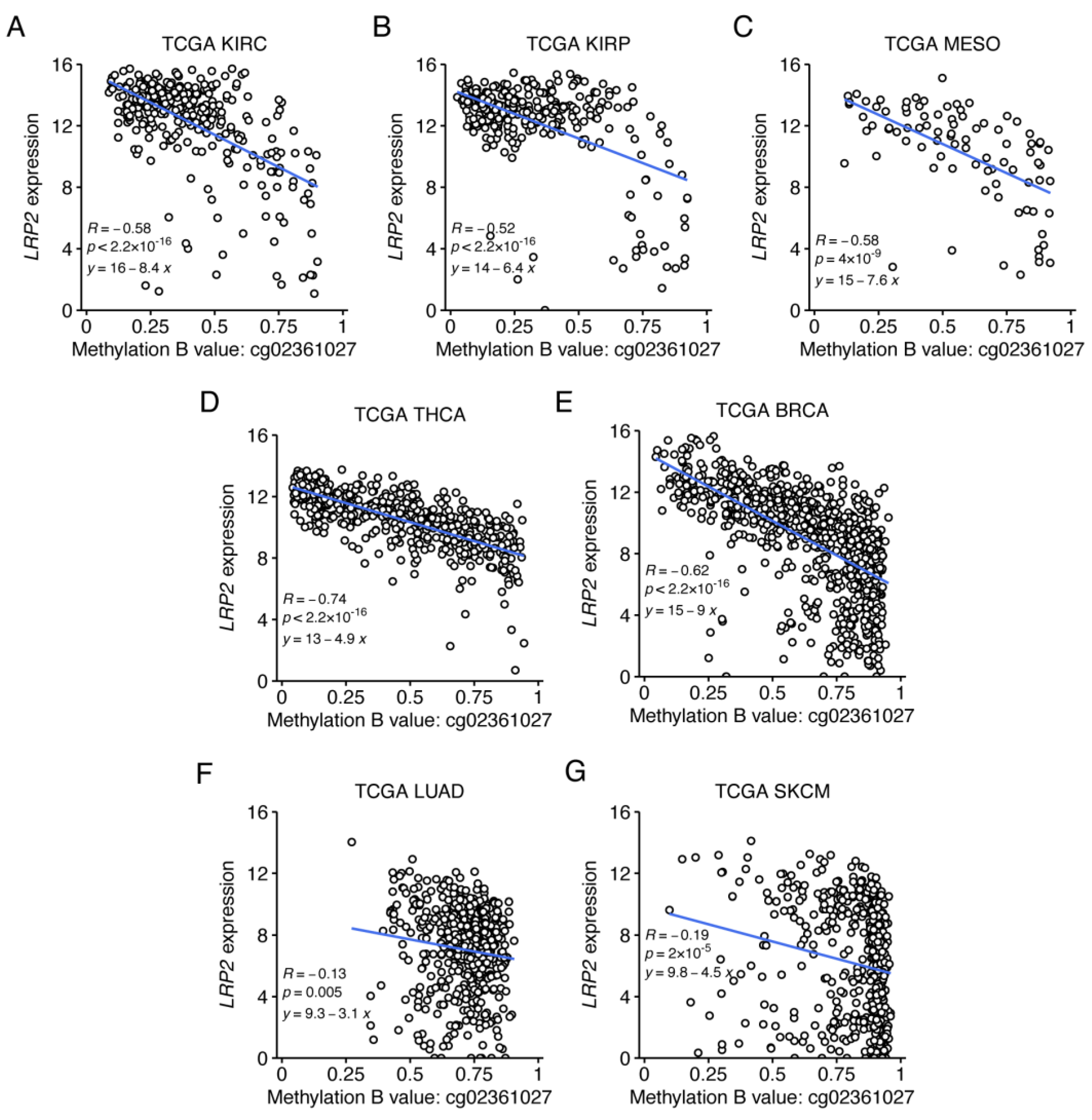
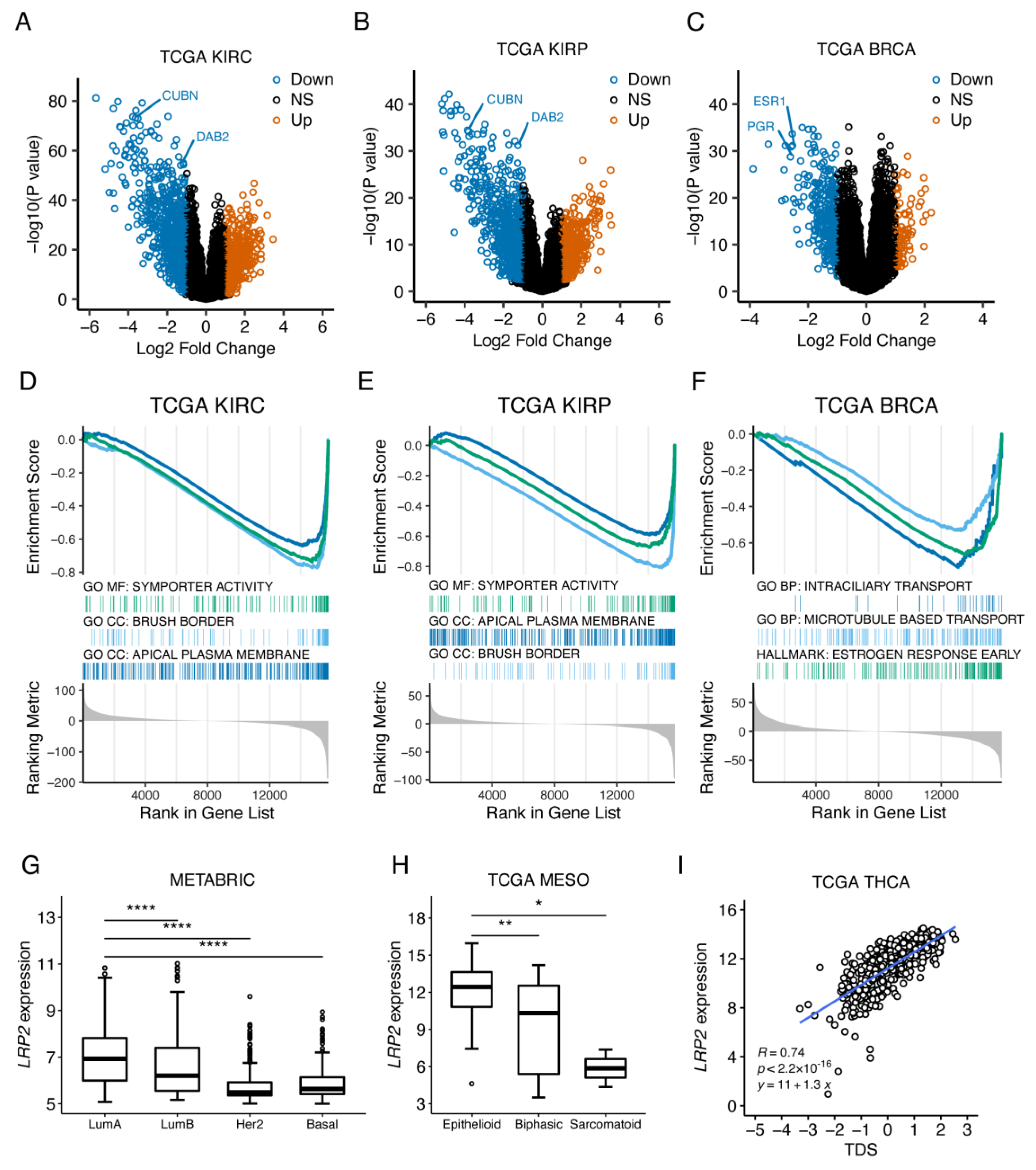
| CpG ID | Chr | CpG Position (Start) | Strand | Location Relative to CpG Island | Location Relative to Gene | Pearson r | p Value |
|---|---|---|---|---|---|---|---|
| cg08580631 | 2 | 169363877 | - | N_Shore | TSS1500 | −0.29 | 4.92 × 10−61 |
| cg01329010 | 2 | 169363731 | + | N_Shore | TSS1500 | −0.28 | 2.49 × 10−57 |
| cg25554917 | 2 | 169363618 | + | S_Shore | TSS1500 | −0.33 | 1.91 × 10−79 |
| cg03019033 | 2 | 169363552 | - | S_Shore | TSS1500 | −0.30 | 1 × 10−63 |
| cg02714065 | 2 | 169362894 | + | Island | TSS1500 | −0.30 | 4.53 × 10−65 |
| cg21645864 | 2 | 169362508 | + | Island | Exon 1 | −0.36 | 2.48 × 10−95 |
| cg13436799 | 2 | 169362486 | + | Island | Exon 1 | −0.34 | 8.43 × 10−83 |
| cg05660179 | 2 | 169362179 | + | Island | Intron 1-2 | −0.38 | <×10−100 |
| cg11403874 | 2 | 169362083 | + | Island | Intron 1-2 | −0.41 | <1×10−100 |
| cg00726174 | 2 | 169361839 | - | Island | Intron 1-2 | −0.33 | 5.3 × 10−78 |
| cg01400477 | 2 | 169361747 | + | N_Shore | Intron 1-2 | −0.42 | <1 × 10−100 |
| cg02361027 | 2 | 169360890 | + | N_Shore | Intron 1-2 | −0.65 | <1 × 10−100 |
| cg03328571 | 2 | 169358292 | - | N_Shelf | Intron 1-2 | 0.13 | 4.95 × 10−14 |
| cg23238677 | 2 | 169300823 | + | OpenSea | Intron 4-5 | 0.38 | <1 × 10−100 |
| cg07069090 | 2 | 169292323 | - | OpenSea | Exon 7 | 0.31 | 2.10 × 10−79 |
| cg05336056 | 2 | 169285877 | + | OpenSea | Intron 9-10 | −0.03 | 3.25 × 10−2 |
| cg19962304 | 2 | 169278848 | + | OpenSea | Intron 12-13 | 0.22 | 9.24 × 10−34 |
| cg26821433 | 2 | 169246977 | + | OpenSea | Exon 21 | 0.35 | 1.63 × 10−90 |
| cg08645980 | 2 | 169128218 | + | OpenSea | Exon 79 | 0.32 | 4.27 × 10−75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rasmussen, M.Q.; Tindbæk, G.; Nielsen, M.M.; Merrild, C.; Steiniche, T.; Pedersen, J.S.; Moestrup, S.K.; Degn, S.E.; Madsen, M. Epigenetic Silencing of LRP2 Is Associated with Dedifferentiation and Poor Survival in Multiple Solid Tumor Types. Cancers 2023, 15, 1830. https://doi.org/10.3390/cancers15061830
Rasmussen MQ, Tindbæk G, Nielsen MM, Merrild C, Steiniche T, Pedersen JS, Moestrup SK, Degn SE, Madsen M. Epigenetic Silencing of LRP2 Is Associated with Dedifferentiation and Poor Survival in Multiple Solid Tumor Types. Cancers. 2023; 15(6):1830. https://doi.org/10.3390/cancers15061830
Chicago/Turabian StyleRasmussen, Martin Q., Gitte Tindbæk, Morten Muhlig Nielsen, Camilla Merrild, Torben Steiniche, Jakob Skou Pedersen, Søren K. Moestrup, Søren E. Degn, and Mette Madsen. 2023. "Epigenetic Silencing of LRP2 Is Associated with Dedifferentiation and Poor Survival in Multiple Solid Tumor Types" Cancers 15, no. 6: 1830. https://doi.org/10.3390/cancers15061830
APA StyleRasmussen, M. Q., Tindbæk, G., Nielsen, M. M., Merrild, C., Steiniche, T., Pedersen, J. S., Moestrup, S. K., Degn, S. E., & Madsen, M. (2023). Epigenetic Silencing of LRP2 Is Associated with Dedifferentiation and Poor Survival in Multiple Solid Tumor Types. Cancers, 15(6), 1830. https://doi.org/10.3390/cancers15061830






