CCR7 Mediates Cell Invasion and Migration in Extrahepatic Cholangiocarcinoma by Inducing Epithelial–Mesenchymal Transition
Abstract
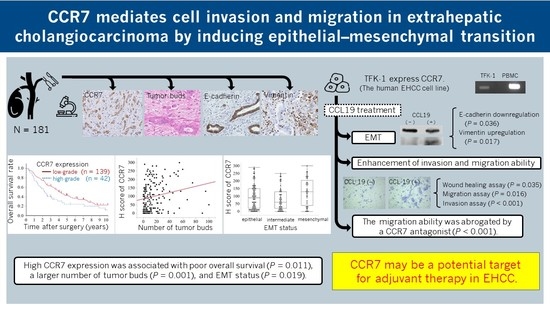
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Tissue Preparation and Histopathology
2.3. Tissue Microarray
2.4. Immunohistochemistry
2.5. Assessment of Tumor Buds
2.6. IHC Scoring in TMA
2.7. EMT Status in IHC Examination
2.8. Cell Culture
2.9. RNA Extraction and Reverse Transcription PCR
2.10. Western Blot Analysis
2.11. Wound Healing Assay
2.12. Cell Proliferation Assay
2.13. Migration and Invasion Assay
2.14. Statistical Analyses
3. Results
3.1. CCR7 Expression Was Observed in PHCC Tumor Tissues
3.2. Clinicopathological Features and Overall Survival Were Associated with CCR7 Expression
3.3. High CCR7 Expression Was Associated with EMT
3.4. CCR7 Expression Was Observed in TFK-1 Cells
3.5. CCR7/CCL19 Axis Affects the Expression of EMT-Related Proteins
3.6. CCL19 Promotes Lateral Migration of TFK-1 Cells
3.7. CCL19 Did Not Affect Cell Proliferation
3.8. CCL19 Promotes the Migration and Invasion of TFK-1 Cells
3.9. CCR7 Antagonist Inhibits CCL19-Mediated Migration of TFK-1 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Amin, M.B.; Edge, S.; Greene, F.; Byrd, D.R.; Washington, M.K.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; et al. AJCC Cancer Staging Manual, 8th ed.; Springer: New York, NY, USA, 2016. [Google Scholar]
- DeOliveira, M.L.; Cunningham, S.C.; Cameron, J.L.; Kamangar, F.; Winter, J.M.; Lillemoe, K.D.; Choti, M.A.; Yeo, C.J.; Schulick, R.D. Cholangiocarcinoma: Thirty-one-year experience with 564 patients at a single institution. Ann. Surg. 2007, 245, 755–762. [Google Scholar] [CrossRef]
- van der Gaag, N.A.; Kloek, J.J.; de Bakker, J.K.; Musters, B.; Geskus, R.B.; Busch, O.R.C.; Bosma, A.; Gouma, D.J.; van Gulik, T.M. Survival analysis and prognostic nomogram for patients undergoing resection of extrahepatic cholangiocarcinoma. Ann. Oncol. 2012, 23, 2642–2649. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, Y.; Okamura, K.; Tsuchikawa, T.; Nakamura, T.; Noji, T.; Asano, T.; Matsui, A.; Tanaka, K.; Murakami, S.; Ebihara, Y.; et al. Time to recurrence after surgical resection and survival after recurrence among patients with perihilar and distal cholangiocarcinomas. Ann. Surg. Oncol. 2020, 27, 4171–4180. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Xu, E.; Liu, H.; Wan, L.; Lai, M. Epithelial-mesenchymal transition in colorectal cancer metastasis: A system review. Pathol. Res. Pract. 2015, 211, 557–569. [Google Scholar] [CrossRef]
- Sung, W.J.; Kim, H.; Park, K.K. The biological role of epithelial-mesenchymal transition in lung cancer (review). Oncol. Rep. 2016, 36, 1199–1206. [Google Scholar] [CrossRef] [Green Version]
- Thiery, J.P.; Sleeman, J.P. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131–142. [Google Scholar] [CrossRef]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef]
- Müller, A.; Homey, B.; Soto, H.; Ge, N.; Catron, D.; Buchanan, M.E.; McClanahan, T.; Murphy, E.; Yuan, W.; Wagner, S.N.; et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410, 50–56. [Google Scholar] [CrossRef]
- Willimann, K.; Legler, D.F.; Loetscher, M.; Roos, R.S.; Delgado, M.B.; Clark-Lewis, I.; Baggiolini, M.; Moser, B. The chemokine Slc is expressed in T cell areas of lymph nodes and mucosal lymphoid tissues and attracts activated T cells via Ccr7. Eur. J. Immunol. 1998, 28, 2025–2034. [Google Scholar] [CrossRef]
- Ott, T.R.; Pahuja, A.; Nickolls, S.A.; Alleva, D.G.; Struthers, R.S. Identification of Cc chemokine Receptor 7 residues important for receptor activation. J. Biol. Chem. 2004, 279, 42383–42392. [Google Scholar] [CrossRef] [Green Version]
- Dieu, M.C.; Vanbervliet, B.; Vicari, A.; Bridon, J.M.; Oldham, E.; Aït-Yahia, S.; Brière, F.; Zlotnik, A.; Lebecque, S.; Caux, C. Selective recruitment of immature and mature dendritic cells by distinct chemokines expressed in different anatomic sites. J. Exp. Med. 1998, 188, 373–386. [Google Scholar] [CrossRef] [Green Version]
- Hirao, M.; Onai, N.; Hiroishi, K.; Watkins, S.C.; Matsushima, K.; Robbins, P.D.; Lotze, M.T.; Tahara, H. Cc chemokine Receptor-7 on dendritic cells is induced after interaction with apoptotic tumor cells: Critical role in migration from the tumor site to draining lymph nodes. Cancer Res. 2000, 60, 2209–2217. [Google Scholar] [PubMed]
- Xu, Y.; Liu, L.; Qiu, X.; Jiang, L.; Huang, B.; Li, H.; Li, Z.; Luo, W.; Wang, E. CCL21/Ccr7 promotes G2/M phase progression via the erk pathway in human non-small cell lung cancer cells. PLoS ONE 2011, 6, e21119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mo, M.; Zhou, M.; Wang, L.; Qi, L.; Zhou, K.; Liu, L.F.; Chen, Z.; Zu, X.B. CCL21/Ccr7 enhances the proliferation, migration, and invasion of human bladder cancer T24 cells. PLoS ONE 2015, 10, e0119506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, B.; Cui, K.; Wang, C.L.; Wang, A.L.; Zhang, B.; Zhou, W.Y.; Zhao, W.H.; Li, S. The chemotactic interaction between CCL21 and its receptor, Ccr7, facilitates the progression of pancreatic cancer via induction of angiogenesis and lymphangiogenesis. J. Hepatobiliary Pancreat. Sci. 2011, 18, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.Y.; Safdar, J.; Li, Z.N.; Fang, Q.G.; Zhang, X.; Xu, Z.F.; Sun, C.F. Ccr7 regulates cell migration and invasion through Jak2/Stat3 in metastatic squamous cell carcinoma of the head and neck. BioMed Res. Int. 2014, 2014, 415375. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.; Zhou, M.; Qiu, W.; Ye, J.; Feng, Q. Ccr7 mediates human breast cancer cell invasion, migration by inducing epithelial-mesenchymal transition and suppressing apoptosis through Akt pathway. Cancer Med. 2017, 6, 1062–1071. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, Y.; Yang, Y. Ccr7 pathway induces epithelial-mesenchymal transition through up-regulation of snail signaling in gastric cancer. Med. Oncol. 2015, 32, 467. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, D.; Li, Y.; Liu, Y.; Xie, X.; Wu, Y.; Zhou, Y.; Ren, J.; Zhang, J.; Zhu, H.; et al. CCL21/Ccr7 axis contributed to CD133+ pancreatic cancer stem-like cell metastasis via emt and erk/NF-κB pathway. PLoS ONE 2016, 11, e0158529. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Gao, L.; Li, S.; Qin, J.; Chen, L.; Liu, X.; Xu, P.; Wang, F.; Xiao, H.; Zhou, S.; et al. Ccr7 enhances Tgf-Beta1-Induced epithelial-mesenchymal transition and is associated with lymph node metastasis and poor overall survival in gastric cancer. Oncotarget 2015, 6, 24348–24360. [Google Scholar] [CrossRef] [Green Version]
- Irino, T.; Takeuchi, H.; Matsuda, S.; Saikawa, Y.; Kawakubo, H.; Wada, N.; Takahashi, T.; Nakamura, R.; Fukuda, K.; Omori, T.; et al. Cc-chemokine receptor Ccr7: A key molecule for lymph node metastasis in esophageal squamous cell carcinoma. BMC Cancer 2014, 14, 291. [Google Scholar] [CrossRef]
- Nakata, B.; Fukunaga, S.; Noda, E.; Amano, R.; Yamada, N.; Hirakawa, K. Chemokine receptor Ccr7 expression correlates with lymph node metastasis in pancreatic cancer. Oncology 2008, 74, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Huang, F.; Li, X.; Chen, Z.; Feng, D.; Jiang, H.; Chen, W.; Zhang, X. CCL21/Ccr7 interaction promotes cellular migration and invasion via modulation of the Mek/ERK1/2 signaling pathway and correlates with lymphatic metastatic spread and poor prognosis in urinary bladder cancer. Int. J. Oncol. 2017, 51, 75–90. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Liu, L.; Xiong, Y.; Bai, Q.; Wang, J.; Xi, W.; Qu, Y.; Xu, J.; Guo, J. Prognostic value of Cc-chemokine receptor seven expression in patients with metastatic renal cell carcinoma treated with tyrosine kinase inhibitor. BMC Cancer 2017, 17, 70. [Google Scholar] [CrossRef] [Green Version]
- Lugli, A.; Kirsch, R.; Ajioka, Y.; Bosman, F.; Cathomas, G.; Dawson, H.; El Zimaity, H.; Fléjou, J.F.; Hansen, T.P.; Hartmann, A.; et al. Recommendations for reporting tumor budding in colorectal cancer based on the International Tumor Budding Consensus Conference (Itbcc) 2016. Mod. Pathol. 2017, 30, 1299–1311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueno, H.; Murphy, J.; Jass, J.R.; Mochizuki, H.; Talbot, I.C. Tumour ‘budding’ as an index to estimate the potential of aggressiveness in rectal cancer. Histopathology 2002, 40, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Ueno, H.; Mochizuki, H.; Hashiguchi, Y.; Shimazaki, H.; Aida, S.; Hase, K.; Matsukuma, S.; Kanai, T.; Kurihara, H.; Ozawa, K.; et al. Risk factors for an adverse outcome in early invasive colorectal carcinoma. Gastroenterology 2004, 127, 385–394. [Google Scholar] [CrossRef]
- Prall, F. Tumour budding in colorectal carcinoma. Histopathology 2007, 50, 151–162. [Google Scholar] [CrossRef]
- Karamitopoulou, E. Tumor budding cells, cancer stem cells and epithelial-mesenchymal transition-type cells in pancreatic cancer. Front. Oncol. 2012, 2, 209. [Google Scholar] [CrossRef] [Green Version]
- Grigore, A.D.; Jolly, M.K.; Jia, D.; Farach-Carson, M.C.; Levine, H. Tumor budding: The name is EMT. Partial EMT. J. Clin. Med. 2016, 5, 51. [Google Scholar] [CrossRef]
- Ogino, M.; Nakanishi, Y.; Mitsuhashi, T.; Hatanaka, Y.; Amano, T.; Marukawa, K.; Nitta, T.; Ueno, T.; Ono, M.; Kuwabara, S.; et al. Impact of tumour budding grade in 310 patients who underwent surgical resection for extrahepatic cholangiocarcinoma. Histopathology 2019, 74, 861–872. [Google Scholar] [CrossRef]
- Bacus, S.; Flowers, J.L.; Press, M.F.; Bacus, J.W.; McCarty, K.S., Jr. The evaluation of estrogen receptor in primary breast carcinoma by computer-assisted image analysis. Am. J. Clin. Pathol. 1988, 90, 233–239. [Google Scholar] [CrossRef] [Green Version]
- Somovilla-Crespo, B.; Alfonso-Pérez, M.; Cuesta-Mateos, C.; Carballo-de Dios, C.; Beltrán, A.E.; Terrón, F.; Pérez-Villar, J.J.; Gamallo-Amat, C.; Pérez-Chacón, G.; Fernández-Ruiz, E.; et al. Anti-Ccr7 therapy exerts a potent anti-tumor activity in a xenograft model of human mantle cell lymphoma. J. Hematol. Oncol. 2013, 6, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Förster, R.; Braun, A.; Worbs, T. Lymph node homing of T cells and dendritic cells via afferent lymphatics. Trends Immunol. 2012, 33, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Förster, R.; Davalos-Misslitz, A.C.; Rot, A. Ccr7 and its ligands: Balancing immunity and tolerance. Nat. Rev. Immunol. 2008, 8, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Winter, S.; Rehm, A.; Wichner, K.; Scheel, T.; Batra, A.; Siegmund, B.; Berek, C.; Lipp, M.; Höpken, U.E. Manifestation of spontaneous and early autoimmune gastritis in Ccr7-deficient mice. Am. J. Pathol. 2011, 179, 754–765. [Google Scholar] [CrossRef]
- Zu, G.; Luo, B.; Yang, Y.; Tan, Y.; Tang, T.; Zhang, Y.; Chen, X.; Sun, D. Meta-analysis of the prognostic value of C-C chemokine receptor type 7 in patients with solid tumors. Cancer Manag. Res. 2019, 11, 1881–1892. [Google Scholar] [CrossRef] [Green Version]
- Kohout, T.A.; Nicholas, S.L.; Perry, S.J.; Reinhart, G.; Junger, S.; Struthers, R.S. Differential desensitization, receptor phosphorylation, beta-arrestin recruitment, and ERK1/2 activation by the two endogenous ligands for the CC chemokine receptor 7. J. Biol. Chem 2004, 279, 23214–23222. [Google Scholar] [CrossRef] [Green Version]
- Steen, A.; Larsen, O.; Thiele, S.; Rosenkilde, M.M. Biased and G protein-independent signaling of chemokine receptors. Front. Immunol. 2014, 5, 277. [Google Scholar] [CrossRef] [Green Version]





| CCR7 Expression Grades | ||||||
|---|---|---|---|---|---|---|
| Low-Grade (n = 139) | High-Grade (n = 42) | |||||
| Clinicopathological feature | n | (%) | n | (%) | p | |
| Age | <70 | 66 | (47.5) | 27 | (64.3) | 0.055 |
| ≥70 | 73 | (52.5) | 15 | (35.7) | ||
| Sex | Male | 108 | (77.7) | 35 | (83.3) | 0.423 |
| Female | 31 | (22.3) | 7 | (16.7) | ||
| Histological grade | G1 | 45 | (32.4) | 4 | (9.5) | 0.003 |
| G2 | 68 | (48.9) | 23 | (54.8) | ||
| G3 | 26 | (18.7) | 15 | (35.7) | ||
| pT classification (AJCC, 8th edition) | T1 | 3 | (2.2) | 2 | (4.8) | 0.242 |
| T2 | 94 | (67.6) | 23 | (54.8) | ||
| T3 | 25 | (18.0) | 7 | (16.7) | ||
| T4 | 17 | (12.2) | 10 | (23.8) | ||
| pN classification (AJCC, 8th edition) | N0 | 75 | (54.0) | 24 | (57.1) | 0.674 |
| N1 | 55 | (39.6) | 14 | (33.3) | ||
| N2 | 9 | (6.5) | 4 | (9.5) | ||
| pM classification (AJCC, 8th edition) | M0 | 137 | (98.6) | 41 | (97.6) | 0.688 |
| M1 | 2 | (1.4) | 1 | (2.4) | ||
| Microscopic lymphatic invasion | Absent | 66 | (47.5) | 18 | (42.9) | 0.598 |
| Present | 73 | (52.5) | 24 | (57.1) | ||
| Microscopic venous invasion | Absent | 61 | (43.9) | 11 | (26.2) | 0.036 |
| Present | 78 | (56.1) | 31 | (73.8) | ||
| Microscopic perineural invasion | Absent | 11 | (7.9) | 6 | (14.3) | 0.235 |
| Present | 128 | (92.1) | 36 | (85.7) | ||
| Invasive carcinoma at resected margin | Negative | 124 | (89.2) | 34 | (81.0) | 0.176 |
| Positive | 15 | (10.8) | 8 | (19.1) | ||
| Median survival time (years) | 3.9 | 2.3 | 0.018 | |||
| No. of Patients | Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|---|
| Variable | n | (%) | Median Survival (Months) | p | Relative Risk (95% CI) | p | |
| Age (years) | <70 | 93 | (51.4) | 46 | 0.491 | ||
| ≥70 | 88 | (48.6) | 44 | ||||
| Sex | Male | 143 | (79.0) | 46 | 0.529 | ||
| Female | 38 | (21.0) | 44 | ||||
| Histological grade | G1 | 49 | (27.1) | 56 | 0.221 | ||
| G2 | 91 | (50.3) | 43 | ||||
| G3 | 41 | (22.7) | 28 | ||||
| pT classification (AJCC, 8th edition) | T1 | 5 | (2.8) | 53 | 0.001 | 1 | 0.026 |
| T2 | 117 | (64.6) | 53 | 1.07 (0.39–4.45) | |||
| T3 | 32 | (17.7) | 30 | 1.70 (0.56–7.36) | |||
| T4 | 27 | (14.9) | 21 | 2.14 (0.71–9.30) | |||
| pN classification (AJCC, 8th edition) | N0 | 99 | (54.7) | 76 | <0.001 | 1 | <0.001 |
| N1 | 69 | (38.1) | 29 | 2.32 (1.60–3.36) | |||
| N2 | 13 | (7.2) | 19 | 3.21 (1.54–6.16) | |||
| pM classification (AJCC, 8th edition) | M0 | 178 | (98.3) | 46 | 0.011 | 1 | 0.265 |
| M1 | 3 | (1.7) | 16 | 2.27 (0.49–7.78) | |||
| Microscopic lymphatic invasion | Absent | 84 | (46.4) | 57 | 0.094 | ||
| Present | 97 | (53.6) | 30 | ||||
| Microscopic venous invasion | Absent | 72 | (39.8) | 53 | 0.009 | 1 | 0.183 |
| Present | 109 | (60.2) | 32 | 1.30 (0.88–1.93) | |||
| Microscopic perineural invasion | Absent | 17 | (9.4) | 58 | 0.195 | ||
| Present | 164 | (90.6) | 43 | ||||
| Invasive carcinoma at resected margin | Negative | 158 | (87.3) | 49 | <0.001 | 1 | 0.056 |
| Positive | 23 | (12.7) | 21 | 1.66 (0.98–2.67) | |||
| CCR7 expression grade | Low-grade | 139 | (76.8) | 50 | 0.011 | 1 | 0.017 |
| High-grade | 42 | (23.2) | 27 | 1.67 (1.10–2.48) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oba, M.; Nakanishi, Y.; Mitsuhashi, T.; Sasaki, K.; Hatanaka, K.C.; Sasaki, M.; Nange, A.; Okumura, A.; Hayashi, M.; Yoshida, Y.; et al. CCR7 Mediates Cell Invasion and Migration in Extrahepatic Cholangiocarcinoma by Inducing Epithelial–Mesenchymal Transition. Cancers 2023, 15, 1878. https://doi.org/10.3390/cancers15061878
Oba M, Nakanishi Y, Mitsuhashi T, Sasaki K, Hatanaka KC, Sasaki M, Nange A, Okumura A, Hayashi M, Yoshida Y, et al. CCR7 Mediates Cell Invasion and Migration in Extrahepatic Cholangiocarcinoma by Inducing Epithelial–Mesenchymal Transition. Cancers. 2023; 15(6):1878. https://doi.org/10.3390/cancers15061878
Chicago/Turabian StyleOba, Mitsunobu, Yoshitsugu Nakanishi, Tomoko Mitsuhashi, Katsunori Sasaki, Kanako C. Hatanaka, Masako Sasaki, Ayae Nange, Asami Okumura, Mariko Hayashi, Yusuke Yoshida, and et al. 2023. "CCR7 Mediates Cell Invasion and Migration in Extrahepatic Cholangiocarcinoma by Inducing Epithelial–Mesenchymal Transition" Cancers 15, no. 6: 1878. https://doi.org/10.3390/cancers15061878
APA StyleOba, M., Nakanishi, Y., Mitsuhashi, T., Sasaki, K., Hatanaka, K. C., Sasaki, M., Nange, A., Okumura, A., Hayashi, M., Yoshida, Y., Nitta, T., Ueno, T., Yamada, T., Ono, M., Kuwabara, S., Okamura, K., Tsuchikawa, T., Nakamura, T., Noji, T., ... Hirano, S. (2023). CCR7 Mediates Cell Invasion and Migration in Extrahepatic Cholangiocarcinoma by Inducing Epithelial–Mesenchymal Transition. Cancers, 15(6), 1878. https://doi.org/10.3390/cancers15061878