Reprogramming of the Genome-Wide DNA Methylation Landscape in Three-Dimensional Cancer Cell Cultures
Abstract
:Simple Summary
Abstract
1. Introduction
2. The Epigenetic Landscape Represents a Platform through Which Multiple Environmental Factors Interact
2.1. Remodeling of Chromatin by Histone Modifications
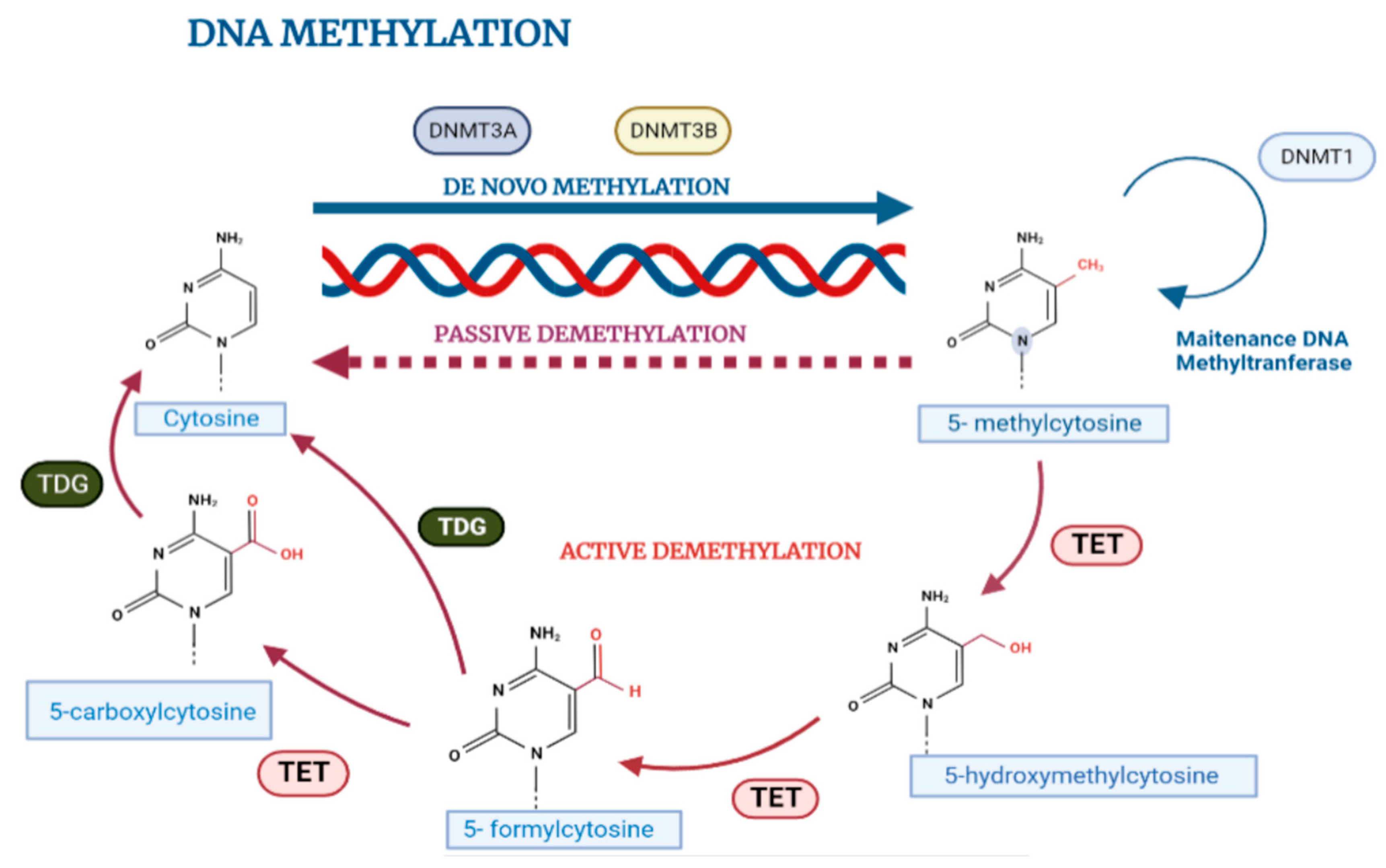
2.2. DNA Methylation in Normal Cell Biology
2.3. Influence of Microenvironment on DNA Methylation
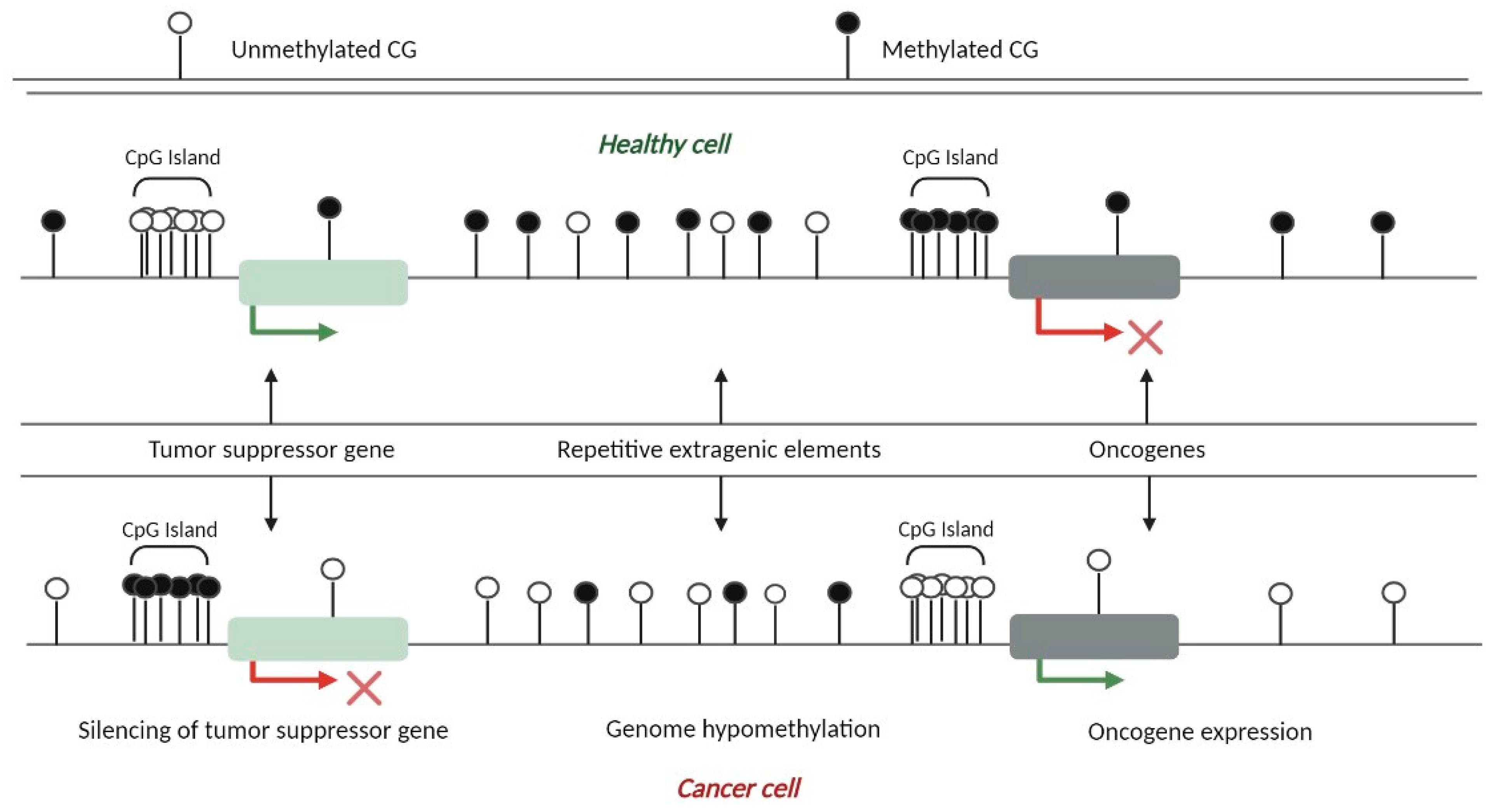
3. Aberrant DNA Methylation Patterns in Cancer Cells
4. Three-Dimensional Cell Cultures in Cancer Research
4.1. Organotypic 3D Cell Cultures
4.2. Scaffold-Free 3D Systems (Spheroids)
4.3. Organoid-Type Culture Systems
5. Three-Dimensional Cell Cultures and Microenvironmental Factors as Key Regulators of Epigenetic Programs in Cancer
6. Three-Dimensional Cell Culture Models Allow a Better Study of the Epigenetic Landscape in Cancer
6.1. Alterations in DNA Methylation in Organotypic 3D Cell Cultures
6.2. Cancer Co-Culture Models for Studying DNA Methylation
6.3. Alterations in DNA Methylation Patterns in Organoid-Type Cell Cultures
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, L.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018, 14, 910–919. [Google Scholar] [CrossRef]
- Duval, K.; Grover, H.; Han, L.-H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joseph, J.S.; Malindisa, S.T.; Ntwasa, M. Two-Dimensional (2D) and Three-Dimensional (3D) Cell Culturing in Drug Discovery. In Cell Culture; IntechOpen: London, UK, 2018. [Google Scholar] [CrossRef] [Green Version]
- Buitrago, D.; Labrador, M.; Arcon, J.P.; Lema, R.; Flores, O.; Esteve-Codina, A.; Blanc, J.; Villegas, N.; Bellido, D.; Gut, M.; et al. Impact of DNA methylation on 3D genome structure. Nat. Commun. 2021, 12, 3243. [Google Scholar] [CrossRef]
- Habanjar, O.; Diab-Assaf, M.; Caldefie-Chezet, F.; Delort, L. 3D Cell Culture Systems: Tumor Application, Advantages, and Disadvantages. Int. J. Mol. Sci. 2021, 22, 12200. [Google Scholar] [CrossRef]
- Sirchia, S.M.; Faversani, A.; Rovina, D.; Russo, M.V.; Paganini, L.; Savi, F.; Augello, C.; Rosso, L.; Del Gobbo, A.; Tabano, S.; et al. Epigenetic effects of chromatin remodeling agents on organotypic cultures. Epigenomics 2016, 8, 341–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DesRochers, T.M.; Shamis, Y.; Alt-Holland, A.; Kudo, Y.; Takata, T.; Wang, G.; Jackson-Grusby, L.; Garlick, J.A. The 3D tissue microenvironment modulates DNA methylation and E-cadherin expression in squamous cell carcinoma. Epigenetics 2012, 7, 34–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef]
- Vásquez-Moctezuma, I.; Luna-Palencia, G.R. Epigenética: La clave de la regulación genética. Rev. Mex. Mastol. 2020, 10, 48–53. [Google Scholar] [CrossRef]
- Greenberg, M.V.C.; Bourc’His, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef]
- Kim, M.; Costello, J. DNA methylation: An epigenetic mark of cellular memory. Exp. Mol. Med. 2017, 49, e322. [Google Scholar] [CrossRef] [Green Version]
- Melamed, P.; Yosefzon, Y.; David, C.; Tsukerman, A.; Pnueli, L. Tet Enzymes, Variants, and Differential Effects on Function. Front. Cell Dev. Biol. 2018, 6, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herceg, Z.; Vaissière, T. Epigenetic mechanisms and cancer: An interface between the environment and the genome. Epigenetics 2011, 6, 804–819. [Google Scholar] [CrossRef] [PubMed]
- Pirola, L.; Ciesielski, O.; Balcerczyk, A. The Methylation Status of the Epigenome: Its Emerging Role in the Regulation of Tumor Angiogenesis and Tumor Growth, and Potential for Drug Targeting. Cancers 2018, 10, 268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antequera, F.; Boyes, J.; Bird, A. High levels of De Novo methylation and altered chromatin structure at CpG islands in cell lines. Cell 1990, 62, 503–514. [Google Scholar] [CrossRef]
- Wu, X.; Su, J.; Wei, J.; Jiang, N.; Ge, X. Recent Advances in Three-Dimensional Stem Cell Culture Systems and Applications. Stem Cells Int. 2021, 2021, 9477332. [Google Scholar] [CrossRef]
- Koivunen, P.; Laukka, T. The TET enzymes. Cell. Mol. Life Sci. 2017, 75, 1339–1348. [Google Scholar] [CrossRef]
- Mahmoud, A.M.; Ali, M.M. Methyl Donor Micronutrients that Modify DNA Methylation and Cancer Outcome. Nutrients 2019, 11, 608. [Google Scholar] [CrossRef] [Green Version]
- Nestor, C.E.; Ottaviano, R.; Reinhardt, D.; Cruickshanks, H.A.; Mjoseng, H.K.; McPherson, R.C.; Lentini, A.; Thomson, J.P.; Dunican, D.S.; Pennings, S.; et al. Rapid reprogramming of epigenetic and transcriptional profiles in mammalian culture systems. Genome Biol. 2015, 16, 11. [Google Scholar] [CrossRef] [Green Version]
- Suarez-Martinez, E.; Suazo-Sanchez, I.; Celis-Romero, M.; Carnero, A. 3D and organoid culture in research: Physiology, hereditary genetic diseases and cancer. Cell Biosci. 2022, 12, 1–19. [Google Scholar] [CrossRef]
- Erdogan, O.S.; Erciyas, S.K.; Bilir, A.; Tuncer, S.B.; Odemis, D.A.; Kurul, S.; Karanlık, H.; Cabıoğlu, N.; Yazıcı, H. Methylation Changes of Primary Tumors, Monolayer, and Spheroid Tissue Culture Environments in Malignant Melanoma and Breast Carcinoma. BioMed Res. Int. 2019, 2019, 1407167. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.-Y.; Meaney, M.J. Epigenetics and the Environmental Regulation of the Genome and Its Function. Annu. Rev. Psychol. 2010, 61, 439–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, N.-E.; Lee, S.-H.; Park, H. Spheroid Culture System Methods and Applications for Mesenchymal Stem Cells. Cells 2019, 8, 1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, M.G.; Kriegstein, A.R. Challenges of Organoid Research. Annu. Rev. Neurosci. 2022, 45, 23–39. [Google Scholar] [CrossRef]
- Huang, Y.; Huang, Z.; Tang, Z.; Chen, Y.; Huang, M.; Liu, H.; Huang, W.; Ye, Q.; Jia, B. Research Progress, Challenges, and Breakthroughs of Organoids as Disease Models. Front. Cell Dev. Biol. 2021, 9, 740574. [Google Scholar] [CrossRef]
- Amatangelo, M.D.; Garipov, A.; Li, H.; Conejo-Garcia, J.R.; Speicher, D.W.; Zhang, R. Three-dimensional culture sensitizes epithelial ovarian cancer cells to EZH2 methyltransferase inhibition. Cell Cycle 2013, 12, 2113–2119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumont, N.; Wilson, M.B.; Crawford, Y.G.; Reynolds, P.A.; Sigaroudinia, M.; Tlsty, T.D. Sustained induction of epithelial to mesenchymal transition activates DNA methylation of genes silenced in basal-like breast cancers. Proc. Natl. Acad. Sci. USA 2008, 105, 14867–14872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brabson, J.P.; Leesang, T.; Mohammad, S.; Cimmino, L. Epigenetic Regulation of Genomic Stability by Vitamin C. Front. Genet. 2021, 12, 675780. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Nirmaladevi, R.; Ilango, S.; Paital, B.; Jayachandran, P.; Padma, P.R. Epigenetic alterations in cancer. Front. Biosci. 2020, 25, 1058–1109. [Google Scholar] [CrossRef]
- Gopinathan, G.; Diekwisch, T.G.H. Epigenetics and Early Development. J. Dev. Biol. 2022, 10, 26. [Google Scholar] [CrossRef]
- Ondeck, M.G.; Kumar, A.; Placone, J.K.; Plunkett, C.M.; Matte, B.F.; Wong, K.C.; Fattet, L.; Yang, J.; Engler, A.J. Dynamically stiffened matrix promotes malignant transformation of mammary epithelial cells via collective mechanical signaling. Proc. Natl. Acad. Sci. USA 2019, 116, 3502–3507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.; Hu, H.; Zhang, Q.; Wang, N.; Yang, X.; Guo, A.-Y. Genome-Wide DNA Methylation Enhances Stemness in the Mechanical Selection of Tumor-Repopulating Cells. Front. Bioeng. Biotechnol. 2020, 8, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stowers, R.S.; Shcherbina, A.; Israeli, J.; Gruber, J.J.; Chang, J.; Nam, S.; Rabiee, A.; Teruel, M.N.; Snyder, M.P.; Kundaje, A.; et al. Matrix stiffness induces a tumorigenic phenotype in mammary epithelium through changes in chromatin accessibility. Nat. Biomed. Eng. 2019, 3, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Li, X.; Huang, W.-P.; Hu, D.-F.; Ren, K.-F.; Ji, J. Dynamically softened substrate regulates malignancy of breast tumor cells. Sci. China Mater. 2021, 64, 2580–2592. [Google Scholar] [CrossRef]
- Kaur, A.; Ecker, B.L.; Douglass, S.M.; Kugel, C.H.; Webster, M.R.; Almeida, F.V.; Somasundaram, R.; Hayden, J.; Ban, E.; Ahmadzadeh, H.; et al. Remodeling of the Collagen Matrix in Aging Skin Promotes Melanoma Metastasis and Affects Immune Cell Motility. Cancer Discov. 2019, 9, 64–81. [Google Scholar] [CrossRef] [Green Version]
- Porter, R.J.; Murray, G.I.; McLean, M.H. Current concepts in tumour-derived organoids. Br. J. Cancer 2020, 123, 1209–1218. [Google Scholar] [CrossRef]
- Joshi, R.S.; De Moura, M.C.; Piñeyro, D.; Alvarez_Errico, D.; Arribas, C.; Esteller, M. The DNA methylation landscape of human cancer organoids available at the American type culture collection. Epigenetics 2020, 15, 1167–1177. [Google Scholar] [CrossRef]
- Alghuwainem, A.; Alshareeda, A.T.; Alsowayan, B. Scaffold-Free 3-D Cell Sheet Technique Bridges the Gap between 2-D Cell Culture and Animal Models. Int. J. Mol. Sci. 2019, 20, 4926. [Google Scholar] [CrossRef] [Green Version]
- Imamura, Y.; Mukohara, T.; Shimono, Y.; Funakoshi, Y.; Chayahara, N.; Toyoda, M.; Kiyota, N.; Takao, S.; Kono, S.; Nakatsura, T.; et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015, 33, 1837–1843. [Google Scholar] [CrossRef] [Green Version]
- Aguilera, O.; Fernández, A.F.; Muñoz, A.; Fraga, M.F. Epigenetics and environment: A complex relationship. J. Appl. Physiol. 2010, 109, 243–251. [Google Scholar] [CrossRef] [Green Version]
- Das, V.; Bruzzese, F.; Konečný, P.; Iannelli, F.; Budillon, A.; Hajdúch, M. Pathophysiologically relevant in vitro tumor models for drug screening. Drug Discov. Today 2015, 20, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Ballav, S.; Deshmukh, A.J.; Siddiqui, S.; Aich, J.; Basu, S. Two-Dimensional and Three-Dimensional Cell Culture and Their Applications. In Cell Culture—Advanced Technology and Applications in Medical and Life Sciences; IntechOpen: London, UK, 2022. [Google Scholar] [CrossRef]
- Białkowska, K.; Komorowski, P.; Bryszewska, M.; Miłowska, K. Spheroids as a Type of Three-Dimensional Cell Cultures—Examples of Methods of Preparation and the Most Important Application. Int. J. Mol. Sci. 2020, 21, 6225. [Google Scholar] [CrossRef] [PubMed]
- Fontoura, J.C.; Viezzer, C.; dos Santos, F.G.; Ligabue, R.A.; Weinlich, R.; Puga, R.D.; Antonow, D.; Severino, P.; Bonorino, C. Comparison of 2D and 3D cell culture models for cell growth, gene expression and drug resistance. Mater. Sci. Eng. C 2019, 107, 110264. [Google Scholar] [CrossRef] [PubMed]
- Hayden, P.J.; Harbell, J.W. Special review series on 3D organotypic culture models: Introduction and historical perspective. Vitr. Cell. Dev. Biol.-Anim. 2020, 57, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning from 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef] [Green Version]
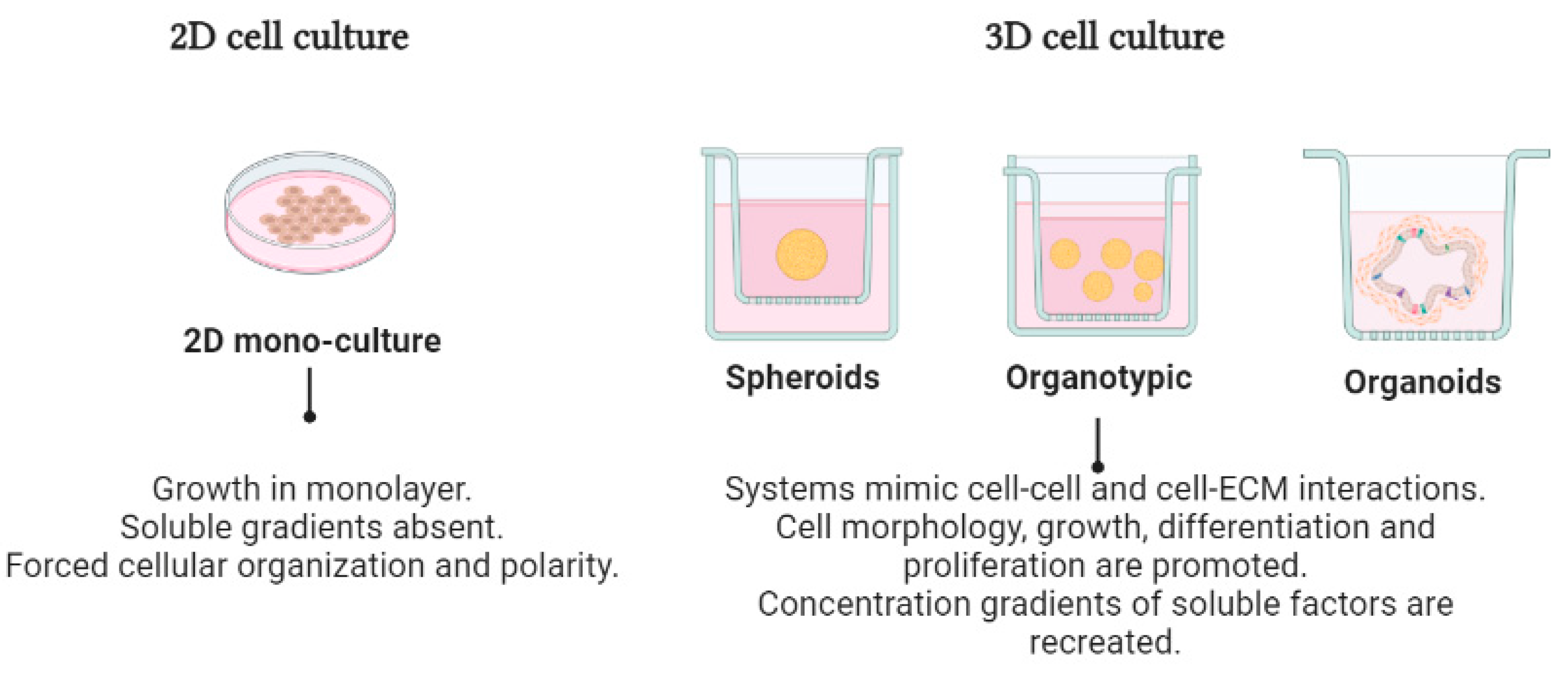

| 2D Cell Culture | 3D Cell Culture | References | |||
|---|---|---|---|---|---|
| Organotypic | Spheroids | Organoids | |||
| Cell morphology | Flattened morphology Monolayer polarized | Including rounder, spread out, and elongated preserved natural cell structures | Rounded spheroids Contain multiple layers | Aggregate form Spherelike structure High polarity | [18] |
| Cell–cell Interactions | Cell–cell and cell–extracellular environment interactions are not represented | Cell–cell and cell–ECM interactions | Strong cell–cell connections Tight intercellular connections | Cell–cell and cell–3D matrix interactions | [19] |
| Culture quality | Highly reproducible Easy to interpret Simplicity of culture | Ability to mimic the ECM High sensitivity to drugs and reproducibility Variety of matrigels with high mechanical-structural differences | High reproducibility and sensitivity to drugs Moderate mechanical-structural integrity of the cells | Highly reproducible. High organotypic differentiation and in vivo-like functions | [20] |
| Molecular mechanisms | Loss of differentiated function, lacking tissue-specific environments and the ECM | Morphology and gene expression patterns, but also migration, cell cycling, and proliferation | Mimic the vascular structure of native tissues. Cells are in contact with the ECM Diffusion gradient of nutrients, waste, oxygen, and drugs | Interactions, mechanical cues, and features such as fluid flow, shear forces, stretching, and organ–organ interactions | [20,21] |
| Drug response | Low sensitivity to drugs | Depending on the 3D environment, the culture might become resistant or susceptible to drugs | Variable resistance to drugs | Depending on the 3D environment, the culture might become resistant or susceptible to drugs | [22] |
| Applications | Therapeutics for diseases | In vitro angiogenesis and drug testing, drug response studies for cancer research | Disease modeling and regenerative medicine Target identification and validation using RNAi | Therapeutic drug screening as well as personalized medicine | [23] |
| Type of System | Description | Advantages | Disadvantages |
|---|---|---|---|
2D cell cultures | Cells grow on flat dishes, regularly made of plastic, where they adhere and spread until they reach confluence. | Inexpensive Ease of observing cells for interpretation Cells can be easily extracted from the medium and used for further experiments Most frequently used method in laboratories | Are not representative of real cellular environments Consist of individual dispersed cells Loss of the original tissue’s heterogeneity Lack of nutrients and oxygen gradients Homogeneous exposition to nutrients and to drugs Lack of ECM–cell interactions and signaling activated by the substrate |
| 3D cell cultures | |||
Spheroids | Spheroids grow over plastic surfaces forming floating 3D structures in which the cells form various layers which mimic some of the physical and biochemical features of solid tumor masses. | Can better to mimic a tumor mass Better at recreating the cell–cell interactions in different types of tumor cells Establishment of barriers between tissues Formation of nutrients and oxygen nutrients | More expensive and time- consuming Few commercially available Lack of nutrients at the core of spheroids affecting cell viability Lack of ECM–cell interactions and signaling activated by the substrate’s compounds |
Organotypic cell cultures in scaffolds | A 3D scaffold provides a way for cells to grow in three dimensions on a 3D plate. It has the ability to mimic the microenvironment in vivo more closely. These are typically provided through biomaterials of animal or vegetable origin called “hydrogels” as an ECM in which cells can survive, grow, and proliferate. | Can be accurately grown and measured An ideal environment for drug discovery and development The high levels of viability in cells can promote cell–cell and cell–ECM interactions and further affect the cells’ shape, metabolism, function, migration, proliferation, differentiation, and adhesion | Natural ECM has poor mechanical properties High sensitivity to enzymes, which limits its potential for application Scaffolds and the topography of cell distribution may cause various behaviors of the cell Microscopic cell observations and cell extraction are restricted for some analyses |
Organoids | The reason for growth in 3D organoid cell cultures is that it is simply a better way of representing human tissues outside the body. | Organoids are a more realistic way to grow and treat cells, imitating the architecture of the parental tissues They are suitable for gene editing and used for simulating host–microbe interactions They can self-renew and maintain the physiological structure and function of tissues Creation of organoid biobanks becomes possible and, in this way, reduces the use of animal models | More time-consuming and very expensive Lack of high-fidelity cell types The limited maturation, atypical physiology, and lack of realization are features that may limit their reliability for certain applications |
| Reported Studies | References |
|---|---|
| Rapid reprogramming of epigenetic and transcriptional profiles in mammalian culture systems | [19] |
| Methylation changes of primary tumors, monolayer, and spheroid tissue culture environments in malignant melanoma and breast carcinoma | [21] |
| Spheroid culture system methods and applications for mesenchymal stem cells | [23] |
| Three-dimensional culture sensitizes epithelial ovarian cancer cells to EZH2 methyltransferase inhibition | [26] |
| Dynamically stiffened matrix promotes malignant transformation of mammary epithelial cells via collective mechanical signaling | [32] |
| Matrix stiffness induces a tumorigenic phenotype in mammary epithelium through changes in chromatin accessibility | [34] |
| Dynamically softened substrate regulates malignancy of breast tumor cells | [35] |
| Remodeling of the collagen matrix in aging skin promotes melanoma metastasis and affects immune cell motility | [36] |
| Current concepts in tumor-derived organoids | [37] |
| The DNA methylation landscape of human cancer organoids available at the American type culture collection | [38] |
| Scaffold-free 3D cell sheet technique bridges the gap between 2D cell culture and animal models | [39] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heredia-Mendez, A.J.; Sánchez-Sánchez, G.; López-Camarillo, C. Reprogramming of the Genome-Wide DNA Methylation Landscape in Three-Dimensional Cancer Cell Cultures. Cancers 2023, 15, 1991. https://doi.org/10.3390/cancers15071991
Heredia-Mendez AJ, Sánchez-Sánchez G, López-Camarillo C. Reprogramming of the Genome-Wide DNA Methylation Landscape in Three-Dimensional Cancer Cell Cultures. Cancers. 2023; 15(7):1991. https://doi.org/10.3390/cancers15071991
Chicago/Turabian StyleHeredia-Mendez, Alma Jaqueline, Gricelda Sánchez-Sánchez, and César López-Camarillo. 2023. "Reprogramming of the Genome-Wide DNA Methylation Landscape in Three-Dimensional Cancer Cell Cultures" Cancers 15, no. 7: 1991. https://doi.org/10.3390/cancers15071991
APA StyleHeredia-Mendez, A. J., Sánchez-Sánchez, G., & López-Camarillo, C. (2023). Reprogramming of the Genome-Wide DNA Methylation Landscape in Three-Dimensional Cancer Cell Cultures. Cancers, 15(7), 1991. https://doi.org/10.3390/cancers15071991








