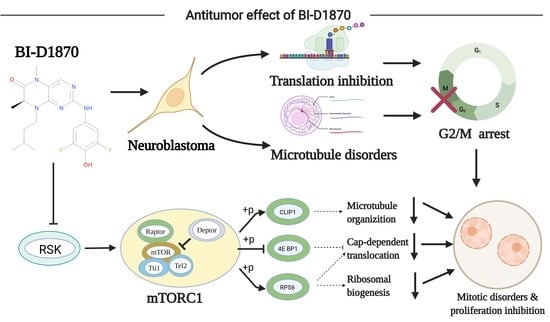
BI-D1870 Induces Mitotic Dysfunction and Apoptosis in Neuroblastoma by Regulating the PI3K-Akt-mTORC1 Signal Axis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. NB Dataset
2.2. Prognostic Gene Screening and PPI Protein Network Analysis
2.3. Drug Forecasting
2.4. Structure of BI-D1870 and Its Target Protein RSK1
2.5. GO-KEGG Enrichment Analysis and GSEA Enrichment Analysis
2.6. Animal Experiments and Histopathology Experiments
2.6.1. In Vivo Experiments
2.6.2. HE Staining
2.6.3. Immunofluorescence Staining
2.6.4. TUNEL
2.7. Cell Lines and Cell Culture
2.8. Cell Proliferation Activity and Morphological Changes
2.9. Calcein/PI Cell Viability/Cytotoxicity Assay
2.10. Cell Invasion Ability Test
2.11. Apoptosis through Flow Cytometry and TUNEL
2.12. Cell Cycle Assay
2.13. RNA Sequencing and Bioinformatics Analysis
2.14. Cell Fluorescence Staining Experiment
2.15. WB Experiment
2.16. Statistics
3. Results
3.1. Bioinformatics Analysis and Drug Prediction of Prognostis-Related Genes
3.2. In Vivo Inhibitory Effect
3.3. Cytotoxicity of BI-D1870
3.4. BI-D1870 Inhibited Cell Migration and Invasion Ability
3.5. BI-D1870-Induced Cell Apoptosis
3.6. BI-D1870 Induces G2/M Arrest and Mitotic Catastrophe
3.7. PI3K-Akt-mTORC1 Signaling Pathway Inhibition
3.8. BI-D1870 Induces Disruption of Cytoskeleton Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shohet, J.; Foster, J. Neuroblastoma. BMJ 2017, 357, j1863. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, J.; Ziegler, D.S.; Trahair, T.; Marshall, G.M.; Haber, M.; Norris, M.D. Too many targets, not enough patients: Rethinking neuroblastoma clinical trials. Nat. Rev. Cancer 2018, 18, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Park, J.R.; Kreissman, S.G.; London, W.B.; Naranjo, A.; Cohn, S.L.; Hogarty, M.D.; Tenney, S.C.; Haas-Kogan, D.; Shaw, P.J.; Kraveka, J.M.; et al. Effect of Tandem Autologous Stem Cell Transplant vs Single Transplant on Event-Free Survival in Patients With High-Risk Neuroblastoma: A Randomized Clinical Trial. JAMA 2019, 322, 746–755. [Google Scholar] [CrossRef] [PubMed]
- Park, J.A.; Cheung, N.K.V. Targets and Antibody Formats for Immunotherapy of Neuroblastoma. J. Clin. Oncol. 2020, 38, 1836–1848. [Google Scholar] [CrossRef]
- Zafar, A.; Wang, W.; Liu, G.; Wang, X.; Xian, W.; McKeon, F.; Foster, J.; Zhou, J.; Zhang, R. Molecular targeting therapies for neuroblastoma: Progress and challenges. Med. Res. Rev. 2021, 41, 961–1021. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Gröbner, S.N.; Worst, B.C.; Weischenfeldt, J.; Buchhalter, I.; Kleinheinz, K.; Rudneva, V.A.; Johann, P.D.; Balasubramanian, G.P.; Segura-Wang, M.; Brabetz, S.; et al. The landscape of genomic alterations across childhood cancers. Nature 2018, 555, 321–327. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.T.W.; Banito, A.; Grünewald, T.G.P.; Haber, M.; Jäger, N.; Kool, M.; Milde, T.; Molenaar, J.J.; Nabbi, A.; Pugh, T.J.; et al. Molecular characteristics and therapeutic vulnerabilities across paediatric solid tumours. Nat. Rev. Cancer 2019, 19, 420–438. [Google Scholar] [CrossRef] [PubMed]
- Gowda, M.; Godder, K.; Kmieciak, M.; Worschech, A.; Ascierto, M.-L.; Wang, E.; Marincola, F.M.; Manjili, M.H. Distinct signatures of the immune responses in low risk versus high risk neuroblastoma. J. Transl. Med. 2011, 9, 170. [Google Scholar] [CrossRef] [Green Version]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.-P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef] [Green Version]
- Lamb, J. The Connectivity Map: A new tool for biomedical research. Nat. Rev. Cancer 2007, 7, 54–60. [Google Scholar] [CrossRef]
- Sapkota, G.P.; Cummings, L.; Newell, F.S.; Armstrong, C.; Bain, J.; Frodin, M.; Grauert, M.; Hoffmann, M.; Schnapp, G.; Steegmaier, M.; et al. BI-D1870 is a specific inhibitor of the p90 RSK (ribosomal S6 kinase) isoforms in vitro and in vivo. Biochem. J. 2007, 401, 29–38. [Google Scholar] [CrossRef] [Green Version]
- Bhatt, R.R.; Ferrell, J.E. The Protein Kinase p90 Rsk as an Essential Mediator of Cytostatic Factor Activity. Science 1999, 286, 1362–1365. [Google Scholar] [CrossRef]
- Anjum, R.; Blenis, J. The RSK family of kinases: Emerging roles in cellular signalling. Nat. Rev. Mol. Cell Biol. 2008, 9, 747–758. [Google Scholar] [CrossRef]
- Shahbazian, D.; Roux, P.P.; Mieulet, V.; Cohen, M.S.; Raught, B.; Taunton, J.; Hershey, J.W.B.; Blenis, J.; Pende, M.; Sonenberg, N. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 2006, 25, 2781–2791. [Google Scholar] [CrossRef]
- Carrière, A.; Cargnello, M.; Julien, L.-A.; Gao, H.; Bonneil, E.; Thibault, P.; Roux, P.P. Oncogenic MAPK Signaling Stimulates mTORC1 Activity by Promoting RSK-Mediated Raptor Phosphorylation. Curr. Biol. 2008, 18, 1269–1277. [Google Scholar] [CrossRef] [Green Version]
- Griner, E.M.; Kazanietz, M.G. Protein kinase C and other diacylglycerol effectors in cancer. Nat. Rev. Cancer 2007, 7, 281–294. [Google Scholar] [CrossRef]
- Salhi, A.; Farhadian, J.A.; Giles, K.M.; de Miera, E.V.-S.; Silva, I.P.; Bourque, C.; Yeh, K.; Chhangawala, S.; Wang, J.; Ye, F.; et al. RSK1 Activation Promotes Invasion in Nodular Melanoma. Am. J. Pathol. 2015, 185, 704–716. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, A.C.; Costa, M.; Zollo, O.; Davis, C.; Feldman, M.E.; Testa, J.R.; Meyuhas, O.; Shokat, K.M.; Ruggero, D. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell 2010, 17, 249–261. [Google Scholar] [CrossRef] [Green Version]
- Milosevic, N.; Kühnemuth, B.; Mühlberg, L.; Ripka, S.; Griesmann, H.; Lölkes, C.; Buchholz, M.; Aust, D.; Pilarsky, C.; Krug, S.; et al. Synthetic Lethality Screen Identifies RPS6KA2 as Modifier of Epidermal Growth Factor Receptor Activity in Pancreatic Cancer. Neoplasia 2013, 15, 1354–1362. [Google Scholar] [CrossRef] [Green Version]
- Fan, L.; Li, P.; Yin, Z.; Fu, G.; Liao, D.J.; Liu, Y.; Zhu, J.; Zhang, Y.; Wang, L.; Yan, Q.; et al. Ribosomal s6 protein kinase 4: A prognostic factor for renal cell carcinoma. Br. J. Cancer 2013, 109, 1137–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiu, C.-F.; Bai, L.-Y.; Kapuriya, N.; Peng, S.-Y.; Wu, C.-Y.; Sargeant, A.M.; Chen, M.Y.; Weng, J.-R. Antitumor effects of BI-D1870 on human oral squamous cell carcinoma. Cancer Chemother. Pharmacol. 2013, 73, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Abdulrahman, N.; Jaballah, M.; Poomakkoth, N.; Riaz, S.; Abdelaziz, S.; Issa, A.; Mraiche, F. Inhibition of p90 ribosomal S6 kinase attenuates cell migration and proliferation of the human lung adenocarcinoma through phospho-GSK-3beta and osteopontin. Mol Cell Biochem. 2016, 418, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Abdulrahman, N.; Siveen, K.S.; Joseph, J.M.; Osman, A.; Yalcin, H.C.; Hasan, A.; Uddin, S.; Mraiche, F. Inhibition of p90 ribosomal S6 kinase potentiates cisplatin activity in A549 human lung adenocarcinoma cells. J. Pharm. Pharmacol. 2020, 72, 1536–1545. [Google Scholar] [CrossRef]
- Theodosakis, N.; Micevic, G.; Langdon, C.G.; Ventura, A.; Means, R.; Stern, D.F.; Bosenberg, M.W. p90RSK Blockade Inhibits Dual BRAF and MEK Inhibitor-Resistant Melanoma by Targeting Protein Synthesis. J. Investig. Dermatol. 2017, 137, 2187–2196. [Google Scholar] [CrossRef] [Green Version]
- Katayama, K.; Nishihata, A. RSK Inhibition Induces Apoptosis by Downregulating Protein Synthesis in a Variety of Acute Myeloid Leukemia Cell Lines. Biol. Pharm. Bull. 2021, 44, 1843–1850. [Google Scholar] [CrossRef]
- Chae, H.-D.; Dutta, R.; Tiu, B.; Hoff, F.W.; Accordi, B.; Serafin, V.; Youn, M.; Huang, M.; Sumarsono, N.; Davis, K.L.; et al. RSK inhibitor BI-D1870 inhibits acute myeloid leukemia cell proliferation by targeting mitotic exit. Oncotarget 2020, 11, 2387–2403. [Google Scholar] [CrossRef]
- Poomakkoth, N.; Issa, A.; Abdulrahman, N.; Abdelaziz, S.G.; Mraiche, F. p90 ribosomal S6 kinase: A potential therapeutic target in lung cancer. J. Transl. Med. 2016, 14, 14. [Google Scholar] [CrossRef] [Green Version]
- Weber, A.; Huesken, C.; Bergmann, E.; Kiess, W.; Christiansen, N.M.; Christiansen, H. Coexpression of insulin receptor-related receptor and insulin-like growth factor 1 receptor correlates with enhanced apoptosis and dedifferentiation in human neuroblastomas. Clin. Cancer Res. 2003, 9, 5683–5692. [Google Scholar]
- Ho, R.; Minturn, J.E.; Hishiki, T.; Zhao, H.; Wang, Q.; Cnaan, A.; Maris, J.; Evans, A.E.; Brodeur, G.M. Proliferation of Human Neuroblastomas Mediated by the Epidermal Growth Factor Receptor. Cancer Res. 2005, 65, 9868–9875. [Google Scholar] [CrossRef] [Green Version]
- Shaw, R.J.; Cantley, L.C. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature 2006, 441, 424–430. [Google Scholar] [CrossRef]
- Opel, D.; Poremba, C.; Simon, T.; Debatin, K.-M.; Fulda, S. Activation of Akt Predicts Poor Outcome in Neuroblastoma. Cancer Res. 2007, 67, 735–745. [Google Scholar] [CrossRef] [Green Version]
- Holz, M.K.; Ballif, B.A.; Gygi, S.P.; Blenis, J. mTOR and S6K1 Mediate Assembly of the Translation Preinitiation Complex through Dynamic Protein Interchange and Ordered Phosphorylation Events. Cell 2005, 123, 569–580. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.-H.; Sarbassov, D.D.; Ali, S.M.; King, J.E.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. mTOR Interacts with Raptor to Form a Nutrient-Sensitive Complex that Signals to the Cell Growth Machinery. Cell 2002, 110, 163–175. [Google Scholar] [CrossRef] [Green Version]
- Fededa, J.P.; Gerlich, D.W. Molecular control of animal cell cytokinesis. Nat. Cell Biol. 2012, 14, 440–447. [Google Scholar] [CrossRef]
- Otegui, M.; Verbrugghe, K.J.; Skop, A.R. Midbodies and phragmoplasts: Analogous structures involved in cytokinesis. Trends Cell Biol. 2005, 15, 404–413. [Google Scholar] [CrossRef] [Green Version]
- Sivakumar, S.; Gorbsky, G.J. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat. Rev. Mol. Cell Biol. 2015, 16, 82–94. [Google Scholar] [CrossRef] [Green Version]
- Dibble, C.C.; Cantley, L.C. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015, 25, 545–555. [Google Scholar] [CrossRef] [Green Version]
- Bohlen, J.; Roiuk, M.; Teleman, A.A. Phosphorylation of ribosomal protein S6 differentially affects mRNA translation based on ORF length. Nucleic Acids Res. 2021, 49, 13062–13074. [Google Scholar] [CrossRef]
- Böhm, R.; Imseng, S.; Jakob, R.P.; Hall, M.N.; Maier, T.; Hiller, S. The dynamic mechanism of 4E-BP1 recognition and phosphorylation by mTORC1. Mol. Cell 2021, 81, 2403–2416.e5. [Google Scholar] [CrossRef]
- She, Q.-B.; Halilovic, E.; Ye, Q.; Zhen, W.; Shirasawa, S.; Sasazuki, T.; Solit, D.B.; Rosen, N. 4E-BP1 Is a Key Effector of the Oncogenic Activation of the AKT and ERK Signaling Pathways that Integrates Their Function in Tumors. Cancer Cell 2010, 18, 39–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinnebusch, A.G. Translational Homeostasis via eIF4E and 4E-BP1. Mol. Cell 2012, 46, 717–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Chen, J.; Dong, Z.; Meyuhas, O.; Chen, J.-K. Phosphorylation of ribosomal protein S6 mediates compensatory renal hypertrophy. Kidney Int. 2015, 87, 543–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bieling, P.; Laan, L.; Schek, H.; Munteanu, E.L.; Sandblad, L.; Dogterom, M.; Brunner, D.; Surrey, T. Reconstitution of a microtubule plus-end tracking system in vitro. Nature 2007, 450, 1100–1105. [Google Scholar] [CrossRef]
- Henty-Ridilla, J.L.; Rankova, A.; Eskin, J.A.; Kenny, K.; Goode, B.L. Accelerated actin filament polymerization from microtubule plus ends. Science 2016, 352, 1004–1009. [Google Scholar] [CrossRef] [Green Version]
- Perez, F.; Diamantopoulos, G.S.; Stalder, R.; Kreis, T.E. CLIP-170 Highlights Growing Microtubule Ends In Vivo. Cell 1999, 96, 517–527. [Google Scholar] [CrossRef] [Green Version]
- Yan, M.; Wang, C.; He, B.; Yang, M.; Tong, M.; Long, Z.; Liu, B.; Peng, F.; Xu, L.; Zhang, Y.; et al. Aurora-A Kinase: A Potent Oncogene and Target for Cancer Therapy. Med. Res. Rev. 2016, 36, 1036–1079. [Google Scholar] [CrossRef]
- Marumoto, T.; Zhang, D.; Saya, H. Aurora-A—A guardian of poles. Nat. Rev. Cancer 2005, 5, 42–50. [Google Scholar] [CrossRef]
- Prigent, C.; Giet, R. Aurora A and Mitotic Commitment. Cell 2003, 114, 531–532. [Google Scholar] [CrossRef]
- Maietta, I.; Del Peschio, F.; Buonocore, P.; Viscusi, E.; Laudati, S.; Iannaci, G.; Minopoli, M.; Motti, M.L.; De Falco, V. p90RSK Regulates p53 Pathway by MDM2 Phosphorylation in Thyroid Tumors. Cancers 2022, 15, 121. [Google Scholar] [CrossRef]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, L.; Mi, T.; Wu, X.; Wang, Z.; Zhang, Z.; Liu, J.; Wang, Z.; Wang, J.; Li, M.; Ren, C.; et al. BI-D1870 Induces Mitotic Dysfunction and Apoptosis in Neuroblastoma by Regulating the PI3K-Akt-mTORC1 Signal Axis. Cancers 2023, 15, 2023. https://doi.org/10.3390/cancers15072023
Jin L, Mi T, Wu X, Wang Z, Zhang Z, Liu J, Wang Z, Wang J, Li M, Ren C, et al. BI-D1870 Induces Mitotic Dysfunction and Apoptosis in Neuroblastoma by Regulating the PI3K-Akt-mTORC1 Signal Axis. Cancers. 2023; 15(7):2023. https://doi.org/10.3390/cancers15072023
Chicago/Turabian StyleJin, Liming, Tao Mi, Xin Wu, Zhang Wang, Zhaoxia Zhang, Jiayan Liu, Zhaoying Wang, Jinkui Wang, Mujie Li, Chunnian Ren, and et al. 2023. "BI-D1870 Induces Mitotic Dysfunction and Apoptosis in Neuroblastoma by Regulating the PI3K-Akt-mTORC1 Signal Axis" Cancers 15, no. 7: 2023. https://doi.org/10.3390/cancers15072023
APA StyleJin, L., Mi, T., Wu, X., Wang, Z., Zhang, Z., Liu, J., Wang, Z., Wang, J., Li, M., Ren, C., Guo, P., & He, D. (2023). BI-D1870 Induces Mitotic Dysfunction and Apoptosis in Neuroblastoma by Regulating the PI3K-Akt-mTORC1 Signal Axis. Cancers, 15(7), 2023. https://doi.org/10.3390/cancers15072023