Transcriptome Meta-Analysis of Triple-Negative Breast Cancer Response to Neoadjuvant Chemotherapy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Workflow
2.2. Gene Expression Datasets
2.3. Data Pre-Processing
2.4. Association Testing for Individual Dataset
2.5. Meta-Analysis
2.6. Pathway and Functional Enrichment Analysis
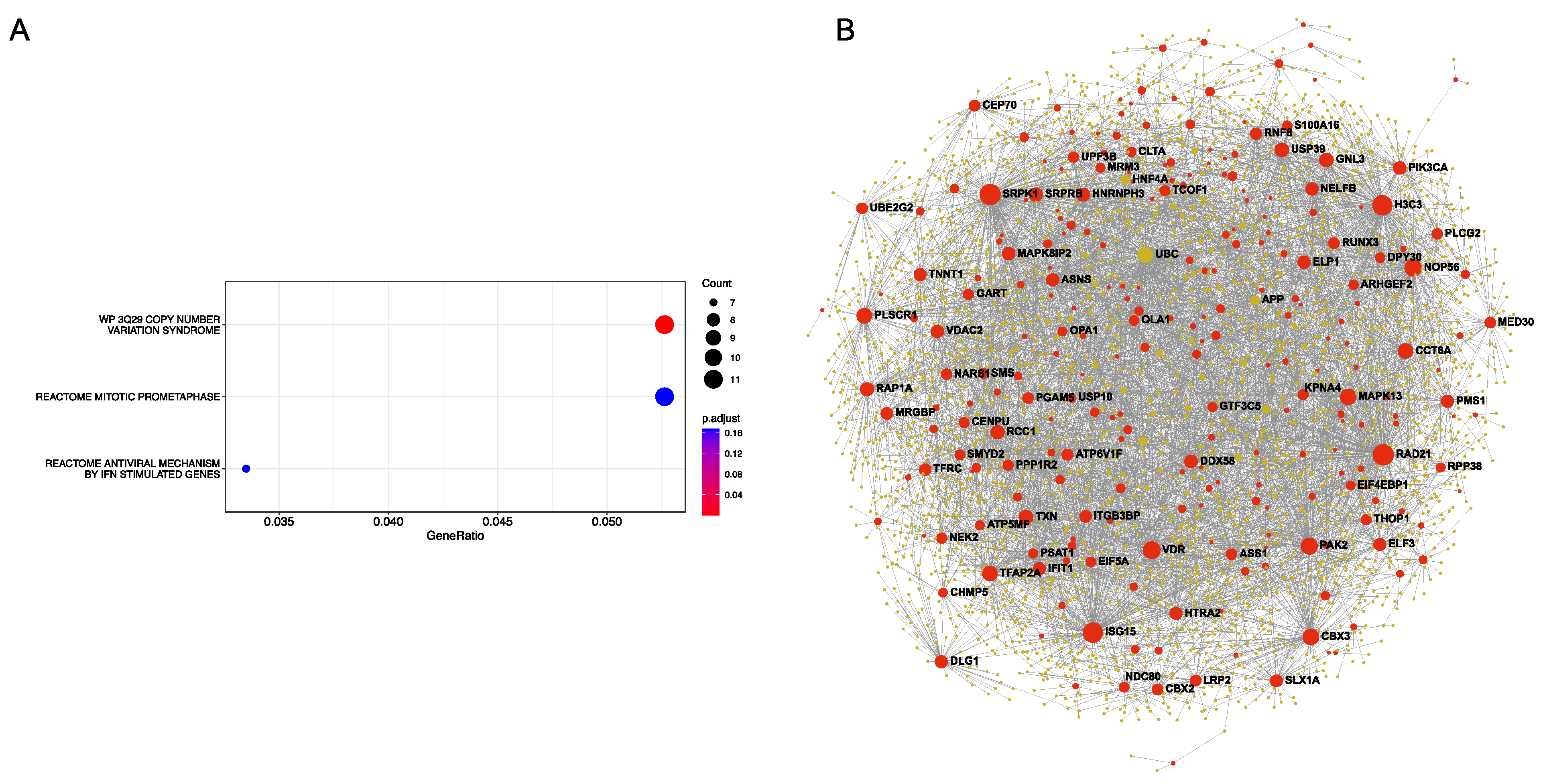
2.7. Protein–Protein Interaction (PPI) Network Analysis
3. Results
3.1. Meta-Analysis Results of NAC Response
3.2. Meta-Analysis Results of DFS
3.3. Integration of pCR and DFS Meta-Analysis Results
3.4. Quadrant I: Genes Associated with pCR and Lower DFS Risk
3.5. Quadrant II: Genes Associated with RD and Lower DFS Risk
3.6. Quadrant III: Genes Associated with RD and Higher DFS Risk
3.7. Quadrant IV: Genes Associated with pCR and Higher DFS Risk
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef] [Green Version]
- Bianchini, G.; Balko, J.M.; Mayer, I.A.; Sanders, M.E.; Gianni, L. Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 2016, 13, 674–690. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Haffty, B.G.; Yang, Q.; Reiss, M.; Kearney, T.; Higgins, S.A.; Weidhaas, J.; Harris, L.; Hait, W.; Toppmeyer, D. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2006, 24, 5652–5657. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, B.D.; Jovanovic, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef]
- Masuda, H.; Baggerly, K.A.; Wang, Y.; Zhang, Y.; Gonzalez-Angulo, A.M.; Meric-Bernstam, F.; Valero, V.; Lehmann, B.D.; Pietenpol, J.A.; Hortobagyi, G.N.; et al. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin. Cancer Res. 2013, 19, 5533–5540. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Li, J.; Gray, W.H.; Lehmann, B.D.; Bauer, J.A.; Shyr, Y.; Pietenpol, J.A. TNBCtype: A Subtyping Tool for Triple-Negative Breast Cancer. Cancer Inform. 2012, 11, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Colaprico, A.; Silva, T.C.; Chen, J.; An, H.; Ban, Y.; Huang, H.; Wang, L.; James, J.L.; Balko, J.M.; et al. Multi-omics analysis identifies therapeutic vulnerabilities in triple-negative breast cancer subtypes. Nat. Commun. 2021, 12, 6276. [Google Scholar] [CrossRef] [PubMed]
- Burstein, M.D.; Tsimelzon, A.; Poage, G.M.; Covington, K.R.; Contreras, A.; Fuqua, S.A.; Savage, M.I.; Osborne, C.K.; Hilsenbeck, S.G.; Chang, J.C.; et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin. Cancer Res. 2015, 21, 1688–1698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bareche, Y.; Venet, D.; Ignatiadis, M.; Aftimos, P.; Piccart, M.; Rothe, F.; Sotiriou, C. Unravelling triple-negative breast cancer molecular heterogeneity using an integrative multiomic analysis. Ann. Oncol. 2018, 29, 895–902. [Google Scholar] [CrossRef]
- Bianchini, G.; De Angelis, C.; Licata, L.; Gianni, L. Treatment landscape of triple-negative breast cancer—Expanded options, evolving needs. Nat. Rev. Clin. Oncol. 2022, 19, 91–113. [Google Scholar] [CrossRef]
- Schmid, P.; Adams, S.; Rugo, H.S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Dieras, V.; Hegg, R.; Im, S.A.; Shaw Wright, G.; et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018, 379, 2108–2121. [Google Scholar] [CrossRef]
- Cortes, J.; Cescon, D.W.; Rugo, H.S.; Nowecki, Z.; Im, S.A.; Yusof, M.M.; Gallardo, C.; Lipatov, O.; Barrios, C.H.; Holgado, E.; et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): A randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet 2020, 396, 1817–1828. [Google Scholar] [CrossRef] [PubMed]
- Robson, M.; Im, S.A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A.; et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N. Engl. J. Med. 2017, 377, 523–533. [Google Scholar] [CrossRef]
- Litton, J.K.; Rugo, H.S.; Ettl, J.; Hurvitz, S.A.; Goncalves, A.; Lee, K.H.; Fehrenbacher, L.; Yerushalmi, R.; Mina, L.A.; Martin, M.; et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N. Engl. J. Med. 2018, 379, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Modi, S.; Saura, C.; Yamashita, T.; Park, Y.H.; Kim, S.B.; Tamura, K.; Andre, F.; Iwata, H.; Ito, Y.; Tsurutani, J.; et al. Trastuzumab Deruxtecan in Previously Treated HER2-Positive Breast Cancer. N. Engl. J. Med. 2020, 382, 610–621. [Google Scholar] [CrossRef]
- Modi, S.; Park, H.; Murthy, R.K.; Iwata, H.; Tamura, K.; Tsurutani, J.; Moreno-Aspitia, A.; Doi, T.; Sagara, Y.; Redfern, C.; et al. Antitumor Activity and Safety of Trastuzumab Deruxtecan in Patients With HER2-Low-Expressing Advanced Breast Cancer: Results From a Phase Ib Study. J. Clin. Oncol. 2020, 38, 1887–1896. [Google Scholar] [CrossRef]
- Mieog, J.S.; van der Hage, J.A.; van de Velde, C.J. Preoperative chemotherapy for women with operable breast cancer. Cochrane Database Syst. Rev. 2007, 2007, CD005002. [Google Scholar] [CrossRef] [PubMed]
- Carey, L.A.; Dees, E.C.; Sawyer, L.; Gatti, L.; Moore, D.T.; Collichio, F.; Ollila, D.W.; Sartor, C.I.; Graham, M.L.; Perou, C.M. The triple negative paradox: Primary tumor chemosensitivity of breast cancer subtypes. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2007, 13, 2329–2334. [Google Scholar] [CrossRef] [Green Version]
- Liedtke, C.; Mazouni, C.; Hess, K.R.; Andre, F.; Tordai, A.; Mejia, J.A.; Symmans, W.F.; Gonzalez-Angulo, A.M.; Hennessy, B.; Green, M.; et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J. Clin. Oncol. 2008, 26, 1275–1281. [Google Scholar] [CrossRef]
- Straver, M.E.; Glas, A.M.; Hannemann, J.; Wesseling, J.; van de Vijver, M.J.; Rutgers, E.J.T.; Peeters, M.J.T.F.D.V.; van Tinteren, H.; van’t Veer, L.J.; Rodenhuis, S. The 70-gene signature as a response predictor for neoadjuvant chemotherapy in breast cancer. Breast Cancer Res. Treat. 2010, 119, 551–558. [Google Scholar] [CrossRef] [Green Version]
- Filho, O.M.; Stover, D.G.; Asad, S.; Ansell, P.J.; Watson, M.; Loibl, S.; Geyer, C.E., Jr.; Bae, J.; Collier, K.; Cherian, M.; et al. Association of Immunophenotype With Pathologic Complete Response to Neoadjuvant Chemotherapy for Triple-Negative Breast Cancer: A Secondary Analysis of the BrighTNess Phase 3 Randomized Clinical Trial. JAMA Oncol. 2021, 7, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Orozco, J.I.J.; Grumley, J.G.; Matsuba, C.; Manughian-Peter, A.O.; Chang, S.C.; Chang, G.; Gago, F.E.; Salomon, M.P.; Marzese, D.M. Clinical Implications of Transcriptomic Changes After Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer. Ann. Surg. Oncol. 2019, 26, 3185–3193. [Google Scholar] [CrossRef]
- Zhao, Y.; Schaafsma, E.; Cheng, C. Gene signature-based prediction of triple-negative breast cancer patient response to Neoadjuvant chemotherapy. Cancer Med. 2020, 9, 6281–6295. [Google Scholar] [CrossRef] [PubMed]
- Perez-Pena, J.; Tibor Fekete, J.; Paez, R.; Baliu-Pique, M.; Garcia-Saenz, J.A.; Garcia-Barberan, V.; Manzano, A.; Perez-Segura, P.; Esparis-Ogando, A.; Pandiella, A.; et al. A Transcriptomic Immunologic Signature Predicts Favorable Outcome in Neoadjuvant Chemotherapy Treated Triple Negative Breast Tumors. Front. Immunol. 2019, 10, 2802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fournier, M.V.; Goodwin, E.C.; Chen, J.; Obenauer, J.C.; Tannenbaum, S.H.; Brufsky, A.M. A Predictor of Pathological Complete Response to Neoadjuvant Chemotherapy Stratifies Triple Negative Breast Cancer Patients with High Risk of Recurrence. Sci. Rep. 2019, 9, 14863. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.K.; Park, A.K.; Ko, E.; Park, W.Y.; Lee, K.M.; Noh, D.Y.; Han, W. Risk stratification of triple-negative breast cancer with core gene signatures associated with chemoresponse and prognosis. Breast Cancer Res. Treat. 2019, 178, 185–197. [Google Scholar] [CrossRef]
- Vishnubalaji, R.; Abdel-Razeq, H.; Gehani, S.; Albagha, O.M.E.; Alajez, N.M. Identification of a Gene Panel Predictive of Triple-Negative Breast Cancer Response to Neoadjuvant Chemotherapy Employing Transcriptomic and Functional Validation. Int. J. Mol. Sci. 2022, 23, 10901. [Google Scholar] [CrossRef]
- Prat, A.; Lluch, A.; Albanell, J.; Barry, W.T.; Fan, C.; Chacon, J.I.; Parker, J.S.; Calvo, L.; Plazaola, A.; Arcusa, A.; et al. Predicting response and survival in chemotherapy-treated triple-negative breast cancer. Br. J. Cancer 2014, 111, 1532–1541. [Google Scholar] [CrossRef] [Green Version]
- Sinn, B.V.; Loibl, S.; Hanusch, C.A.; Zahm, D.M.; Sinn, H.P.; Untch, M.; Weber, K.; Karn, T.; Becker, C.; Marme, F.; et al. Immune-related Gene Expression Predicts Response to Neoadjuvant Chemotherapy but not Additional Benefit from PD-L1 Inhibition in Women with Early Triple-negative Breast Cancer. Clin. Cancer Res. 2021, 27, 2584–2591. [Google Scholar] [CrossRef]
- Jovanovic, B.; Mayer, I.A.; Mayer, E.L.; Abramson, V.G.; Bardia, A.; Sanders, M.E.; Kuba, M.G.; Estrada, M.V.; Beeler, J.S.; Shaver, T.M.; et al. A Randomized Phase II Neoadjuvant Study of Cisplatin, Paclitaxel With or Without Everolimus in Patients with Stage II/III Triple-Negative Breast Cancer (TNBC): Responses and Long-term Outcome Correlated with Increased Frequency of DNA Damage Response Gene Mutations, TNBC Subtype, AR Status, and Ki67. Clin. Cancer Res. 2017, 23, 4035–4045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCall, M.N.; Bolstad, B.M.; Irizarry, R.A. Frozen robust multiarray analysis (fRMA). Biostatistics 2010, 11, 242–253. [Google Scholar] [CrossRef] [Green Version]
- Karn, T.; Metzler, D.; Ruckhaberle, E.; Hanker, L.; Gatje, R.; Solbach, C.; Ahr, A.; Schmidt, M.; Holtrich, U.; Kaufmann, M.; et al. Data-driven derivation of cutoffs from a pool of 3,030 Affymetrix arrays to stratify distinct clinical types of breast cancer. Breast Cancer Res. Treat. 2010, 120, 567–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riley, R.D.; Higgins, J.P.; Deeks, J.J. Interpretation of random effects meta-analyses. BMJ 2011, 342, d549. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Boyle, E.I.; Weng, S.; Gollub, J.; Jin, H.; Botstein, D.; Cherry, J.M.; Sherlock, G. GO::TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 2004, 20, 3710–3715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef] [Green Version]
- Orchard, S.; Kerrien, S.; Abbani, S.; Aranda, B.; Bhate, J.; Bidwell, S.; Bridge, A.; Briganti, L.; Brinkman, F.S.; Cesareni, G.; et al. Protein interaction data curation: The International Molecular Exchange (IMEx) consortium. Nat. Methods 2012, 9, 345–350. [Google Scholar] [CrossRef]
- von Minckwitz, G.; Untch, M.; Blohmer, J.U.; Costa, S.D.; Eidtmann, H.; Fasching, P.A.; Gerber, B.; Eiermann, W.; Hilfrich, J.; Huober, J.; et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J. Clin. Oncol. 2012, 30, 1796–1804. [Google Scholar] [CrossRef] [Green Version]
- Denkert, C.; von Minckwitz, G.; Brase, J.C.; Sinn, B.V.; Gade, S.; Kronenwett, R.; Pfitzner, B.M.; Salat, C.; Loi, S.; Schmitt, W.D.; et al. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J. Clin. Oncol. 2015, 33, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Asad, S.; Weber, Z.; Tallman, D.; Nock, W.; Wyse, M.; Bey, J.F.; Dean, K.L.; Adams, E.J.; Stockard, S.; et al. Genomic features of rapid versus late relapse in triple negative breast cancer. BMC Cancer 2021, 21, 568. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, J.H.; Ballman, K.; Polley, M.C.; Campbell, J.D.; Fan, C.; Selitsky, S.; Fernandez-Martinez, A.; Parker, J.S.; Hoadley, K.A.; Hu, Z.; et al. CALGB 40603 (Alliance): Long-Term Outcomes and Genomic Correlates of Response and Survival After Neoadjuvant Chemotherapy With or Without Carboplatin and Bevacizumab in Triple-Negative Breast Cancer. J. Clin. Oncol. 2022, 40, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Koedoot, E.; van Steijn, E.; Vermeer, M.; Gonzalez-Prieto, R.; Vertegaal, A.C.O.; Martens, J.W.M.; Le Devedec, S.E.; van de Water, B. Splicing factors control triple-negative breast cancer cell mitosis through SUN2 interaction and sororin intron retention. J. Exp. Clin. Cancer Res. 2021, 40, 82. [Google Scholar] [CrossRef] [PubMed]
- Frigyesi, I.; Adolfsson, J.; Ali, M.; Christophersen, M.K.; Johnsson, E.; Turesson, I.; Gullberg, U.; Hansson, M.; Nilsson, B. Robust isolation of malignant plasma cells in multiple myeloma. Blood 2014, 123, 1336–1340. [Google Scholar] [CrossRef] [Green Version]
- Vishnubalaji, R.; Alajez, N.M. Transcriptional landscape associated with TNBC resistance to neoadjuvant chemotherapy revealed by single-cell RNA-seq. Mol. Ther. Oncolytics 2021, 23, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Huntoon, K.; Lee, D.; Wang, Y.; Ha, J.; Qie, Y.; Li, X.; Schrank, B.R.; Dong, S.; Gallup, T.D.; et al. Immunological conversion of solid tumours using a bispecific nanobioconjugate for cancer immunotherapy. Nat. Nanotechnol. 2022, 17, 1332–1341. [Google Scholar] [CrossRef]
- Roy, P.G.; Thompson, A.M. Cyclin D1 and breast cancer. Breast 2006, 15, 718–727. [Google Scholar] [CrossRef]
- Montalto, F.I.; De Amicis, F. Cyclin D1 in Cancer: A Molecular Connection for Cell Cycle Control, Adhesion and Invasion in Tumor and Stroma. Cells 2020, 9, 2648. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Huang, Y.; Gao, X.; Li, Y.; Lin, J.; Chen, L.; Chang, L.; Chen, G.; Guan, Y.; Pan, L.K.; et al. CCND1 Amplification Contributes to Immunosuppression and Is Associated With a Poor Prognosis to Immune Checkpoint Inhibitors in Solid Tumors. Front. Immunol. 2020, 11, 1620. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, A.; Lindstrom, L.S.; Li, J.; Harrell, J.C.; Darai-Ramqvist, E.; Sifakis, E.G.; Foukakis, T.; Perou, C.M.; Czene, K.; Bergh, J.; et al. The long-term prognostic and predictive capacity of cyclin D1 gene amplification in 2305 breast tumours. Breast Cancer Res. 2019, 21, 34. [Google Scholar] [CrossRef] [Green Version]
- Reis-Filho, J.S.; Savage, K.; Lambros, M.B.; James, M.; Steele, D.; Jones, R.L.; Dowsett, M. Cyclin D1 protein overexpression and CCND1 amplification in breast carcinomas: An immunohistochemical and chromogenic in situ hybridisation analysis. Mod. Pathol. 2006, 19, 999–1009. [Google Scholar] [CrossRef] [Green Version]
- Jeffreys, S.A.; Becker, T.M.; Khan, S.; Soon, P.; Neubauer, H.; de Souza, P.; Powter, B. Prognostic and Predictive Value of CCND1/Cyclin D1 Amplification in Breast Cancer With a Focus on Postmenopausal Patients: A Systematic Review and Meta-Analysis. Front. Endocrinol. 2022, 13, 895729. [Google Scholar] [CrossRef]
- Valla, M.; Klaestad, E.; Ytterhus, B.; Bofin, A.M. CCND1 Amplification in Breast Cancer -associations With Proliferation, Histopathological Grade, Molecular Subtype and Prognosis. J. Mammary Gland. Biol. Neoplasia 2022, 27, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Hoogstraat, M.; Lips, E.H.; Mayayo-Peralta, I.; Mulder, L.; Kristel, P.; van der Heijden, I.; Annunziato, S.; van Seijen, M.; Nederlof, P.M.; Sonke, G.S.; et al. Comprehensive characterization of pre- and post-treatment samples of breast cancer reveal potential mechanisms of chemotherapy resistance. NPJ Breast Cancer 2022, 8, 60. [Google Scholar] [CrossRef]
- Echavarria, I.; Lopez-Tarruella, S.; Picornell, A.; Garcia-Saenz, J.A.; Jerez, Y.; Hoadley, K.; Gomez, H.L.; Moreno, F.; Monte-Millan, M.D.; Marquez-Rodas, I.; et al. Pathological Response in a Triple-Negative Breast Cancer Cohort Treated with Neoadjuvant Carboplatin and Docetaxel According to Lehmann’s Refined Classification. Clin. Cancer Res. 2018, 24, 1845–1852. [Google Scholar] [CrossRef] [Green Version]
- Santonja, A.; Sanchez-Munoz, A.; Lluch, A.; Chica-Parrado, M.R.; Albanell, J.; Chacon, J.I.; Antolin, S.; Jerez, J.M.; de la Haba, J.; de Luque, V.; et al. Triple negative breast cancer subtypes and pathologic complete response rate to neoadjuvant chemotherapy. Oncotarget 2018, 9, 26406–26416. [Google Scholar] [CrossRef] [Green Version]
- Qu, Q.; Mao, Y.; Fei, X.C.; Shen, K.W. The impact of androgen receptor expression on breast cancer survival: A retrospective study and meta-analysis. PLoS ONE 2013, 8, e82650. [Google Scholar] [CrossRef] [Green Version]
- Vera-Badillo, F.E.; Templeton, A.J.; de Gouveia, P.; Diaz-Padilla, I.; Bedard, P.L.; Al-Mubarak, M.; Seruga, B.; Tannock, I.F.; Ocana, A.; Amir, E. Androgen receptor expression and outcomes in early breast cancer: A systematic review and meta-analysis. J. Natl. Cancer Inst. 2014, 106, djt319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutta, S.W.; Volaric, A.; Morgan, J.T.; Chinn, Z.; Atkins, K.A.; Janowski, E.M. Pathologic Evaluation and Prognostic Implications of Nodal Micrometastases in Breast Cancer. Semin. Radiat. Oncol. 2019, 29, 102–110. [Google Scholar] [CrossRef]
- Houvenaeghel, G.; Sabatier, R.; Reyal, F.; Classe, J.M.; Giard, S.; Charitansky, H.; Rouzier, R.; Faure, C.; Garbay, J.R.; Darai, E.; et al. Axillary lymph node micrometastases decrease triple-negative early breast cancer survival. Br. J. Cancer 2016, 115, 1024–1031. [Google Scholar] [CrossRef] [Green Version]
- Prat, A.; Cruz, C.; Hoadley, K.A.; Diez, O.; Perou, C.M.; Balmana, J. Molecular features of the basal-like breast cancer subtype based on BRCA1 mutation status. Breast Cancer Res. Treat. 2014, 147, 185–191. [Google Scholar] [CrossRef] [Green Version]
- Paul, M.R.; Pan, T.C.; Pant, D.K.; Shih, N.N.; Chen, Y.; Harvey, K.L.; Solomon, A.; Lieberman, D.; Morrissette, J.J.; Soucier-Ernst, D.; et al. Genomic landscape of metastatic breast cancer identifies preferentially dysregulated pathways and targets. J. Clin. Investig. 2020, 130, 4252–4265. [Google Scholar] [CrossRef]
- Loibl, S.; Untch, M.; Burchardi, N.; Huober, J.; Sinn, B.V.; Blohmer, J.U.; Grischke, E.M.; Furlanetto, J.; Tesch, H.; Hanusch, C.; et al. A randomised phase II study investigating durvalumab in addition to an anthracycline taxane-based neoadjuvant therapy in early triple-negative breast cancer: Clinical results and biomarker analysis of GeparNuevo study. Ann. Oncol. 2019, 30, 1279–1288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agostinetto, E.; Gligorov, J.; Piccart, M. Systemic therapy for early-stage breast cancer: Learning from the past to build the future. Nat. Rev. Clin. Oncol. 2022, 19, 763–774. [Google Scholar] [CrossRef]
- Symmans, W.F.; Wei, C.; Gould, R.; Yu, X.; Zhang, Y.; Liu, M.; Walls, A.; Bousamra, A.; Ramineni, M.; Sinn, B.; et al. Long-Term Prognostic Risk After Neoadjuvant Chemotherapy Associated With Residual Cancer Burden and Breast Cancer Subtype. J. Clin. Oncol. 2017, 35, 1049–1060. [Google Scholar] [CrossRef] [Green Version]
- Sammut, S.J.; Crispin-Ortuzar, M.; Chin, S.F.; Provenzano, E.; Bardwell, H.A.; Ma, W.; Cope, W.; Dariush, A.; Dawson, S.J.; Abraham, J.E.; et al. Multi-omic machine learning predictor of breast cancer therapy response. Nature 2022, 601, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Vanguri, R.S.; Luo, J.; Aukerman, A.T.; Egger, J.V.; Fong, C.J.; Horvat, N.; Pagano, A.; Araujo-Filho, J.A.B.; Geneslaw, L.; Rizvi, H.; et al. Multimodal integration of radiology, pathology and genomics for prediction of response to PD-(L)1 blockade in patients with non-small cell lung cancer. Nat. Cancer 2022, 3, 1151–1164. [Google Scholar] [CrossRef]
- Chen, R.J.; Lu, M.Y.; Williamson, D.F.K.; Chen, T.Y.; Lipkova, J.; Noor, Z.; Shaban, M.; Shady, M.; Williams, M.; Joo, B.; et al. Pan-cancer integrative histology-genomic analysis via multimodal deep learning. Cancer Cell 2022, 40, 865–878. [Google Scholar] [CrossRef] [PubMed]
- Boehm, K.M.; Aherne, E.A.; Ellenson, L.; Nikolovski, I.; Alghamdi, M.; Vazquez-Garcia, I.; Zamarin, D.; Long Roche, K.; Liu, Y.; Patel, D.; et al. Multimodal data integration using machine learning improves risk stratification of high-grade serous ovarian cancer. Nat. Cancer 2022, 3, 723–733. [Google Scholar] [CrossRef]
- Lara-Medina, F.; Perez-Sanchez, V.; Saavedra-Perez, D.; Blake-Cerda, M.; Arce, C.; Motola-Kuba, D.; Villarreal-Garza, C.; Gonzalez-Angulo, A.M.; Bargallo, E.; Aguilar, J.L.; et al. Triple-negative breast cancer in Hispanic patients: High prevalence, poor prognosis, and association with menopausal status, body mass index, and parity. Cancer 2011, 117, 3658–3669. [Google Scholar] [CrossRef]
- Lin, N.U.; Vanderplas, A.; Hughes, M.E.; Theriault, R.L.; Edge, S.B.; Wong, Y.N.; Blayney, D.W.; Niland, J.C.; Winer, E.P.; Weeks, J.C. Clinicopathologic features, patterns of recurrence, and survival among women with triple-negative breast cancer in the National Comprehensive Cancer Network. Cancer 2012, 118, 5463–5472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Killelea, B.K.; Yang, V.Q.; Wang, S.Y.; Hayse, B.; Mougalian, S.; Horowitz, N.R.; Chagpar, A.B.; Pusztai, L.; Lannin, D.R. Racial Differences in the Use and Outcome of Neoadjuvant Chemotherapy for Breast Cancer: Results From the National Cancer Data Base. J. Clin. Oncol. 2015, 33, 4267–4276. [Google Scholar] [CrossRef] [PubMed]
- Dietze, E.C.; Sistrunk, C.; Miranda-Carboni, G.; O’Regan, R.; Seewaldt, V.L. Triple-negative breast cancer in African-American women: Disparities versus biology. Nat. Rev. Cancer 2015, 15, 248–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]

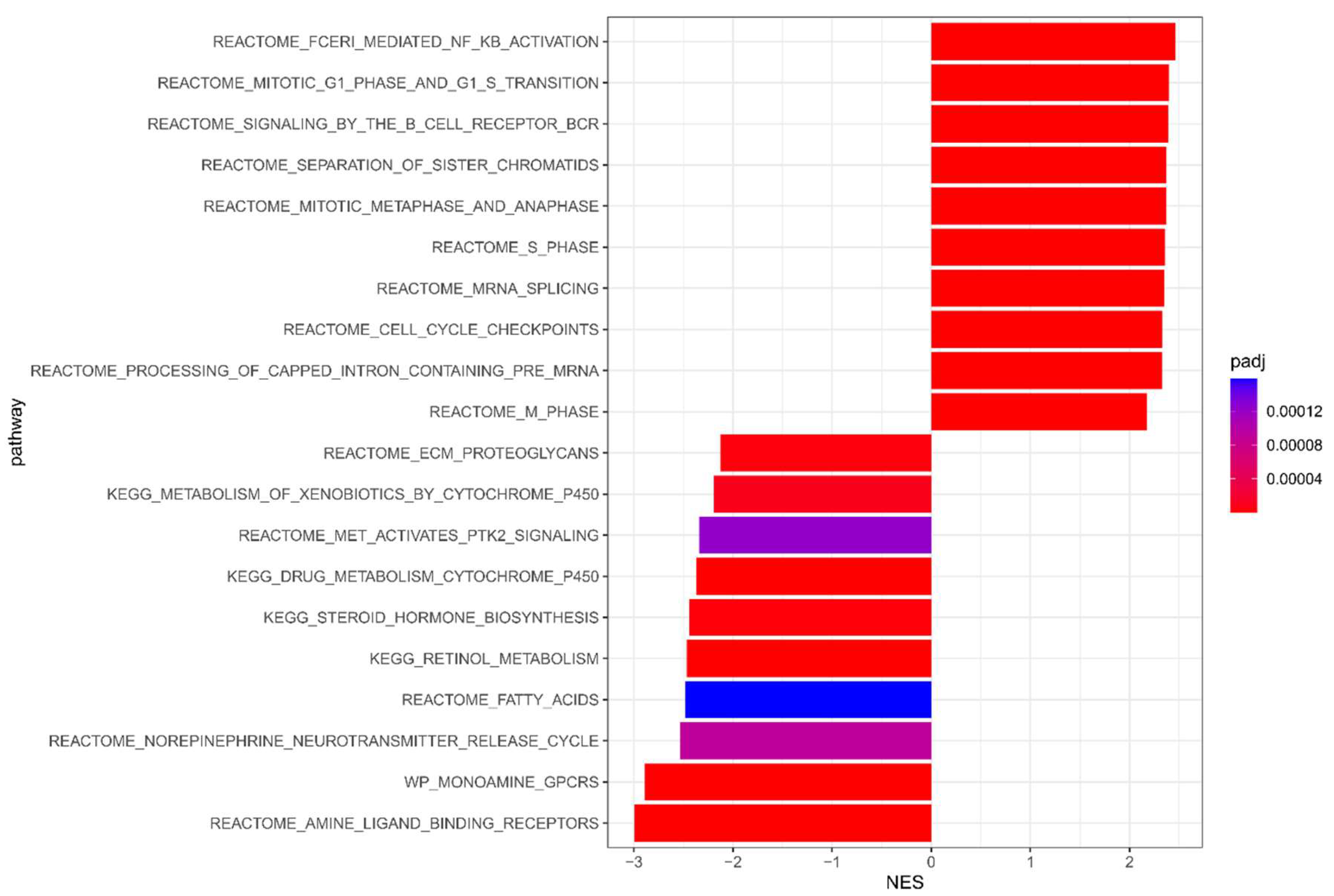
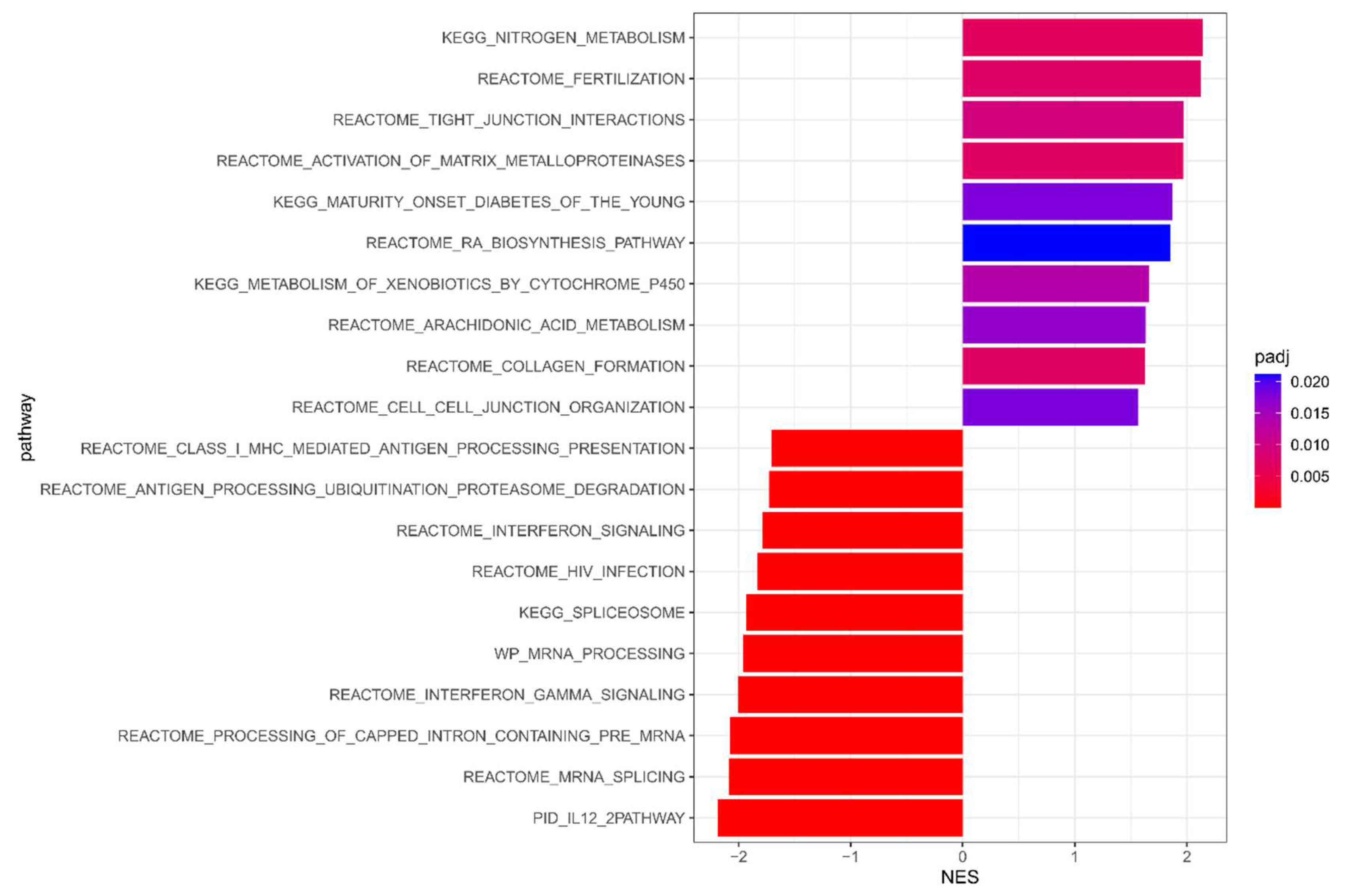
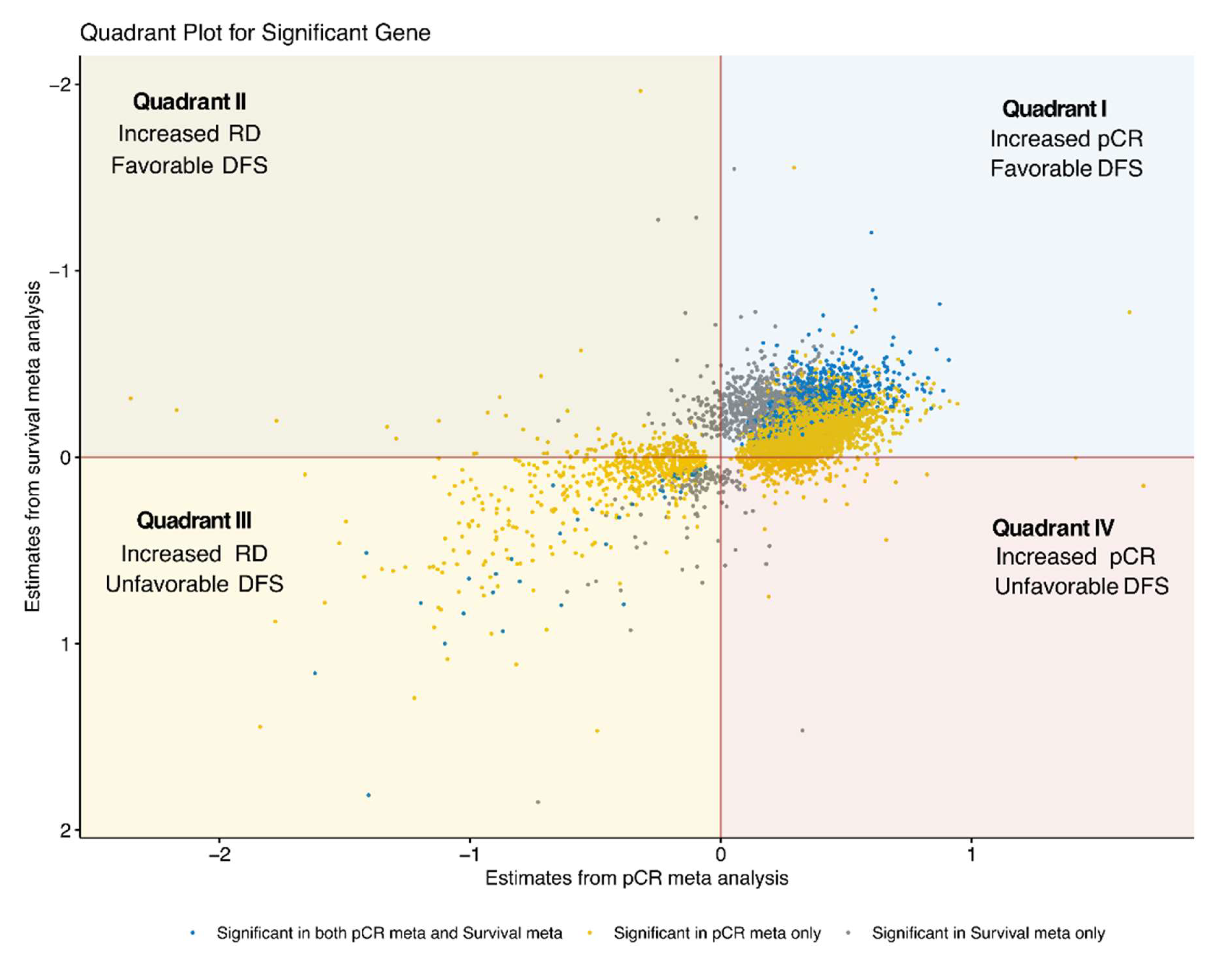

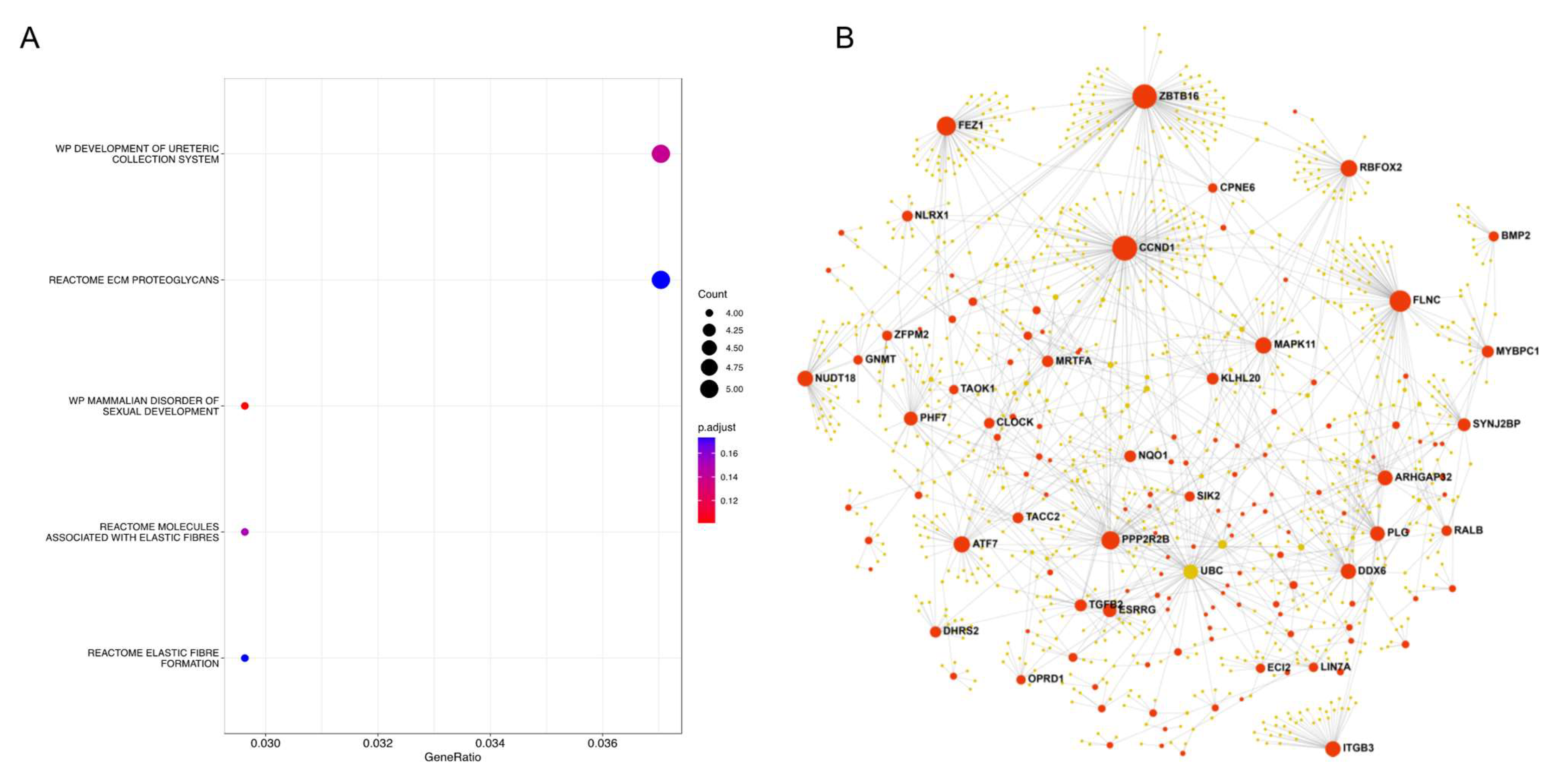

| Datasets | Type | Numbers of Sample | pCR | DSF Event | Median Follow-Up (sd, Range), Years | Mean Age (sd, Range) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | TNBC | Yes | No | NA | Yes | No | ||||
| GSE16446 | Microarray | 120 | 86 | 10 | 73 | 3 | 18 | 68 | 2.9 (1.5, 0.2–5.9) | / |
| GSE20271 | Microarray | 178 | 59 | 13 | 46 | / | / | / | / | 51 (10.8, 29–74) |
| GSE22093 | Microarray | 103 | 39 | 16 | 23 | / | / | / | / | 46.7 (10.2, 31–67) |
| GSE18864 | Microarray | 84 | 24 | 8 | 16 | / | / | / | / | 49.8 (10.3, 29–68) |
| GSE20194 | Microarray | 278 | 71 | 25 | 46 | / | / | / | / | 50.4 (10.8, 29–75) |
| GSE25055 | Microarray | 310 | 119 | 40 | 78 | 1 | 37 | 82 | 2.1 (1.7, 0.1–7.4) | 49.6 (10.8, 28–75) |
| GSE25065 | Microarray | 198 | 63 | 22 | 36 | 5 | 22 | 41 | 2.8 (1.5, 0.4–6.1) | 49.1 (11.2, 24–72) |
| GSE18728 | Microarray | 61 | 22 | 6 | 16 | / | / | / | / | / |
| GSE41998 | Microarray | 279 | 145 | 47 | 83 | 15 | / | / | / | / |
| GSE32646 | Microarray | 115 | 26 | 10 | 16 | / | / | / | / | 54.4 (12.5, 28–70) |
| GSE164458 | RNA-Seq | 482 | 482 | 236 | 246 | / | / | / | / | / |
| VUMC | RNA-Seq | 145 | 45 | 29 | 16 | / | 8 | 37 | 2.6 (0.8, 0.7–5.8) | 50.6 (10.8, 29–75) |
| GSE154524 | RNA-Seq | 389 | 389 | 210 | 179 | / | 115 | 274 | 5.3 (2, 0.1–8.1) | / |
| GSE22226 | RNA-Seq | 150 | 27 | 7 | 20 | / | 11 | 16 | 3.6 (2.2, 0.5–6.7) | 47 (8, 33.5–63.2) |
| GSE192341 | RNA-Seq | 87 | 52 | 21 | 29 | 2 | 10 | 42 | 1.8 (1.2, 0.4–4.2) | 44.6 (11.7, 26–73) |
| GSE163882 | RNA-Seq | 222 | 90 | 38 | 52 | / | / | / | / | 54.1 (11.5, 23–74) |
| Gene | Odds Ratio | Estimate | se | z-Value | p-Value | FDR | Direction * | Number of Datasets |
|---|---|---|---|---|---|---|---|---|
| Positive associated genes | ||||||||
| SLAMF7 | 1.419 | 0.350 | 0.045 | 7.792 | 6.62 × 10−15 | 2.95 × 10−10 | +++++-++++-+-+++ | 16 |
| GBP5 | 1.398 | 0.335 | 0.043 | 7.736 | 1.02 × 10−14 | 2.95 × 10−10 | +++++-+-++ | 10 |
| GZMB | 1.356 | 0.305 | 0.041 | 7.461 | 8.59 × 10−14 | 1.65 × 10−9 | ++++-+++++++-+++ | 16 |
| CD274 | 1.590 | 0.464 | 0.063 | 7.404 | 1.32 × 10−13 | 1.91 × 10−9 | +++++++-++ | 10 |
| GBP4 | 1.474 | 0.388 | 0.054 | 7.189 | 6.54 × 10−13 | 7.56 × 10−9 | +++++++-++ | 10 |
| NUP210 | 1.599 | 0.470 | 0.067 | 6.972 | 3.14 × 10−12 | 3.02 × 10−8 | ++++--++++++++++ | 16 |
| NOL7 | 2.017 | 0.702 | 0.101 | 6.947 | 3.73 × 10−12 | 3.08 × 10−8 | ++++-+++++++++++ | 16 |
| MCM5 | 1.749 | 0.559 | 0.082 | 6.843 | 7.76 × 10−12 | 5.61 × 10−8 | +++-+++++++++++ | 15 |
| NKG7 | 1.348 | 0.298 | 0.044 | 6.809 | 9.84 × 10−12 | 6.32 × 10−8 | ++++++++++--+++ | 15 |
| CXCL10 | 1.244 | 0.218 | 0.032 | 6.775 | 1.25 × 10−11 | 7.20 × 10−8 | +++++-+++++-+++ | 15 |
| Negative associated genes | ||||||||
| CCND1 | 0.770 | −0.261 | 0.046 | −5.695 | 1.24 × 10−8 | 7.85 × 10−6 | -+-+-+---------- | 16 |
| LINC00622 | 0.635 | −0.454 | 0.086 | −5.260 | 1.44 × 10−7 | 4.49 × 10−5 | ---+---+ | 8 |
| CTSF | 0.683 | −0.381 | 0.074 | −5.124 | 2.98 × 10−7 | 8.13 × 10−5 | ----+---++-++--+ | 16 |
| SLC4A11 | 0.814 | −0.206 | 0.044 | −4.638 | 3.51 × 10−6 | 5.23 × 10−4 | ----+--+-- | 10 |
| ZNF697 | 0.651 | −0.429 | 0.095 | −4.535 | 5.77 × 10−6 | 7.57 × 10−4 | --------++ | 10 |
| AHNAK2 | 0.825 | −0.193 | 0.043 | −4.500 | 6.80 × 10−6 | 8.68 × 10−4 | -+--+--+-------+ | 16 |
| OPLAH | 0.800 | −0.223 | 0.052 | −4.260 | 2.05 × 10−5 | 2.04 × 10−3 | ----++---------+ | 16 |
| MAPRE3 | 0.702 | −0.353 | 0.086 | −4.089 | 4.34 × 10−5 | 3.69 × 10−3 | -+--+----------+ | 16 |
| MGAM | 0.767 | −0.266 | 0.066 | −4.047 | 5.19 × 10−5 | 4.26 × 10−3 | --------------+ | 15 |
| MSX1 | 0.755 | −0.282 | 0.071 | −3.971 | 7.15 × 10−5 | 5.29 × 10−3 | ---+-----------+ | 16 |
| Gene | Hazard Ratio | Estimate | se | z-Value | p-Value | FDR | Direction * | Number of Datasets |
|---|---|---|---|---|---|---|---|---|
| Positive associated genes | ||||||||
| MST1L | 1.367 | 0.312 | 0.078 | 3.985 | 6.74 × 10−5 | 8.70 × 10−2 | +-++ | 4 |
| LINC00905 | 2.043 | 0.715 | 0.212 | 3.369 | 7.56 × 10−4 | 2.55 × 10−1 | +++ | 3 |
| MYBPH | 1.165 | 0.153 | 0.048 | 3.163 | 1.56 × 10−3 | 3.13 × 10−1 | +-+-+++ | 7 |
| FAM20C | 1.372 | 0.316 | 0.100 | 3.151 | 1.63 × 10−3 | 3.16 × 10−1 | +++-+ | 5 |
| CLDN16 | 1.145 | 0.135 | 0.043 | 3.120 | 1.81 × 10−3 | 3.35 × 10−1 | +-++++ | 6 |
| SCGB2A1 | 1.098 | 0.093 | 0.030 | 3.090 | 2.00 × 10−3 | 3.46 × 10−1 | +++++++ | 7 |
| TGM5 | 1.132 | 0.124 | 0.041 | 3.045 | 2.33 × 10−3 | 3.46 × 10−1 | ++++++ | 6 |
| VPS50 | 2.202 | 0.790 | 0.265 | 2.985 | 2.84 × 10−3 | 3.46 × 10−1 | +++- | 4 |
| LINC00032 | 1.302 | 0.264 | 0.090 | 2.933 | 3.35 × 10−3 | 3.69 × 10−1 | +-+ | 3 |
| TBC1D21 | 4.330 | 1.466 | 0.505 | 2.902 | 3.70 × 10−3 | 3.77 × 10−1 | +-+ | 3 |
| Negative associated genes | ||||||||
| PPP1R12A | 0.628 | −0.465 | 0.107 | −4.331 | 1.48 × 10−5 | 8.70 × 10−2 | ------- | 7 |
| AKAP5 | 0.627 | −0.467 | 0.110 | −4.236 | 2.28 × 10−5 | 8.70 × 10−2 | ---+-- | 6 |
| SLAMF7 | 0.818 | −0.200 | 0.049 | −4.077 | 4.57 × 10−5 | 8.70 × 10−2 | ------- | 7 |
| VEZF1 | 0.640 | −0.446 | 0.110 | −4.075 | 4.61 × 10−5 | 8.70 × 10−2 | ------- | 7 |
| OGG1 | 0.586 | −0.534 | 0.132 | −4.060 | 4.90 × 10−5 | 8.70 × 10−2 | -----+- | 7 |
| LRRK2 | 0.651 | −0.429 | 0.106 | −4.048 | 5.16 × 10−5 | 8.70 × 10−2 | ----- | 5 |
| RBMXL1 | 0.596 | −0.517 | 0.131 | −3.943 | 8.03 × 10−5 | 8.70 × 10−2 | ---+- | 5 |
| HERPUD1 | 0.670 | −0.401 | 0.104 | −3.837 | 1.25 × 10−4 | 9.95 × 10−2 | ------ | 6 |
| PDCD1LG2 | 0.729 | −0.316 | 0.083 | −3.795 | 1.48 × 10−4 | 1.12 × 10−1 | ---+--- | 7 |
| TXNDC5 | 0.639 | −0.448 | 0.118 | −3.792 | 1.49 × 10−4 | 1.12 × 10−1 | --- | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Li, E.; Wang, L.; Lehmann, B.D.; Chen, X.S. Transcriptome Meta-Analysis of Triple-Negative Breast Cancer Response to Neoadjuvant Chemotherapy. Cancers 2023, 15, 2194. https://doi.org/10.3390/cancers15082194
Zhang W, Li E, Wang L, Lehmann BD, Chen XS. Transcriptome Meta-Analysis of Triple-Negative Breast Cancer Response to Neoadjuvant Chemotherapy. Cancers. 2023; 15(8):2194. https://doi.org/10.3390/cancers15082194
Chicago/Turabian StyleZhang, Wei, Emma Li, Lily Wang, Brian D. Lehmann, and X. Steven Chen. 2023. "Transcriptome Meta-Analysis of Triple-Negative Breast Cancer Response to Neoadjuvant Chemotherapy" Cancers 15, no. 8: 2194. https://doi.org/10.3390/cancers15082194
APA StyleZhang, W., Li, E., Wang, L., Lehmann, B. D., & Chen, X. S. (2023). Transcriptome Meta-Analysis of Triple-Negative Breast Cancer Response to Neoadjuvant Chemotherapy. Cancers, 15(8), 2194. https://doi.org/10.3390/cancers15082194







