High VEGFA Expression Is Associated with Improved Progression-Free Survival after Bevacizumab Treatment in Recurrent Glioblastoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Immunohistochemistry Analysis in Human Tissue Samples
2.2.1. MMP-2, MMP-9 and VEGFA Immunohistochemistry
2.2.2. YKL40 Immunohistochemistry
2.2.3. Immunohistochemistry Evaluation and Scoring Method
2.3. Data and Statistical Analysis
3. Results
3.1. Evaluation of the Expression of Secretome-Associated Proteins in GB Patient Samples
3.1.1. Clinicopathological Features of GB Patients
3.1.2. IHC Analysis of MMP-2, MMP-9, VEGFA, and YKL40 Expression in GB Patients’ Tumor Tissues
3.1.3. Association of Protein Expression in GB Tissues with Patients’ Clinicopathologic Features and Survival Data
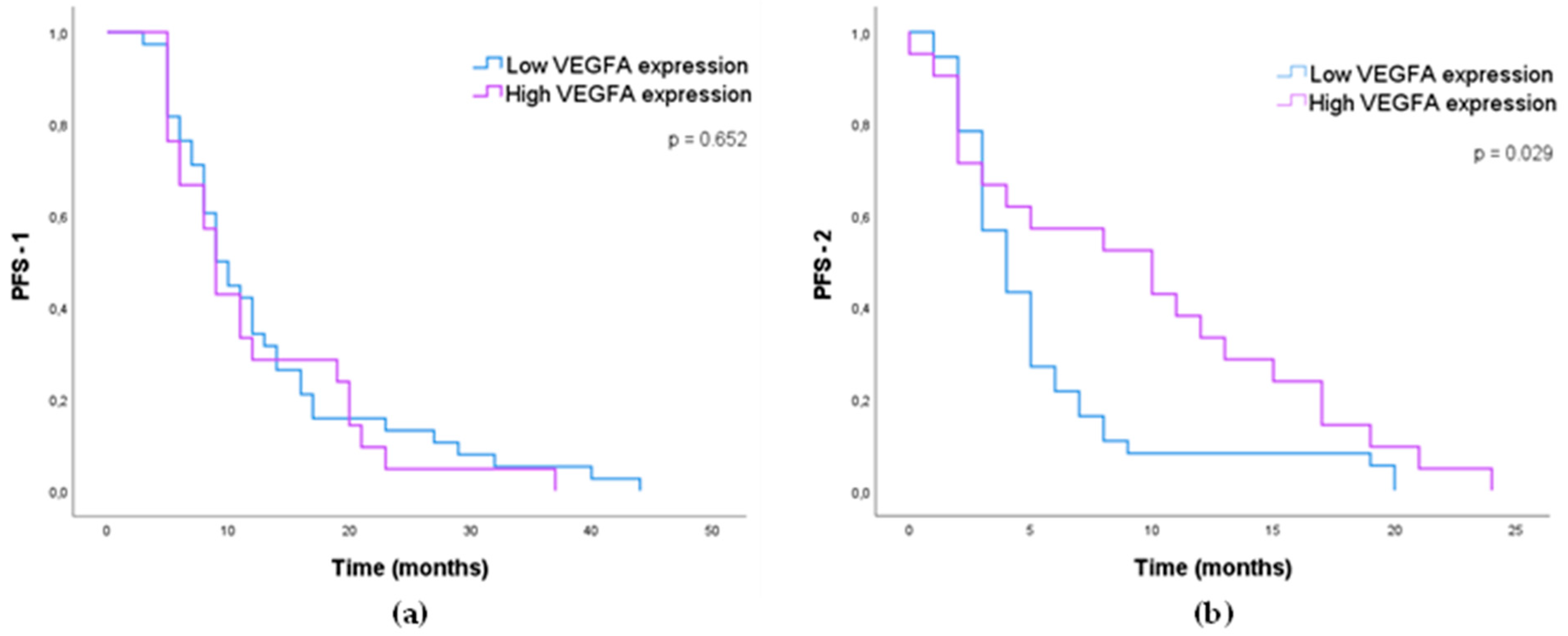
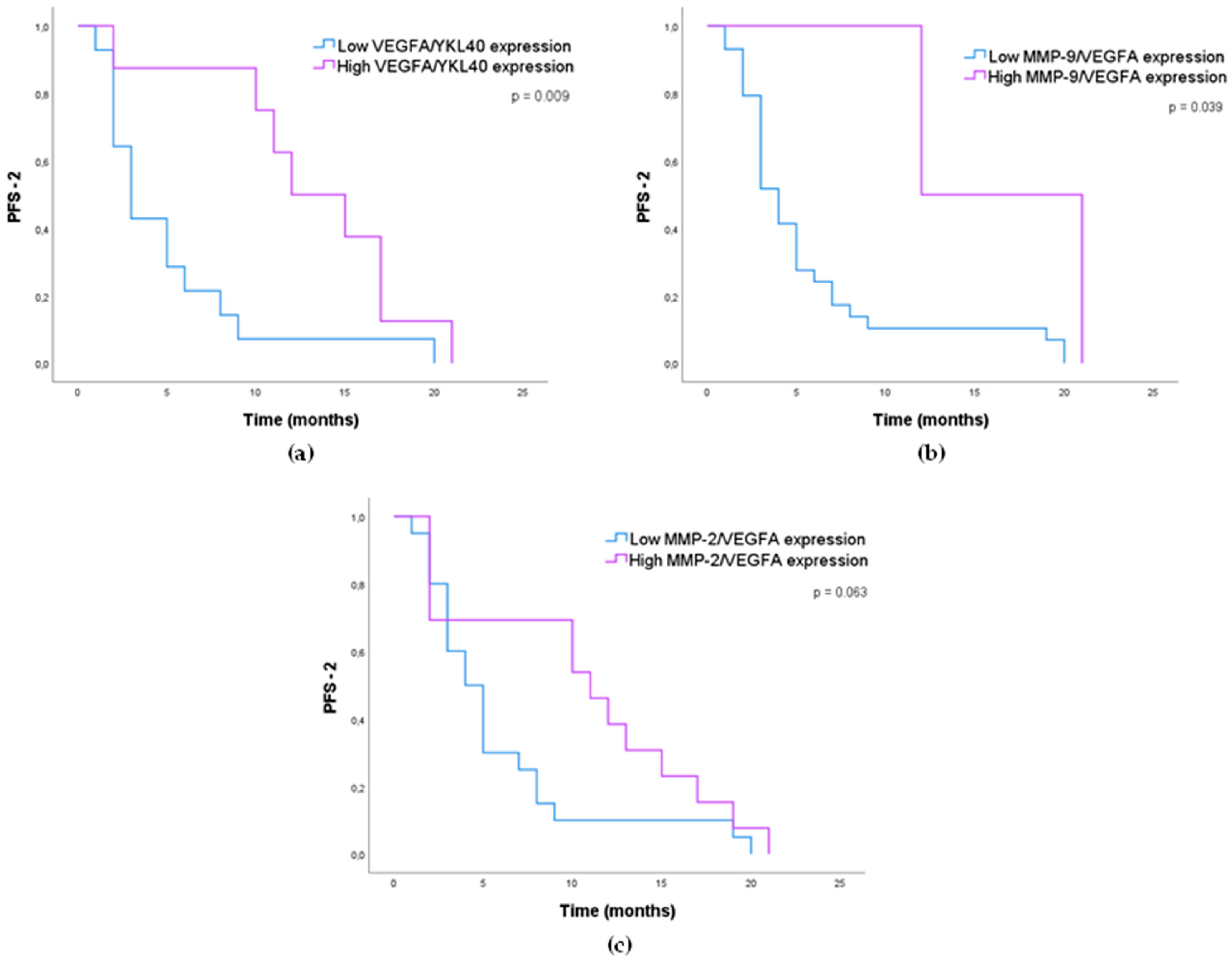
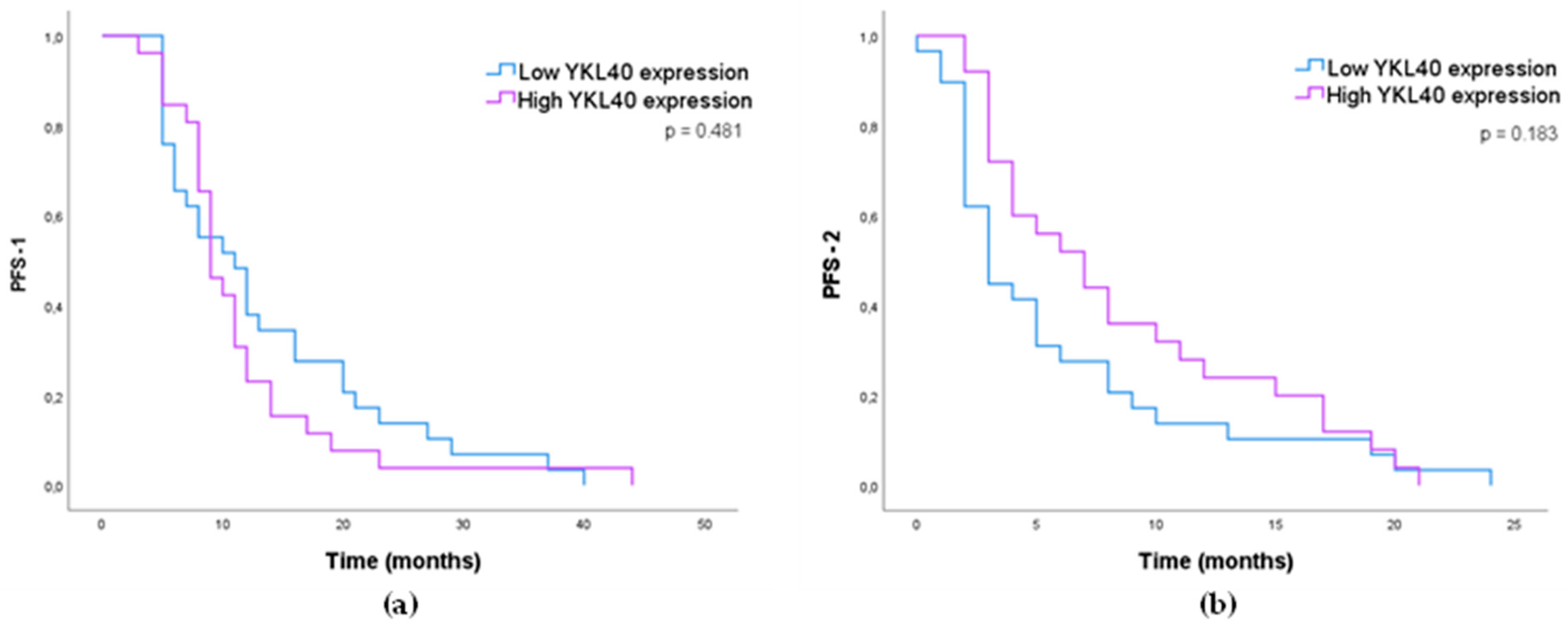
3.1.4. Impact of MMP-2, MMP-9, VEGFA, and YKL40 Expression in GB Patients’ Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hatoum, A.; Mohammed, R.; Zakieh, O. The Unique Invasiveness of Glioblastoma and Possible Drug Targets on Extracellular Matrix. Cancer Manag. Res. 2019, 11, 1843–1855. [Google Scholar] [CrossRef] [Green Version]
- Ostrom, Q.T.; Bauchet, L.; Davis, F.G.; Deltour, I.; Fisher, J.L.; Langer, C.E.; Pekmezci, M.; Schwartzbaum, J.A.; Turner, M.C.; Walsh, K.M.; et al. The Epidemiology of Glioma in Adults: A “State of the Science” Review. Neuro Oncol. 2014, 16, 896–913. [Google Scholar] [CrossRef] [Green Version]
- Wen, P.Y.; Weller, M.; Lee, E.Q.; Alexander, B.M.; Barnholtz-Sloan, J.S.; Barthel, F.P.; Batchelor, T.T.; Bindra, R.S.; Chang, S.M.; Chiocca, E.A.; et al. Glioblastoma in Adults: A Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) Consensus Review on Current Management and Future Directions. Neuro Oncol. 2020, 22, 1073–1113. [Google Scholar] [CrossRef]
- Kan, L.K.; Drummond, K.; Hunn, M.; Williams, D.; O’Brien, T.J.; Monif, M. Potential Biomarkers and Challenges in Glioma Diagnosis, Therapy and Prognosis. BMJ Neurol. Open 2020, 2, e000069. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [Green Version]
- Arora, A.; Somasundaram, K. Glioblastoma vs. Temozolomide: Can the Red Queen Race Be Won? Cancer Biol. Ther. 2019, 20, 1083–1090. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Ai, D.; Li, T.; Xia, L.; Sun, L. Effectiveness of Lomustine Combined with Bevacizumab in Glioblastoma: A Meta-Analysis. Front. Neurol. 2021, 11, 603947. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; van den Bent, M.; Preusser, M.; le Rhun, E.; Tonn, J.C.; Minniti, G.; Bendszus, M.; Balana, C.; Chinot, O. EANO Guidelines on the Diagnosis and Treatment of Diffuse Gliomas of Adulthood. Nat. Rev. Clin. Oncol. 2021, 18, 170–186. [Google Scholar] [CrossRef] [PubMed]
- Chinot, O.L.; Wick, W.; Mason, W.; Henriksson, R.; Saran, F.; Nishikawa, R.; Carpentier, A.F.; Hoang-Xuan, K.; Kavan, P.; Cernea, D.; et al. Bevacizumab plus Radiotherapy–Temozolomide for Newly Diagnosed Glioblastoma. N. Engl. J. Med. 2014, 370, 709–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eder, K.; Kalman, B. Molecular Heterogeneity of Glioblastoma and Its Clinical Relevance. Pathol. Oncol. Res. 2014, 20, 777–787. [Google Scholar] [CrossRef]
- Davis, M.E. Glioblastoma: Overview of Disease and Treatment. Clin. J. Oncol. Nurs. 2016, 20, S2–S8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birzu, C.; French, P.; Caccese, M.; Cerretti, G.; Idbaih, A.; Zagonel, V.; Lombardi, G. Recurrent Glioblastoma: From Molecular Landscape to New Treatment Perspectives. Cancers 2021, 13, 47. [Google Scholar] [CrossRef]
- Mayer, T.M. Can We Predict Bevacizumab Responders in Patients with Glioblastoma? J. Clin. Oncol. 2015, 33, 2721–2722. [Google Scholar] [CrossRef]
- Shieh, L.T.; Guo, H.R.; Ho, C.H.; Lin, L.C.; Chang, C.H.; Ho, S.Y. Survival of Glioblastoma Treated with a Moderately Escalated Radiation Dose—Results of a Retrospective Analysis. PLoS ONE 2020, 15, e0233188. [Google Scholar] [CrossRef]
- Xu, C.; Wu, X.; Zhu, J. VEGF Promotes Proliferation of Human Glioblastoma Multiforme Stem-Like Cells through VEGF Receptor 2. Sci. World J. 2013, 2013, 417413. [Google Scholar] [CrossRef] [Green Version]
- Nørøxe, D.S.; Poulsen, H.S.; Lassen, U. Hallmarks of Glioblastoma: A Systematic Review. ESMO Open 2016, 1, e000144. [Google Scholar] [CrossRef]
- Pullen, N.; Pickford, A.; Perry, M.; Jaworski, D.; Loveson, K.; Arthur, D.; Holliday, J.; van Meter, T.; Peckham, R.; Younas, W.; et al. Current Insights into Matrix Metalloproteinases and Glioma Progression: Transcending the Degradation Boundary. Met. Med. 2018, 5, 13–30. [Google Scholar] [CrossRef] [Green Version]
- Dahlberg, D.; Rummel, J.; Distante, S.; de Souza, G.A.; Stensland, M.E.; Mariussen, E.; Rootwelt, H.; Voie, Ø.; Hassel, B. Glioblastoma Microenvironment Contains Multiple Hormonal and Non-Hormonal Growth-Stimulating Factors. Fluids Barriers CNS 2022, 19, 45. [Google Scholar] [CrossRef]
- Chen, W.; He, D.; Li, Z.; Zhang, X.; Pan, D.; Chen, G. Overexpression of Vascular Endothelial Growth Factor Indicates Poor Outcomes of Glioma: A Systematic Review and Meta-Analysis. Int. J. Clin. Exp. Med. 2015, 8, 8709–8719. [Google Scholar]
- Linhares, P.; Carvalho, B.; Vaz, R.; Costa, B.M. Glioblastoma: Is There Any Blood Biomarker with True Clinical Relevance? Int. J. Mol. Sci. 2020, 21, 5809. [Google Scholar] [CrossRef]
- Sareen, H.; Ma, Y.; Becker, T.M.; Roberts, T.L.; de Souza, P.; Powter, B. Molecular Biomarkers in Glioblastoma: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2022, 23, 8835. [Google Scholar] [CrossRef]
- Chen, W.-J.; Zhang, X.; Han, H.; Lv, J.-N.; Kang, E.-M.; Zhang, Y.-L.; Liu, W.-P.; He, X.-S.; Wang, J.; Wang, G.-H.; et al. The Different Role of YKL-40 in Glioblastoma Is a Function of MGMT Promoter Methylation Status. Cell Death Dis. 2020, 11, 668. [Google Scholar] [CrossRef] [PubMed]
- Sincevičiūtė, R.; Vaitkienė, P.; Urbanavičiūtė, R.; Steponaitis, G.; Tamašauskas, A.; Skiriutė, D. MMP2 Is Associated with Glioma Malignancy and Patient Outcome. Int. J. Clin. Exp. Pathol. 2018, 11, 3010–3018. [Google Scholar] [PubMed]
- Ramachandran, R.K.; Sørensen, M.D.; Aaberg-Jessen, C.; Hermansen, S.K.; Kristensen, B.W. Expression and Prognostic Impact of Matrix Metalloproteinase-2 (MMP-2) in Astrocytomas. PLoS ONE 2017, 12, e0172234. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Chen, B.; Cai, J.; Sun, Y.; Wang, G.; Li, Y.; Li, R.; Feng, Y.; Han, B.; Li, J.; et al. Comparative Analysis of Matrix Metalloproteinase Family Members Reveals That MMP9 Predicts Survival and Response to Temozolomide in Patients with Primary Glioblastoma. PLoS ONE 2016, 11, e0151815. [Google Scholar] [CrossRef] [Green Version]
- Hormigo, A.; Gu, B.; Karimi, S.; Riedel, E.; Panageas, K.S.; Edgar, M.A.; Tanwar, M.K.; Rao, J.S.; Fleisher, M.; DeAngelis, L.M.; et al. YKL-40 and Matrix Metalloproteinase-9 as Potential Serum Biomarkers for Patients with High-Grade Gliomas. Clin. Cancer Res. 2006, 12, 5698–5704. [Google Scholar] [CrossRef] [Green Version]
- Bernardi, D.; Padoan, A.; Ballin, A.; Sartori, M.T.; Manara, R.; Scienza, R.; Plebani, M.; della Puppa, A. Serum YKL-40 Following Resection for Cerebral Glioblastoma. J. Neurooncol. 2012, 107, 299–305. [Google Scholar] [CrossRef]
- Crocker, M.; Ashley, S.; Giddings, I.; Petrik, V.; Hardcastle, A.; Aherne, W.; Pearson, A.; Bell, B.A.; Zacharoulis, S.; Papadopoulos, M.C. Serum Angiogenic Profile of Patients with Glioblastoma Identifies Distinct Tumor Subtypes and Shows That TIMP-1 Is a Prognostic Factor. Neuro Oncol. 2011, 13, 99–108. [Google Scholar] [CrossRef]
- Ahmadipour, Y.; Gembruch, O.; Pierscianek, D.; Sure, U.; Jabbarli, R. Does the Expression of Glial Fibrillary Acid Protein (GFAP) Stain in Glioblastoma Tissue Have a Prognostic Impact on Survival? Neurochirurgie 2020, 66, 150–154. [Google Scholar] [CrossRef]
- Tabouret, E.; Boudouresque, F.; Barrie, M.; Matta, M.; Boucard, C.; Loundou, A.; Carpentier, A.; Sanson, M.; Metellus, P.; Figarella-Branger, D.; et al. Association of Matrix Metalloproteinase 2 Plasma Level with Response and Survival in Patients Treated with Bevacizumab for Recurrent High-Grade Glioma. Neuro Oncol. 2014, 16, 392–399. [Google Scholar] [CrossRef] [Green Version]
- Jiguet-Jiglaire, C.; Boissonneau, S.; Denicolai, E.; Hein, V.; Lasseur, R.; Garcia, J.; Romain, S.; Appay, R.; Graillon, T.; Mason, W.; et al. Plasmatic MMP9 Released from Tumor-Infiltrating Neutrophils Is Predictive for Bevacizumab Efficacy in Glioblastoma Patients: An AVAglio Ancillary Study. Acta Neuropathol. Commun. 2022, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.; Ohgaki, H.; Wiestler, O.; Kleihues, P.; Ellison, D. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, B.; Lopes, J.M.; Silva, R.; Peixoto, J.; Leitão, D.; Soares, P.; Fernandes, A.C.; Linhares, P.; Vaz, R.; Lima, J. The Role of C-Met and VEGFR2 in Glioblastoma Resistance to Bevacizumab. Sci. Rep. 2021, 11, 6067. [Google Scholar] [CrossRef]
- Virga, J.; Szivos, L.; Hortobágyi, T.; Chalsaraei, M.K.; Zahuczky, G.; Steiner, L.; Tóth, J.; Reményi-Puskár, J.; Bognár, L.; Klekner, A. Extracellular Matrix Differences in Glioblastoma Patients with Different Prognoses. Oncol. Lett. 2019, 17, 797–806. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Yu, X.; Sun, S.; Zhang, X.; Yang, W.; Zhang, J.; Zhang, X.; Jiang, Z. Increased Expression of MMP-2 and MMP-9 Indicates Poor Prognosis in Glioma Recurrence. Biomed. Pharmacother. 2019, 118, 109369. [Google Scholar] [CrossRef]
- Horbinski, C.; Wang, G.; Wiley, C.A. YKL-40 Is Directly Produced by Tumor Cells and Is Inversely Linked to EGFR in Glioblastomas. Int. J. Clin. Exp. Pathol. 2010, 3, 226. [Google Scholar]
- Kazakova, M.H.; Staneva, D.N.; Koev, I.G.; Staikov, D.G.; Mateva, N.; Timonov, P.T.; Miloshev, G.A.; Sarafian, V.S.; Sarafian, V.S. Protein and MRNA Levels of YKL-40 in High-Grade Glioma. Folia Biol. 2014, 60, 261–267. [Google Scholar]
- Pelloski, C.E.; Mahajan, A.; Maor, M.; Chang, E.L.; Woo, S.; Gilbert, M.; Colman, H.; Yang, H.; Ledoux, A.; Blair, H.; et al. YKL-40 Expression Is Associated with Poorer Response to Radiation and Shorter Overall Survival in Glioblastoma. Clin. Cancer Res. 2005, 11, 3326–3334. [Google Scholar] [CrossRef] [Green Version]
- Francescone, R.A.; Scully, S.; Faibish, M.; Taylor, S.L.; Oh, D.; Moral, L.; Yan, W.; Bentley, B.; Shao, R. Role of YKL-40 in the Angiogenesis, Radioresistance, and Progression of Glioblastoma. J. Biol. Chem. 2011, 286, 15332–15343. [Google Scholar] [CrossRef] [Green Version]
- Gong, J.; Zhu, S.; Zhang, Y.; Wang, J. Interplay of VEGFa and MMP2 Regulates Invasion of Glioblastoma. Tumor Biol. 2014, 35, 11879–11885. [Google Scholar] [CrossRef]
- Holst, C.B.; Pedersen, H.; Obara, E.A.A.; Vitting-Seerup, K.; Jensen, K.E.; Skjøth-Rasmussen, J.; Lund, E.L.; Poulsen, H.S.; Johansen, J.S.; Hamerlik, P. Perspective: Targeting VEGF-A and YKL-40 in Glioblastoma—Matter Matters. Cell Cycle 2021, 20, 702–715. [Google Scholar] [CrossRef] [PubMed]
- Ku, B.M.; Lee, Y.K.; Ryu, J.; Jeong, J.Y.; Choi, J.; Eun, K.M.; Shin, H.Y.; Kim, D.G.; Hwang, E.M.; Yoo, J.C.; et al. CHI3L1 (YKL-40) Is Expressed in Human Gliomas and Regulates the Invasion, Growth and Survival of Glioma Cells. Int. J. Cancer 2011, 128, 1316–1326. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, Y.; Zhang, X.S.; Shen, B.Z. Expression of VEGF and MMP-9 and MRI Imaging Changes in Cerebral Glioma. Oncol. Lett. 2011, 2, 1171–1175. [Google Scholar] [CrossRef] [Green Version]
- Lu, K.V.; Chang, J.P.; Parachoniak, C.A.; Pandika, M.M.; Aghi, M.K.; Meyronet, D.; Isachenko, N.; Fouse, S.D.; Phillips, J.J.; Cheresh, D.A.; et al. VEGF Inhibits Tumor Cell Invasion and Mesenchymal Transition through a MET/VEGFR2 Complex. Cancer Cell 2012, 22, 21–35. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Li, Y.; Zhang, X.; Zhao, G.; Xu, H. Levels of Vascular Endothelial Growth Factor and Matrix Metalloproteinase-9 Proteins in Patients with Glioma. J. Int. Med. Res. 2014, 42, 198–204. [Google Scholar] [CrossRef] [Green Version]
- Saidi, A.; Javerzat, S.; Bellahcène, A.; de Vos, J.; Bello, L.; Castronovo, V.; Deprez, M.; Loiseau, H.; Bikfalvi, A.; Hagedorn, M. Experimental Anti-Angiogenesis Causes Upregulation of Genes Associated with Poor Survival in Glioblastoma. Int. J. Cancer 2008, 122, 2187–2198. [Google Scholar] [CrossRef]
- Lamborn, K.R.; Yung, W.K.A.; Chang, S.M.; Wen, P.Y.; Cloughesy, T.F.; DeAngelis, L.M.; Robins, H.I.; Lieberman, F.S.; Fine, H.A.; Fink, K.L.; et al. Progression-Free Survival: An Important End Point in Evaluating Therapy for Recurrent High-Grade Gliomas. Neuro Oncol. 2008, 10, 162–170. [Google Scholar] [CrossRef]
- Ballman, K.V.; Buckner, J.C.; Brown, P.D.; Giannini, C.; Flynn, P.J.; LaPlant, B.R.; Jaeckle, K.A. The Relationship between Six-Month Progression-Free Survival and 12-Month Overall Survival End Points for Phase II Trials in Patients with Glioblastoma Multiforme. Neuro Oncol. 2007, 9, 29. [Google Scholar] [CrossRef]
- Boisen, M.K.; Holst, C.B.; Consalvo, N.; Chinot, O.L.; Johansen, J.S. Plasma YKL-40 as a Biomarker for Bevacizumab Efficacy in Patients with Newly Diagnosed Glioblastoma in the Phase 3 Randomized AVAglio Trial. Oncotarget 2018, 9, 6752. [Google Scholar] [CrossRef] [Green Version]
- Güttler, A.; Giebler, M.; Cuno, P.; Wichmann, H.; Keßler, J.; Ostheimer, C.; Söling, A.; Strauss, C.; Illert, J.; Kappler, M.; et al. Osteopontin and Splice Variant Expression Level in Human Malignant Glioma: Radiobiologic Effects and Prognosis after Radiotherapy. Radiother. Oncol. 2013, 108, 535–540. [Google Scholar] [CrossRef]
- Pérez-Larraya, J.G.; Paris, S.; Idbaih, A.; Dehais, C.; Laigle-Donadey, F.; Navarro, S.; Capelle, L.; Mokhtari, K.; Marie, Y.; Sanson, M.; et al. Diagnostic and Prognostic Value of Preoperative Combined GFAP, IGFBP-2, and YKL-40 Plasma Levels in Patients with Glioblastoma. Cancer 2014, 120, 3972–3980. [Google Scholar] [CrossRef]
- Reynés, G.; Vila, V.; Martín, M.; Parada, A.; Fleitas, T.; Reganon, E.; Martínez-Sales, V. Circulating Markers of Angiogenesis, Inflammation, and Coagulation in Patients with Glioblastoma. J. Neurooncol. 2011, 102, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Baumgarten, P.; Blank, A.-E.; Franz, K.; Hattingen, E.; Dunst, M.; Zeiner, P.; Hoffmann, K.; Bähr, O.; Mäder, L.; Goeppert, B.; et al. Differential Expression of Vascular Endothelial Growth Factor A, Its Receptors VEGFR-1,-2, and-3 and Co-Receptors Neuropilin-1 and-2 Does Not Predict Bevacizumab Response in Human Astrocytomas. Neuro Oncol. 2016, 18, 173–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, I.; Cunliffe, C.H.; Bollo, R.J.; Raza, S.; Monoky, D.; Chiriboga, L.; Parker, E.C.; Golfinos, J.G.; Kelly, P.J.; Knopp, E.A.; et al. High-Grade Glioma before and after Treatment with Radiation and Avastin: Initial Observations. Neuro Oncol. 2008, 10, 700–708. [Google Scholar] [CrossRef] [Green Version]
- Johansson, M.; Brännström, T.; Tommy Bergenheim, A.; Henriksson, R. Spatial Expression of VEGF-A in Human Glioma. J. Neurooncol. 2002, 59, 1–6. [Google Scholar] [CrossRef]
- Tamura, R.; Ohara, K.; Sasaki, H.; Morimoto, Y.; Kosugi, K.; Yoshida, K.; Toda, M. Difference in Immunosuppressive Cells between Peritumoral Area and Tumor Core in Glioblastoma. World Neurosurg. 2018, 120, e601–e610. [Google Scholar] [CrossRef]
- Clara, C.A.; Marie, S.K.N.; de Almeida, J.R.W.; Wakamatsu, A.; Oba-Shinjo, S.M.; Uno, M.; Neville, M.; Rosemberg, S. Angiogenesis and Expression of PDGF-C, VEGF, CD105 and HIF-1α in Human Glioblastoma. Neuropathology 2014, 34, 343–352. [Google Scholar] [CrossRef]
- Plate, K.H.; Breier, G.; Weich, H.A.; Risau, W. Vascular Endothelial Growth Factor Is a Potential Tumour Angiogenesis Factor in Human Gliomas in Vivo. Nature 1992, 359, 845–848. [Google Scholar] [CrossRef] [PubMed]
- Wojtukiewicz, M.Z.; Mysliwiec, M.; Matuszewska, E.; Sulkowski, S.; Zimnoch, L.; Politynska, B.; Wojtukiewicz, A.M.; Tucker, S.C.; Honn, K.V. Heterogeneous Expression of Proangiogenic and Coagulation Proteins in Gliomas of Different Histopathological Grade. Pathol. Oncol. Res. 2021, 27, 18. [Google Scholar] [CrossRef]
- Gilbert, M.R.; Dignam, J.J.; Armstrong, T.S.; Wefel, J.S.; Blumenthal, D.T.; Vogelbaum, M.A.; Colman, H.; Chakravarti, A.; Pugh, S.; Won, M.; et al. A Randomized Trial of Bevacizumab for Newly Diagnosed Glioblastoma. N. Engl. J. Med. 2014, 370, 699–708. [Google Scholar] [CrossRef] [Green Version]
- García-Romero, N.; Palacín-Aliana, I.; Madurga, R.; Carrión-Navarro, J.; Esteban-Rubio, S.; Jiménez, B.; Collazo, A.; Pérez-Rodríguez, F.; Ortiz de Mendivil, A.; Fernández-Carballal, C.; et al. Bevacizumab Dose Adjustment to Improve Clinical Outcomes of Glioblastoma. BMC Med. 2020, 18, 142. [Google Scholar] [CrossRef] [PubMed]
- Kazazi-Hyseni, F.; Beijnen, J.H.; Schellens, J.H.M. Bevacizumab. Oncologist 2010, 15, 819. [Google Scholar] [CrossRef] [PubMed]
- Sathornsumetee, S.; Cao, Y.; Marcello, J.E.; Herndon, J.E.; McLendon, R.E.; Desjardins, A.; Friedman, H.S.; Dewhirst, M.W.; Vredenburgh, J.J.; Rich, J.N. Tumor Angiogenic and Hypoxic Profiles Predict Radiographic Response and Survival in Malignant Astrocytoma Patients Treated with Bevacizumab and Irinotecan. J. Clin. Oncol. 2008, 26, 271. [Google Scholar] [CrossRef] [PubMed]
- Pouyafar, A.; Heydarabad, M.Z.; Mahboob, S.; Mokhtarzadeh, A.; Rahbarghazi, R. Angiogenic Potential of YKL-40 in the Dynamics of Tumor Niche. Biomed. Pharmacother. 2018, 100, 478–485. [Google Scholar] [CrossRef]
- Akiyama, Y.; Ashizawa, T.; Komiyama, M.; Miyata, H.; Oshita, C.; Omiya, M.; Iizuka, A.; Kume, A.; Sugino, T.; Hayashi, N.; et al. YKL-40 Downregulation Is a Key Factor to Overcome Temozolomide Resistance in a Glioblastoma Cell Line. Oncol. Rep. 2014, 32, 159–166. [Google Scholar] [CrossRef] [Green Version]
- Boisen, M.K.; Madsen, C.V.; Dehlendorff, C.; Jakobsen, A.; Johansen, J.S.; Steffensen, K.D. The Prognostic Value of Plasma YKL-40 in Patients with Chemotherapy-Resistant Ovarian Cancer Treated with Bevacizumab. Int. J. Gynecol. Cancer 2016, 26, 1390–1398. [Google Scholar] [CrossRef]
- Suo, F.; Zhong, B.; Lu, F.; Dong, Z. The Combined Use of EphA2/MMP-2 Expression and MRI Findings Contributes to the Determination of Cerebral Glioma Grade. Oncol. Lett. 2019, 18, 5607–5613. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Wang, T.; Liu, S.; Yoshida, D.; Teramoto, A. The Expression of Matrix Metalloproteinase-2 and -9 in Human Gliomas of Different Pathological Grades. Brain Tumor Pathol. 2003, 20, 65–72. [Google Scholar] [CrossRef]
- Ricci, S.; Guadagno, E.; Bruzzese, D.; del Basso De Caro, M.; Peca, C.; Sgulò, F.G.; Maiuri, F.; di Carlo, A. Evaluation of Matrix Metalloproteinase Type IV-Collagenases in Serum of Patients with Tumors of the Central Nervous System. J. Neurooncol. 2017, 131, 223–232. [Google Scholar] [CrossRef] [Green Version]
- Xue, Q.; Li, C.; Chen, X.Y.; Zhao, J.; Gao, L.; Li, S.Z.; Fei, Z. High Expression of MMP9 in Glioma Affects Cell Proliferation and Is Associated with Patient Survival Rates. Oncol. Lett. 2017, 13, 1325–1330. [Google Scholar] [CrossRef] [Green Version]
- Hu, F.; Ku, M.C.; Markovic, D.; Dzaye, O.D.; Lehnardt, S.; Synowitz, M.; Wolf, S.A.; Kettenmann, H. Glioma Associated Microglial MMP9 Expression Is up Regulated by TLR2 Signalling and Sensitive to Minocycline. Int. J. Cancer 2014, 135, 2569. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, V.L.; Landry, R.P.; Liu, Y.; Romero-Sandoval, E.A.; de Leo, J.A. Propentofylline Decreases Tumor Growth in a Rodent Model of Glioblastoma Multiforme by a Direct Mechanism on Microglia. Neuro Oncol. 2012, 14, 119. [Google Scholar] [CrossRef]
- Könnecke, H.; Bechmann, I. The Role of Microglia and Matrix Metalloproteinases Involvement in Neuroinflammation and Gliomas. Clin. Dev. Immunol. 2013, 2013, 914104. [Google Scholar] [CrossRef] [Green Version]
- Mondal, S.; Adhikari, N.; Banerjee, S.; Amin, S.A.; Jha, T. Matrix Metalloproteinase-9 (MMP-9) and Its Inhibitors in Cancer: A Minireview. Eur. J. Med. Chem. 2020, 194, 112260. [Google Scholar] [CrossRef]

| (a) | (b) | |||
|---|---|---|---|---|
| Staining Intensity | Percentage of Positive Cells | |||
| MMP-2 and MMP-9 | VEGFA and YKL40 | |||
| Absent Weak Moderate Strong | 0 1 2 3 | 0% 1−5% 6−50% 51−75% >75% | 0 1 2 3 4 | 0% 1−24% 25−49% 50−75% >75% |
| Clinical Features | n (%) |
|---|---|
| Sex [n = 63] | |
| Male | 34 (54.0%) |
| Female | 29 (46.0%) |
| Age at diagnosis (years) [n = 63] | |
| Median (range) | 56 (26–73) |
| Type of surgery [n = 57] | |
| Biopsy | 4 (7.0%) |
| Partial or subtotal resection | 26 (45.6%) |
| Total resection | 27 (47.4%) |
| Tumor location [n = 63] | |
| Frontal | 12 (19.0%) |
| Temporal | 21 (33.3%) |
| Parietal | 9 (14.3%) |
| Multiple | 17 (27.0%) |
| Other | 4 (6.3%) |
| Tumor laterality [n = 62] | |
| Unilateral | 60 (96.8%) |
| Bilateral | 2 (3.2%) |
| Tumor focality [n = 60] | |
| Unifocal | 55 (91.7%) |
| Multifocal or multicentric | 5 (8.3%) |
| ECOG [n = 61] | |
| 0 | 13 (21.3%) |
| 1 | 42 (68.9%) |
| 2 | 5 (8.2%) |
| 3 | 1 (1.6%) |
| KPS [n = 61] | |
| <70 | 1 (1.6%) |
| ≥70 | 60 (98.4%) |
| MGMT Status [n = 40] | |
| Methylated | 20 (50.0%) |
| Non-Methylated | 20 (50.0%) |
| Number of temozolomide cycles [n = 63] | |
| Median (range) | 6 (0–42) |
| Number of bevacizumab doses [n = 63] | |
| Median (range) | 9 (1–41) |
| Progression-free survival after temozolomide (months) [n = 63] | |
| Median (range) | 9 (3–44) |
| Progression-free survival after bevacizumab (months) [n = 62] | |
| Median (range) | 4 (0–24) |
| Overall survival (months) [n = 63] | |
| Median (range) | 23 (1–63) |
| Patient status [n = 63] | |
| Dead | 52 (82.5%) |
| Alive or Unknown | 11 (17.5%) |
| MMP-2 (n = 59) | MMP-9 (n = 61) | |
|---|---|---|
| Percentage of positive stained cells | ||
| 0% | 0 (0.0%) | 52 (85.2%) |
| 1–5% | 1 (1.7%) | 8 (13.1%) |
| 6–50% | 7 (11.9%) | 1 (1.6%) |
| 51–75% | 10 (16.9%) | 0 (0.0%) |
| >75% | 41 (69.5%) | 0 (0.0%) |
| Staining intensity | ||
| Absent | 0 (0.0%) | 52 (85.2%) |
| Weak | 3 (5.1%) | 2 (3.3%) |
| Moderate | 23 (39.0%) | 5 (8.2%) |
| Strong | 33 (55.9%) | 2 (3.3%) |
| IHS | ||
| 0 | 0 (0.0%) | 52 (85.2%) |
| 1 | 1 (1.7%) | 2 (3.3%) |
| 2 | 1 (1.7%) | 4 (6.6%) |
| 3 | 1 (1.7%) | 2 (3.3%) |
| 4 | 5 (8.5%) | 1 (1.6%) |
| 6 | 9 (15.3%) | 0 (0.0%) |
| 8 | 10 (16.9%) | 0 (0.0%) |
| 9 | 1 (1.7%) | 0 (0.0%) |
| 12 | 31 (52.5%) | 0 (0.0%) |
| VEGFA (n = 59) | YKL40 (n = 55) | |
|---|---|---|
| Percentage of positive stained cells | ||
| 0% | 10 (16.9%) | 8 (14.5%) |
| 1–24% | 10 (16.9%) | 20 (36.4%) |
| 25–49% | 8 (13.6%) | 6 (10.9%) |
| 50–75% | 15 (25.4%) | 9 (16.4%) |
| >75% | 16 (27.1%) | 12 (21.8%) |
| Staining intensity | ||
| Absent | 10 (16.9%) | 8 (14.5%) |
| Weak | 21 (35.6%) | 1 (1.8%) |
| Moderate | 23 (39.0%) | 7 (12.7%) |
| Strong | 5 (8.5%) | 39 (70.9%) |
| IHS | ||
| 0 | 10 (16.9%) | 8 (14.5%) |
| 1 | 8 (13.6%) | 1 (1.8%) |
| 2 | 8 (13.6%) | 5 (9.1%) |
| 3 | 4 (6.8%) | 14 (25.5%) |
| 4 | 4 (6.8%) | 1 (1.8%) |
| 6 | 10 (16.9%) | 6 (10.9%) |
| 8 | 11 (18.6%) | 0 (0.0%) |
| 9 | 2 (3.4%) | 8 (14.5%) |
| 12 | 2 (3.4%) | 12 (21.8%) |
| Protein | IHS Threshold | |
|---|---|---|
| MMP-2 | <12 | 12 |
| 28 (47.5%) | 31 (52.5%) | |
| MMP-9 | 0 | ≥1 |
| 52 (85.2%) | 9 (14.8%) | |
| VEGFA | ≤4 | >4 |
| 38 (64.4%) | 21 (35.6%) | |
| YKL40 | ≤4 | >4 |
| 29 (52.7%) | 26 (47.3%) | |
| Clinical Features | YKL40 | ||
|---|---|---|---|
| IHS ≤ 4 | IHS > 4 | p Value | |
| Tumor Location Frontal Temporal Parietal Multiple Others | n = 57 | 0.007 | |
| 5 (17.2%) 8 (27.6%) 1(3.4%) 12 (41.4%) 3 (10.3%) | 5 (19.2%) 10 (38.5%) 8 (30.8%) 3 (11.5%) 0 (0.0%) | ||
| Number of Bevacizumab Doses Median (range) | n = 55 | 0.035 | |
| 6 (1–37) | 9.5 (2–41) | ||
| VEGFA Expression | YKL40 Expression | |
|---|---|---|
| Mann–Whitney U | 252.500 | 258.000 |
| p value | 0.035 | 0.069 |
| Variable | Multivariate Analysis | ||
|---|---|---|---|
| HR | 95% CI | p Value | |
| Age <65 years versus ≥65 years | 1.37 | 0.76–2.45 | 0.294 |
| ECOG 0–1 versus ≥ 2 | 1.16 | 0.48–2.83 | 0.745 |
| Type of surgery | 0.77 | 0.23–2.64 | 0.682 |
| VEGFA expression | 0.57 | 0.31–1.06 | 0.075 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alves, B.; Peixoto, J.; Macedo, S.; Pinheiro, J.; Carvalho, B.; Soares, P.; Lima, J.; Lima, R.T. High VEGFA Expression Is Associated with Improved Progression-Free Survival after Bevacizumab Treatment in Recurrent Glioblastoma. Cancers 2023, 15, 2196. https://doi.org/10.3390/cancers15082196
Alves B, Peixoto J, Macedo S, Pinheiro J, Carvalho B, Soares P, Lima J, Lima RT. High VEGFA Expression Is Associated with Improved Progression-Free Survival after Bevacizumab Treatment in Recurrent Glioblastoma. Cancers. 2023; 15(8):2196. https://doi.org/10.3390/cancers15082196
Chicago/Turabian StyleAlves, Bárbara, Joana Peixoto, Sofia Macedo, Jorge Pinheiro, Bruno Carvalho, Paula Soares, Jorge Lima, and Raquel T. Lima. 2023. "High VEGFA Expression Is Associated with Improved Progression-Free Survival after Bevacizumab Treatment in Recurrent Glioblastoma" Cancers 15, no. 8: 2196. https://doi.org/10.3390/cancers15082196






