An Aptamer That Rapidly Internalizes into Cancer Cells Utilizes the Transferrin Receptor Pathway
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Aptamer Synthesis and Labeling
2.3. Binding Curves on Recombinant Protein and Canine Cancer Cells
2.4. Aptamer and Transferrin Competition Assays
2.5. siRNA Knockdown
2.6. Molecular Modeling
3. Results
3.1. Characterization of E3 Binding to Recombinant Human TfR
3.2. E3 Competes with C2.min but Not Waz for Cell Binding and Internalization
3.3. hTfR Is Sufficient to Mediate E3 Binding to Cells
3.4. Knockdown of TfR Abolished E3 Binding to Cells
3.5. Molecular Modeling of E3 Binding to Transferrin Receptor
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Debela, D.T.; Muzazu, S.G.; Heraro, K.D.; Ndalama, M.T.; Mesele, B.W.; Haile, D.C.; Kitui, S.K.; Manyazewal, T. New approaches and procedures for cancer treatment: Current perspectives. SAGE Open Med. 2021, 9, 20503121211034366. [Google Scholar] [CrossRef] [PubMed]
- Perez-Herrero, E.; Fernandez-Medarde, A. Advanced targeted therapies in cancer: Drug nanocarriers, the future of chemotherapy. Eur. J. Pharm. Biopharm. 2015, 93, 52–79. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, J.H.; Ballman, K.; Polley, M.C.; Campbell, J.D.; Fan, C.; Selitsky, S.; Fernandez-Martinez, A.; Parker, J.S.; Hoadley, K.A.; Hu, Z.; et al. CALGB 40603 (Alliance): Long-Term Outcomes and Genomic Correlates of Response and Survival After Neoadjuvant Chemotherapy With or Without Carboplatin and Bevacizumab in Triple-Negative Breast Cancer. J. Clin. Oncol. 2022, 40, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Bardia, A.; Mayer, I.A.; Diamond, J.R.; Moroose, R.L.; Isakoff, S.J.; Starodub, A.N.; Shah, N.C.; O’Shaughnessy, J.; Kalinsky, K.; Guarino, M.; et al. Efficacy and Safety of Anti-Trop-2 Antibody Drug Conjugate Sacituzumab Govitecan (IMMU-132) in Heavily Pretreated Patients With Metastatic Triple-Negative Breast Cancer. J. Clin. Oncol. 2017, 35, 2141–2148. [Google Scholar] [CrossRef] [PubMed]
- Hafeez, U.; Parakh, S.; Gan, H.K.; Scott, A.M. Antibody-Drug Conjugates for Cancer Therapy. Molecules 2020, 25, 4746. [Google Scholar] [CrossRef] [PubMed]
- Shastry, M.; Jacob, S.; Rugo, H.S.; Hamilton, E. Antibody-drug conjugates targeting TROP-2: Clinical development in metastatic breast cancer. Breast 2022, 66, 169–177. [Google Scholar] [CrossRef]
- Dean, A.Q.; Luo, S.; Twomey, J.D.; Zhang, B. Targeting cancer with antibody-drug conjugates: Promises and challenges. MAbs 2021, 13, 1951427. [Google Scholar] [CrossRef]
- Beck, A.; Goetsch, L.; Dumontet, C.; Corvaia, N. Strategies and challenges for the next generation of antibody-drug conjugates. Nat. Rev. Drug Discov. 2017, 16, 315–337. [Google Scholar] [CrossRef]
- Anami, Y.; Otani, Y.; Xiong, W.; Ha, S.Y.Y.; Yamaguchi, A.; Rivera-Caraballo, K.A.; Zhang, N.; An, Z.; Kaur, B.; Tsuchikama, K. Homogeneity of antibody-drug conjugates critically impacts the therapeutic efficacy in brain tumors. Cell Rep. 2022, 39, 110839. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Nimjee, S.M.; White, R.R.; Becker, R.C.; Sullenger, B.A. Aptamers as Therapeutics. Annu. Rev. Pharmacol. 2017, 57, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Kelly, L.; Maier, K.E.; Yan, A.; Levy, M. A comparative analysis of cell surface targeting aptamers. Nat. Commun. 2021, 12, 6275. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.; Cheek, M.A.; Sharaf, M.L.; Li, N.; Ellington, A.D.; Sullenger, B.A.; Shaw, B.R.; White, R.R. Aptamer-mediated delivery of chemotherapy to pancreatic cancer cells. Nucleic Acid Ther. 2012, 22, 295–305. [Google Scholar] [CrossRef]
- Zhou, J.; Soontornworajit, B.; Martin, J.; Sullenger, B.A.; Gilboa, E.; Wang, Y. A hybrid DNA aptamer-dendrimer nanomaterial for targeted cell labeling. Macromol. Biosci. 2009, 9, 831–835. [Google Scholar] [CrossRef]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Bawab, A.A.; Ismail, S.I. Aptamers Chemistry: Chemical Modifications and Conjugation Strategies. Molecules 2019, 25, 3. [Google Scholar] [CrossRef]
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent Advances in Aptamer Discovery and Applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef] [PubMed]
- Padilla, R.; Sousa, R. A Y639F/H784A T7 RNA polymerase double mutant displays superior properties for synthesizing RNAs with non-canonical NTPs. Nucleic Acids Res. 2002, 30, e138. [Google Scholar] [CrossRef]
- Kratschmer, C.; Levy, M. Effect of Chemical Modifications on Aptamer Stability in Serum. Nucleic Acid Ther. 2017, 27, 335–344. [Google Scholar] [CrossRef]
- Drolet, D.W.; Green, L.S.; Gold, L.; Janjic, N. Fit for the Eye: Aptamers in Ocular Disorders. Nucleic Acid Ther. 2016, 26, 127–146. [Google Scholar] [CrossRef]
- Pernar, C.H.; Ebot, E.M.; Wilson, K.M.; Mucci, L.A. The Epidemiology of Prostate Cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a030361. [Google Scholar] [CrossRef]
- Yan, A.; Levy, M. Cell internalization SELEX: In vitro selection for molecules that internalize into cells. Methods Mol. Biol. 2014, 1103, 241–265. [Google Scholar]
- Powell Gray, B.; Kelly, L.; Ahrens, D.P.; Barry, A.P.; Kratschmer, C.; Levy, M.; Sullenger, B.A. Tunable cytotoxic aptamer-drug conjugates for the treatment of prostate cancer. Proc. Natl. Acad. Sci. USA 2018, 115, 4761–4766. [Google Scholar] [CrossRef]
- Powell Gray, B.; Song, X.; Hsu, D.S.; Kratschmer, C.; Levy, M.; Barry, A.P.; Sullenger, B.A. An Aptamer for Broad Cancer Targeting and Therapy. Cancers 2020, 12, 3217. [Google Scholar] [CrossRef] [PubMed]
- Mayle, K.M.; Le, A.M.; Kamei, D.T. The intracellular trafficking pathway of transferrin. Biochim. Biophys. Acta 2012, 1820, 264–281. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.; Heuser, J.; Stahl, P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983, 97, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Gammella, E.; Buratti, P.; Cairo, G.; Recalcati, S. The transferrin receptor: The cellular iron gate. Metallomics 2017, 9, 1367–1375. [Google Scholar] [CrossRef]
- Milto, I.V.; Suhodolo, I.V.; Prokopieva, V.D.; Klimenteva, T.K. Molecular and Cellular Bases of Iron Metabolism in Humans. Biochemistry 2016, 81, 549–564. [Google Scholar] [CrossRef]
- Wang, Y.F.; Zhang, J.; Su, Y.; Shen, Y.Y.; Jiang, D.X.; Hou, Y.Y.; Geng, M.Y.; Ding, J.; Chen, Y. G9a regulates breast cancer growth by modulating iron homeostasis through the repression of ferroxidase hephaestin. Nat. Commun. 2017, 8, 274. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Ramakrishnan, S.K.; Weisz, K.; Triner, D.; Xie, L.; Attili, D.; Pant, A.; Gyorffy, B.; Zhan, M.; Carter-Su, C.; et al. Iron Uptake via DMT1 Integrates Cell Cycle with JAK-STAT3 Signaling to Promote Colorectal Tumorigenesis. Cell Metab. 2016, 24, 447–461. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.M.; Hwang, S.; Seong, R.H. Transferrin receptor regulates pancreatic cancer growth by modulating mitochondrial respiration and ROS generation. Biochem. Biophys. Res. Commun. 2016, 471, 373–379. [Google Scholar] [CrossRef]
- Shen, Y.; Li, X.; Dong, D.; Zhang, B.; Xue, Y.; Shang, P. Transferrin receptor 1 in cancer: A new sight for cancer therapy. Am. J. Cancer Res. 2018, 8, 916–931. [Google Scholar] [PubMed]
- Leoh, L.S.; Kim, Y.K.; Candelaria, P.V.; Martinez-Maza, O.; Daniels-Wells, T.R.; Penichet, M.L. Efficacy and Mechanism of Antitumor Activity of an Antibody Targeting Transferrin Receptor 1 in Mouse Models of Human Multiple Myeloma. J. Immunol. 2018, 200, 3485–3494. [Google Scholar] [CrossRef]
- Soule, E.E.; Yu, H.; Olson, L.; Naqvi, I.; Kumar, S.; Krishnaswamy, S.; Sullenger, B.A. Generation of an anticoagulant aptamer that targets factor V/Va and disrupts the FVa-membrane interaction in normal and COVID-19 patient samples. Cell Chem. Biol. 2022, 29, 215–225.e215. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Boniecki, M.J.; Lach, G.; Dawson, W.K.; Tomala, K.; Lukasz, P.; Soltysinski, T.; Rother, K.M.; Bujnicki, J.M. SimRNA: A coarse-grained method for RNA folding simulations and 3D structure prediction. Nucleic Acids Res. 2016, 44, e63. [Google Scholar] [CrossRef]
- Eckenroth, B.E.; Steere, A.N.; Chasteen, N.D.; Everse, S.J.; Mason, A.B. How the binding of human transferrin primes the transferrin receptor potentiating iron release at endosomal pH. Proc. Natl. Acad. Sci. USA 2011, 108, 13089–13094. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Maier, K.E.; Jangra, R.K.; Shieh, K.R.; Cureton, D.K.; Xiao, H.; Snapp, E.L.; Whelan, S.P.; Chandran, K.; Levy, M. A New Transferrin Receptor Aptamer Inhibits New World Hemorrhagic Fever Mammarenavirus Entry. Mol. Ther. Nucleic Acids 2016, 5, e321. [Google Scholar] [CrossRef]
- Wilner, S.E.; Wengerter, B.; Maier, K.; de Lourdes Borba Magalhaes, M.; Del Amo, D.S.; Pai, S.; Opazo, F.; Rizzoli, S.O.; Yan, A.; Levy, M. An RNA alternative to human transferrin: A new tool for targeting human cells. Mol. Ther. Nucleic Acids 2012, 1, e21. [Google Scholar] [CrossRef]
- Wengerter, B.C.; Katakowski, J.A.; Rosenberg, J.M.; Park, C.G.; Almo, S.C.; Palliser, D.; Levy, M. Aptamer-targeted antigen delivery. Mol. Ther. 2014, 22, 1375–1387. [Google Scholar] [CrossRef]
- Johnsen, K.B.; Burkhart, A.; Melander, F.; Kempen, P.J.; Vejlebo, J.B.; Siupka, P.; Nielsen, M.S.; Andresen, T.L.; Moos, T. Targeting transferrin receptors at the blood-brain barrier improves the uptake of immunoliposomes and subsequent cargo transport into the brain parenchyma. Sci. Rep. 2017, 7, 10396. [Google Scholar] [CrossRef] [PubMed]
- Stocki, P.; Szary, J.; Rasmussen, C.L.M.; Demydchuk, M.; Northall, L.; Logan, D.B.; Gauhar, A.; Thei, L.; Moos, T.; Walsh, F.S.; et al. Blood-brain barrier transport using a high affinity, brain-selective VNAR antibody targeting transferrin receptor 1. FASEB J. 2021, 35, e21172. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Bing, T.; Shen, L.; Feng, L.; Liu, X.; Shangguan, D. A DNA Aptameric Ligand of Human Transferrin Receptor Generated by Cell-SELEX. Int. J. Mol. Sci. 2021, 22, 8923. [Google Scholar] [CrossRef] [PubMed]
- Cheng, E.L.; Cardle, I.I.; Kacherovsky, N.; Bansia, H.; Wang, T.; Zhou, Y.; Raman, J.; Yen, A.; Gutierrez, D.; Salipante, S.J.; et al. Discovery of a Transferrin Receptor 1-Binding Aptamer and Its Application in Cancer Cell Depletion for Adoptive T-Cell Therapy Manufacturing. J. Am. Chem. Soc. 2022, 144, 13851–13864. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; El-Khoueiry, A.; Hafez, N.; Lakhani, N.; Mamdani, H.; Rodon, J.; Sanborn, R.E.; Garcia-Corbacho, J.; Boni, V.; Stroh, M.; et al. Phase I, First-in-Human Study of the Probody Therapeutic CX-2029 in Adults with Advanced Solid Tumor Malignancies. Clin. Cancer Res. 2021, 27, 4521–4530. [Google Scholar] [CrossRef]
- Okuyama, T.; Eto, Y.; Sakai, N.; Minami, K.; Yamamoto, T.; Sonoda, H.; Yamaoka, M.; Tachibana, K.; Hirato, T.; Sato, Y. Iduronate-2-Sulfatase with Anti-human Transferrin Receptor Antibody for Neuropathic Mucopolysaccharidosis II: A Phase 1/2 Trial. Mol. Ther. 2019, 27, 456–464. [Google Scholar] [CrossRef]
- Okuyama, T.; Eto, Y.; Sakai, N.; Nakamura, K.; Yamamoto, T.; Yamaoka, M.; Ikeda, T.; So, S.; Tanizawa, K.; Sonoda, H.; et al. A Phase 2/3 Trial of Pabinafusp Alfa, IDS Fused with Anti-Human Transferrin Receptor Antibody, Targeting Neurodegeneration in MPS-II. Mol. Ther. 2021, 29, 671–679. [Google Scholar] [CrossRef]
- Xiang, W.; Peng, Y.; Zeng, H.; Yu, C.; Zhang, Q.; Liu, B.; Liu, J.; Hu, X.; Wei, W.; Deng, M.; et al. Targeting treatment of bladder cancer using PTK7 aptamer-gemcitabine conjugate. Biomater. Res. 2022, 26, 74. [Google Scholar] [CrossRef]
- Liu, J.; Liu, X.; Yi, M.; Sun, Y.; Bing, T.; Zhang, N.; Shangguan, D. A photo-activated aptamer-drug conjugate for targeted drug delivery. Chem. Commun. 2022, 58, 10797–10800. [Google Scholar] [CrossRef]
- Heidari, F.; Mohajeri, N.; Zarghami, N. Targeted design of green carbon dot-CA-125 aptamer conjugate for the fluorescence imaging of ovarian cancer cell. Cell Biochem. Biophys 2022, 80, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Cai, X.; Sun, Q.; Guo, X.; Liang, C.; Tang, H.; Huang, H.; Luo, H.; Chen, L.; Chen, J. Design and synthesis of aptamer-cyclometalated iridium(III) complex conjugate targeting cancer cells. Eur. J. Med. Chem. 2022, 236, 114335. [Google Scholar] [CrossRef]
- Puzzo, F.; Zhang, C.; Powell Gray, B.; Zhang, F.; Sullenger, B.A.; Kay, M.A. Aptamer-programmable adeno-associated viral vectors as a novel platform for cell-specific gene transfer. Mol. Ther. Nucleic Acids 2023, 31, 383–397. [Google Scholar] [CrossRef]
- Macdonald, J.; Denoyer, D.; Henri, J.; Jamieson, A.; Burvenich, I.J.G.; Pouliot, N.; Shigdar, S. Bifunctional Aptamer-Doxorubicin Conjugate Crosses the Blood-Brain Barrier and Selectively Delivers Its Payload to EpCAM-Positive Tumor Cells. Nucleic Acid Ther. 2020, 30, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Xuan, W.; Xia, Y.; Li, T.; Wang, L.; Liu, Y.; Tan, W. Molecular Self-Assembly of Bioorthogonal Aptamer-Prodrug Conjugate Micelles for Hydrogen Peroxide and pH-Independent Cancer Chemodynamic Therapy. J. Am. Chem. Soc. 2020, 142, 937–944. [Google Scholar] [CrossRef]
- Russo, V.; Paciocco, A.; Affinito, A.; Roscigno, G.; Fiore, D.; Palma, F.; Galasso, M.; Volinia, S.; Fiorelli, A.; Esposito, C.L.; et al. Aptamer-miR-34c Conjugate Affects Cell Proliferation of Non-Small-Cell Lung Cancer Cells. Mol. Ther. Nucleic Acids 2018, 13, 334–346. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Lu, J.; Liu, J.; Liang, C.; Wang, M.; Wang, L.; Li, D.; Yao, H.; Zhang, Q.; Wen, J.; et al. A water-soluble nucleolin aptamer-paclitaxel conjugate for tumor-specific targeting in ovarian cancer. Nat. Commun. 2017, 8, 1390. [Google Scholar] [CrossRef] [PubMed]
- Kotula, J.W.; Sun, J.; Li, M.; Pratico, E.D.; Fereshteh, M.P.; Ahrens, D.P.; Sullenger, B.A.; Kovacs, J.J. Targeted disruption of beta-arrestin 2-mediated signaling pathways by aptamer chimeras leads to inhibition of leukemic cell growth. PLoS ONE 2014, 9, e93441. [Google Scholar] [CrossRef]
- Kotula, J.W.; Pratico, E.D.; Ming, X.; Nakagawa, O.; Juliano, R.L.; Sullenger, B.A. Aptamer-mediated delivery of splice-switching oligonucleotides to the nuclei of cancer cells. Nucleic Acid Ther. 2012, 22, 187–195. [Google Scholar] [CrossRef]
- Huang, Y.F.; Shangguan, D.; Liu, H.; Phillips, J.A.; Zhang, X.; Chen, Y.; Tan, W. Molecular assembly of an aptamer-drug conjugate for targeted drug delivery to tumor cells. Chembiochem 2009, 10, 862–868. [Google Scholar] [CrossRef]
- Dollins, C.M.; Nair, S.; Sullenger, B.A. Aptamers in immunotherapy. Hum. Gene Ther. 2008, 19, 443–450. [Google Scholar] [CrossRef] [PubMed]
- McNamara, J.O., II; Andrechek, E.R.; Wang, Y.; Viles, K.D.; Rempel, R.E.; Gilboa, E.; Sullenger, B.A.; Giangrande, P.H. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006, 24, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Sullenger, B.A.; Cech, T.R. Tethering ribozymes to a retroviral packaging signal for destruction of viral RNA. Science 1993, 262, 1566–1569. [Google Scholar] [CrossRef] [PubMed]


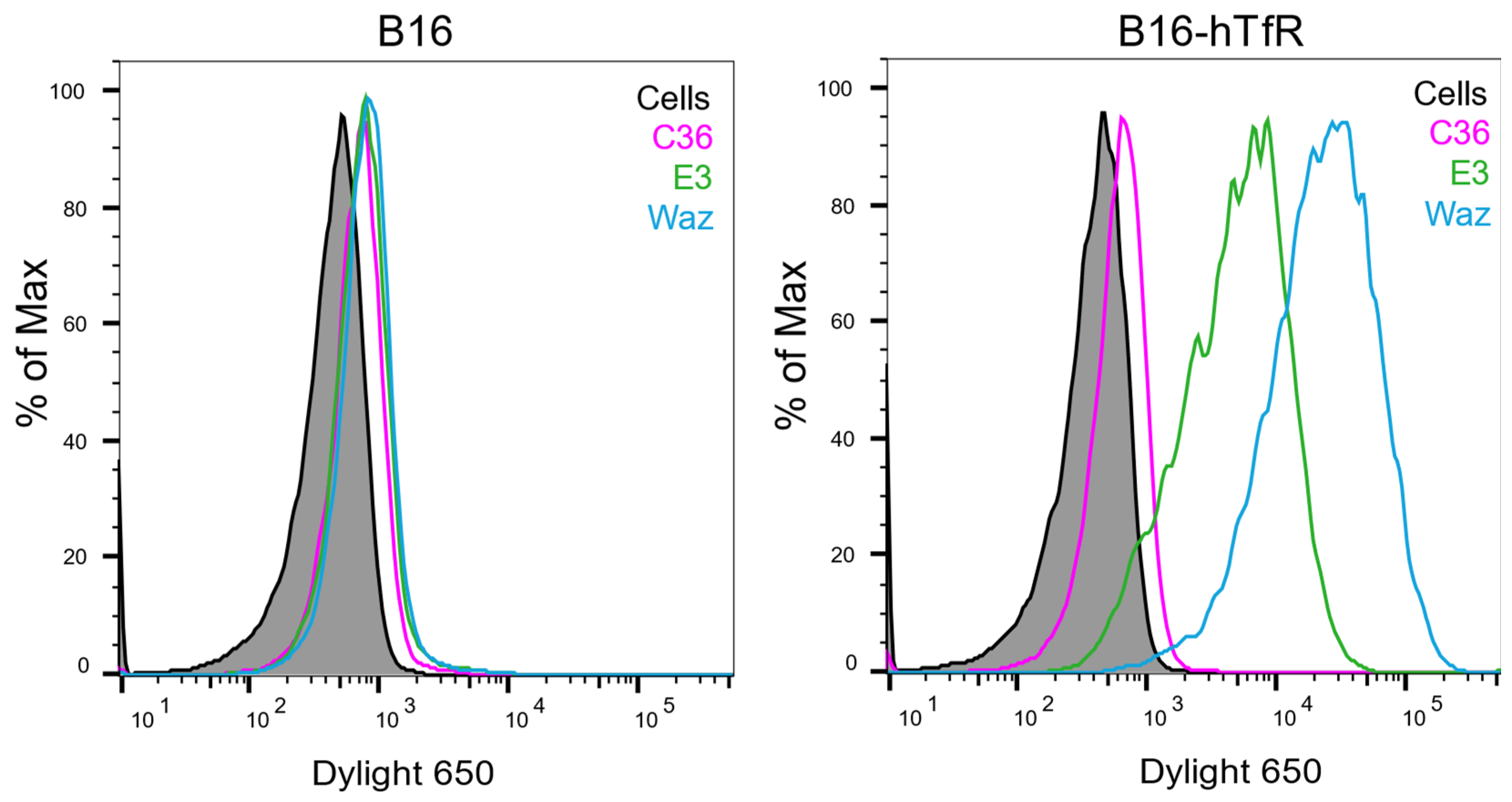

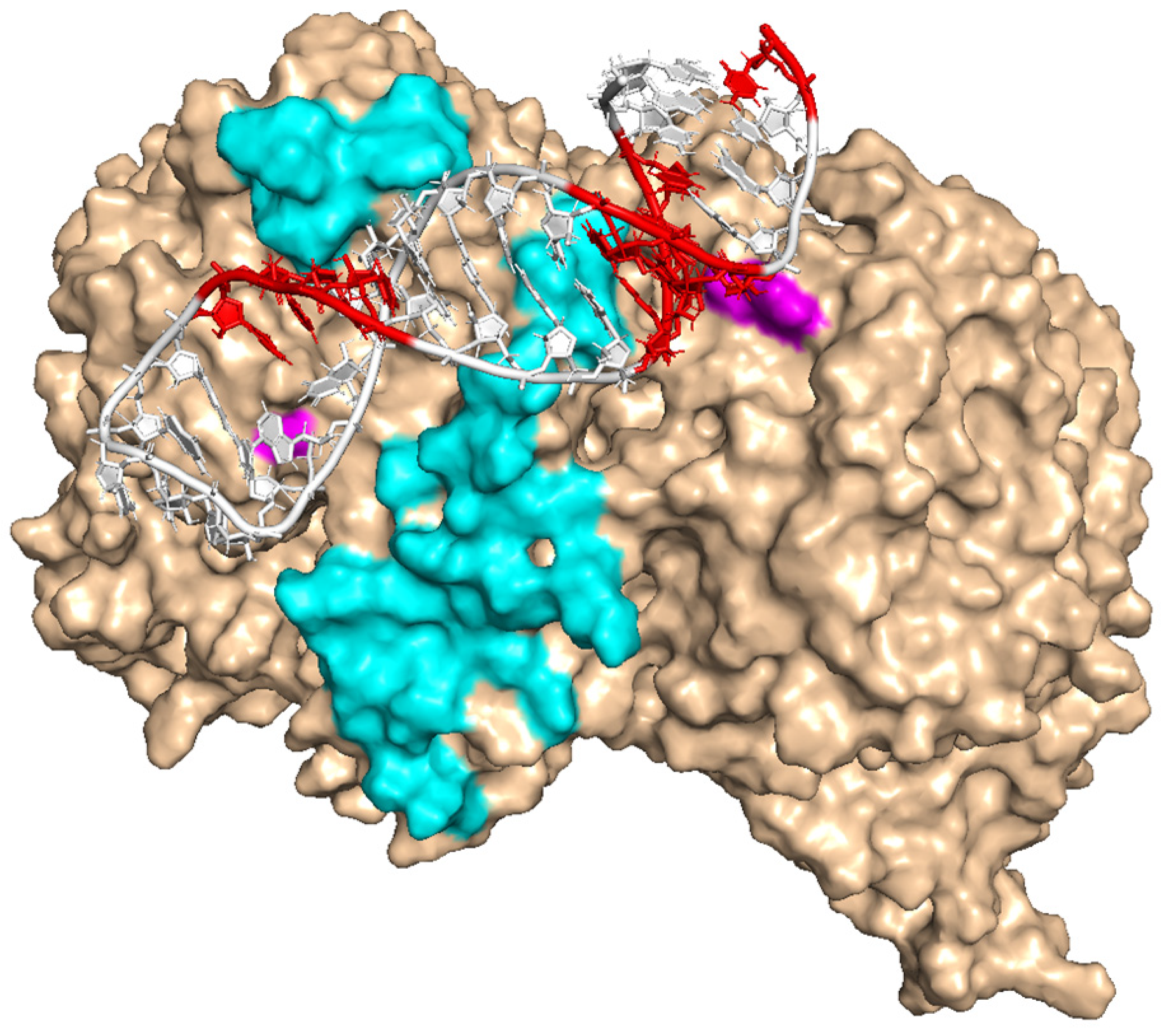
| Aptamer | Apparent KD (95% CI) (nM) | Bmax (95% CI) |
|---|---|---|
| E3 | 4.23 (3.55–5.04) | 197.0 (190.4–203.9) |
| C2.min | 7.44 (4.46–13.06) | 355.3 (319.8–395.7) |
| Waz | 22.09 (16.05–30.62) | 198.8 (177.8–224.4) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, X.; Yu, H.; Sullenger, C.; Gray, B.P.; Yan, A.; Kelly, L.; Sullenger, B. An Aptamer That Rapidly Internalizes into Cancer Cells Utilizes the Transferrin Receptor Pathway. Cancers 2023, 15, 2301. https://doi.org/10.3390/cancers15082301
Song X, Yu H, Sullenger C, Gray BP, Yan A, Kelly L, Sullenger B. An Aptamer That Rapidly Internalizes into Cancer Cells Utilizes the Transferrin Receptor Pathway. Cancers. 2023; 15(8):2301. https://doi.org/10.3390/cancers15082301
Chicago/Turabian StyleSong, Xirui, Haixiang Yu, Cynthia Sullenger, Bethany Powell Gray, Amy Yan, Linsley Kelly, and Bruce Sullenger. 2023. "An Aptamer That Rapidly Internalizes into Cancer Cells Utilizes the Transferrin Receptor Pathway" Cancers 15, no. 8: 2301. https://doi.org/10.3390/cancers15082301
APA StyleSong, X., Yu, H., Sullenger, C., Gray, B. P., Yan, A., Kelly, L., & Sullenger, B. (2023). An Aptamer That Rapidly Internalizes into Cancer Cells Utilizes the Transferrin Receptor Pathway. Cancers, 15(8), 2301. https://doi.org/10.3390/cancers15082301






