Occurrence of Hepatoblastomas in Patients with Beckwith–Wiedemann Spectrum (BWSp)
Abstract
:Simple Summary
Abstract
1. Introduction
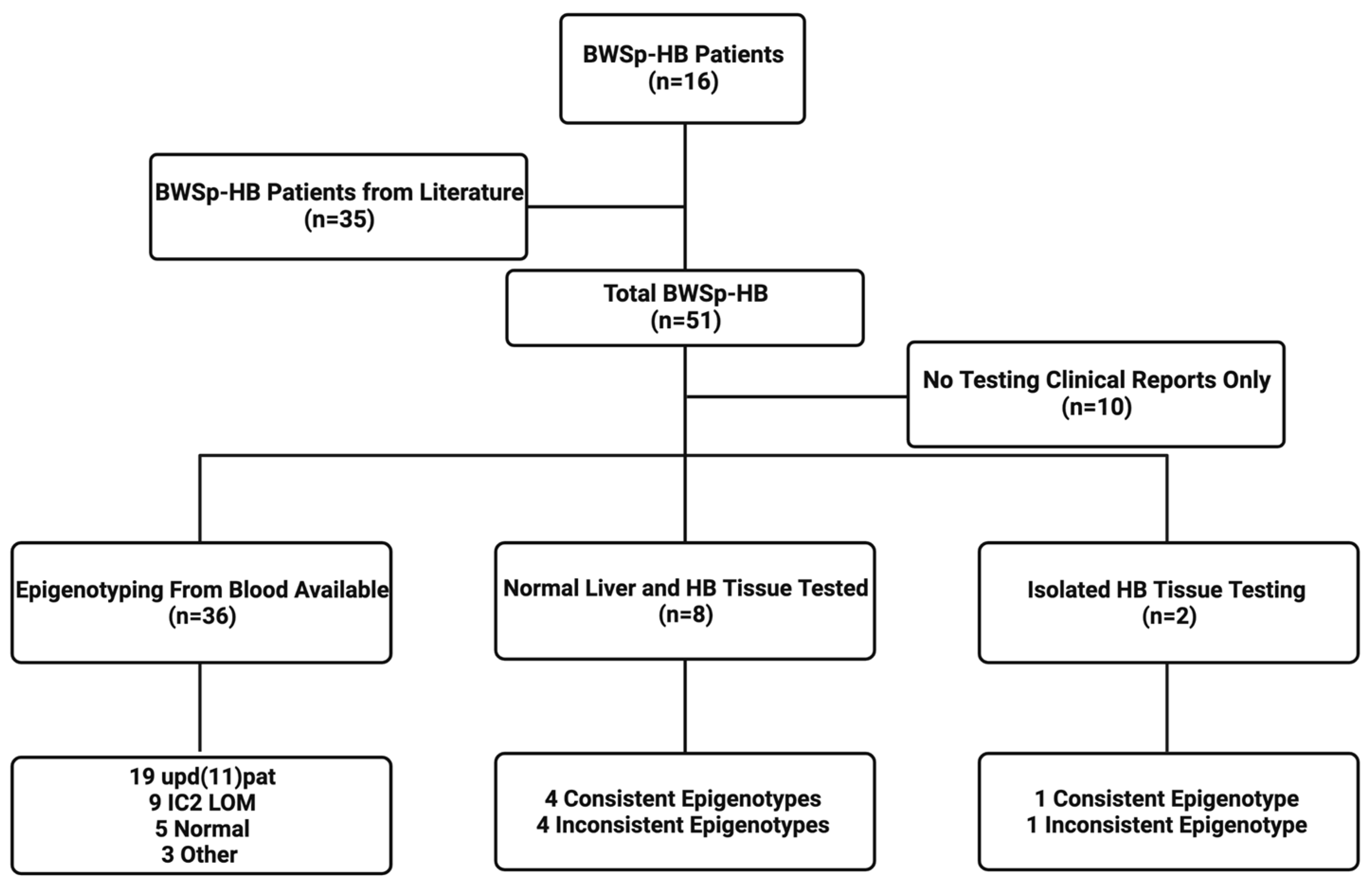
2. Materials and Methods
2.1. Patient Cohort
2.2. Molecular Testing
3. Results
3.1. Demographics, Epigenotypes, and Pathologic Types of Patients with BWSp-HB
3.2. BWS Clinical Features
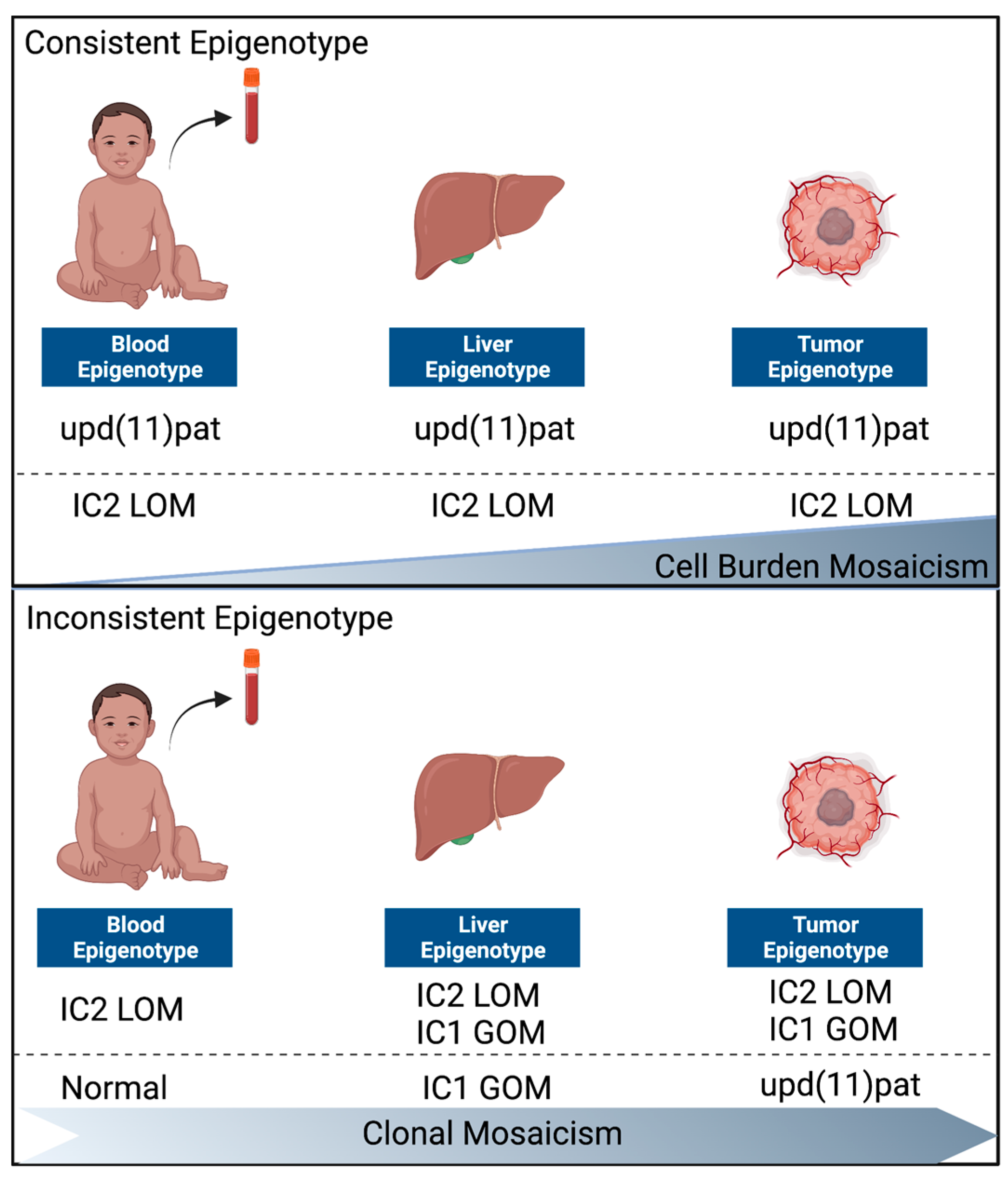
3.3. Normal Liver and Tumor Methylation Analysis
3.4. Solid Tumor Panel Analysis
4. Discussion
4.1. Risk Stratification Based on Blood Genotypes in BWS-HBs
4.2. Two Distinct Groups of BWS-HBs
4.3. The Role of CTNNB1 in BWS-HBs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kahla, J.A.; Siegel, D.A.; Dai, S.; Lupo, P.J.; Foster, J.H.; Scheurer, M.E.; Heczey, A.A. Incidence and 5-year survival of children and adolescents with hepatoblastoma in the United States. Pediatr. Blood Cancer 2022, 10, e29763. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Guan, Q.; Guo, H.; Miao, L.; Zhuo, Z. The Genetic Changes of Hepatoblastoma. Front. Oncol. 2021, 11, 690641. [Google Scholar] [CrossRef] [PubMed]
- Spector, L.G.; Birch, J. The epidemiology of hepatoblastoma. Pediatr. Blood Cancer 2012, 59, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, G.E.; Kappler, R. Genetics and epigenetics of hepatoblastoma. Pediatr. Blood Cancer 2012, 59, 785–792. [Google Scholar] [CrossRef]
- Goudie, C.; Witkowski, L.; Cullinan, N.; Reichman, L.; Schiller, I.; Tachdjian, M.; Armstrong, L.; Blood, K.A.; Brossard, J.; Brunga, L.; et al. Performance of the McGill Interactive Pediatric OncoGenetic Guidelines for Identifying Cancer Predisposition Syndromes. JAMA Oncol. 2021, 7, 1806–1814. [Google Scholar] [CrossRef]
- Grant, C.N.; Rhee, D.; Tracy, E.T.; Aldrink, J.H.; Baertschiger, R.M.; Lautz, T.B.; Glick, R.D.; Rodeberg, D.A.; Ehrlich, P.F.; Christison-Lagay, E. Pediatric solid tumors and associated cancer predisposition syndromes: Workup, management, and surveillance. A summary from the APSA Cancer Committee. J. Pediatr. Surg. 2022, 57, 430–442. [Google Scholar] [CrossRef]
- Huber, S.; Schimmel, M.; Dunstheimer, D.; Nemes, K.; Richter, M.; Streble, J.; Vollert, K.; Walden, U.; Frühwald, M.C.; Kuhlen, M. The need for tumor surveillance of children and adolescents with cancer predisposition syndromes: A retrospective cohort study in a tertiary-care children’s hospital. Eur. J. Pediatr. 2022, 181, 1585–1596. [Google Scholar] [CrossRef]
- Kalish, J.M.; Doros, L.; Helman, L.J.; Hennekam, R.C.; Kuiper, R.P.; Maas, S.M.; Maher, E.; Nichols, K.E.; Plon, S.E.; Porter, C.C.; et al. Surveillance Recommendations for Children with Overgrowth Syndromes and Predisposition to Wilms Tumors and Hepatoblastoma. Clin. Cancer Res. 2017, 23, e115–e122. [Google Scholar] [CrossRef]
- Achatz, M.I.; Porter, C.C.; Brugières, L.; Druker, H.; Frebourg, T.; Foulkes, W.D.; Kratz, C.P.; Kuiper, R.P.; Hansford, J.R.; Hernandez, H.S.; et al. Cancer Screening Recommendations and Clinical Management of Inherited Gastrointestinal Cancer Syndromes in Childhood. Clin. Cancer Res. 2017, 23, e107–e114. [Google Scholar] [CrossRef]
- Brioude, F.; Kalish, J.M.; Mussa, A.; Foster, A.C.; Bliek, J.; Ferrero, G.B.; E Boonen, S.; Cole, T.; Baker, R.; Bertoletti, M.; et al. Expert consensus document: Clinical and molecular diagnosis, screening and management of Beckwith-Wiedemann syndrome: An international consensus statement. Nat. Rev. Endocrinol. 2018, 14, 229–249. [Google Scholar] [CrossRef]
- Duffy, K.A.; Cielo, C.M.; Cohen, J.L.; Gonzalez-Gandolfi, C.X.; Griff, J.R.; Hathaway, E.R.; Kupa, J.; Taylor, J.A.; Wang, K.H.; Ganguly, A.; et al. Characterization of the Beckwith-Wiedemann spectrum: Diagnosis and management. Am. J. Med Genet. Part C: Semin. Med Genet. 2019, 181, 693–708. [Google Scholar] [CrossRef]
- Fiala, E.M.; Ortiz, M.V.; Kennedy, J.A.; Glodzik, D.; Fleischut, M.H.; Duffy, K.A.; Hathaway, E.R.; Heaton, T.; Gerstle, J.T.; Steinherz, P.; et al. 11p15.5 epimutations in children with Wilms tumor and hepatoblastoma detected in peripheral blood. Cancer 2020, 126, 3114–3121. [Google Scholar] [CrossRef]
- Duffy, K.A.; Getz, K.D.; Hathaway, E.R.; Byrne, M.E.; MacFarland, S.P.; Kalish, J.M. Characteristics Associated with Tumor Development in Individuals Diagnosed with Beckwith–Wiedemann Spectrum: Novel Tumor-(epi)Genotype-Phenotype Associations in the BWSp Population. Genes 2021, 12, 1839. [Google Scholar] [CrossRef]
- Sotelo-Avila, C.; Gonzalez-Crussi, F.; Fowler, J.W. Complete and incomplete forms of Beckwith-Wiedemann syndrome: Their oncogenic potential. J. Pediatr. 1980, 96, 47–50. [Google Scholar] [CrossRef]
- DeBaun, M.R.; Tucker, M.A. Risk of cancer during the first four years of life in children from The Beckwith-Wiedemann Syndrome Registry. J. Pediatr. 1998, 132, 398–400. [Google Scholar] [CrossRef]
- Bliek, J.; Gicquel, C.; Maas, S.; Gaston, V.; le Bouc, Y.; Mannens, M. Epigenotyping as a tool for the prediction of tumor risk and tumor type in patients with Beckwith-Wiedemann syndrome (BWS). J. Pediatr. 2004, 145, 796–799. [Google Scholar] [CrossRef]
- Maas, S.M.; Vansenne, F.; Kadouch, D.J.M.; Ibrahim, A.; Bliek, J.; Hopman, S.; Mannens, M.M.; Merks, J.H.M.; Maher, E.R.; Hennekam, R.C. Phenotype, cancer risk, and surveillance in Beckwith-Wiedemann syndrome depending on molecular genetic subgroups. Am. J. Med Genet. Part A 2016, 170, 2248–2260. [Google Scholar] [CrossRef]
- Weksberg, R.; Nishikawa, J.; Caluseriu, O.; Fei, Y.-L.; Shuman, C.; Wei, C.; Steele, L.; Cameron, J.; Smith, A.; Ambus, I.; et al. Tumor development in the Beckwith-Wiedemann syndrome is associated with a variety of constitutional molecular 11p15 alterations including imprinting defects of KCNQ1OT1. Hum. Mol. Genet. 2001, 10, 2989–3000. [Google Scholar] [CrossRef]
- Mussa, A.; Molinatto, C.; Baldassarre, G.; Riberi, E.; Russo, S.; Larizza, L.; Riccio, A.; Ferrero, G.B. Cancer Risk in Beckwith-Wiedemann Syndrome: A Systematic Review and Meta-Analysis Outlining a Novel (Epi)Genotype Specific Histotype Targeted Screening Protocol. J. Pediatr. 2016, 176, 142–149.e1. [Google Scholar] [CrossRef]
- Duffy, K.A.; Deardorff, M.A.; Kalish, J.M. The utility of alpha-fetoprotein screening in Beckwith-Wiedemann syndrome. Am. J. Med Genet. Part A 2017, 173, 581–584. [Google Scholar] [CrossRef]
- Mussa, A.; Russo, S.; de Crescenzo, A.; Freschi, A.; Calzari, L.; Maitz, S.; Macchiaiolo, M.; Molinatto, C.; Baldassarre, G.; Mariani, M.; et al. Fetal growth patterns in Beckwith-Wiedemann syndrome. Clin. Genet. 2016, 90, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.W.; Duffy, K.A.; Richards-Yutz, J.; Deardorff, M.A.; Kalish, J.M.; Ganguly, A. Improved molecular detection of mosaicism in Beckwith-Wiedemann Syndrome. J. Med Genet. 2021, 58, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Conlin, L.K.; Thiel, B.D.; Bonnemann, C.G.; Medne, L.; Ernst, L.; Zackai, E.H.; Deardorff, M.A.; Krantz, I.D.; Hakonarson, H.; Spinner, N.B. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum. Mol. Genet. 2010, 19, 1263–1275. [Google Scholar] [CrossRef] [PubMed]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef]
- Calton, E.A.; Temple, I.K.; Mackay, D.J.; Lever, M.; Ellard, S.; Flanagan, S.E.; Davies, J.H.; Hussain, K.; Gray, J.C. Hepatoblastoma in a child with a paternally-inherited ABCC8 mutation and mosaic paternal uniparental disomy 11p causing focal congenital hyperinsulinism. Eur. J. Med Genet. 2013, 56, 114–117. [Google Scholar] [CrossRef]
- Little, M.; Thomson, D.B.; Hayward, N.K.; Smith, P.J. Loss of alleles on the short arm of chromosome 11 in a hepatoblastoma from a child with Beckwith-Wiedemann syndrome. Hum. Genet. 1988, 79, 186–189. [Google Scholar] [CrossRef]
- Bachmann, N.; Crazzolara, R.; Bohne, F.; Kotzot, D.; Maurer, K.; Enklaar, T.; Prawitt, D.; Bergmann, C. Novel deletion in 11p15.5 imprinting center region 1 in a patient with Beckwith-Wiedemann syndrome provides insight into distal enhancer regulation and tumorigenesis. Pediatr. Blood Cancer 2017, 64, e26241. [Google Scholar] [CrossRef]
- Haas, O.A.; Zoubek, A.; Grümayer, E.R.; Gadner, H. Constitutional interstitial deletion of 11p11 and pericentric inversion of chromosome 9 in a patient with Wiedemann-Beckwith syndrome and hepatoblastoma. Cancer Genet. Cytogenet. 1986, 23, 95–104. [Google Scholar] [CrossRef]
- Kiruthiga, K.G.; Ramakrishna, B.; Saha, S.; Sen, S. Histological and immunohistochemical study of hepatoblastoma: Correlation with tumour behaviour and survival. J. Gastrointest. Oncol. 2018, 9, 326–337. [Google Scholar] [CrossRef]
- Aronson, D.C.; Meyers, R.L. Malignant tumors of the liver in children. Semin. Pediatr. Surg. 2016, 25, 265–275. [Google Scholar] [CrossRef]
- López-Terrada, D.; Alaggio, R.; de Dávila, M.T.; Czauderna, P.; Hiyama, E.; Katzenstein, H.; Leuschner, I.; Malogolowkin, M.; Meyers, R.; Ranganathan, S.; et al. Towards an international pediatric liver tumor consensus classification: Proceedings of the Los Angeles COG liver tumors symposium. Mod. Pathol. 2014, 27, 472–491. [Google Scholar] [CrossRef]
- Naveh, N.S.S.; Traxler, E.M.; Duffy, K.A.; Kalish, J.M. Molecular networks of hepatoblastoma predisposition and oncogenesis in Beckwith-Wiedemann syndrome. Hepatol. Commun. 2022, 6, 2132–2146. [Google Scholar] [CrossRef]
- Kim, S.; Jeong, S. Mutation Hotspots in the beta-Catenin Gene: Lessons from the Human Cancer Genome Databases. Mol. Cells 2019, 42, 8–16. [Google Scholar]
- Larsen, L.J.; Møller, L.B. Crosstalk of Hedgehog and mTORC1 Pathways. Cells 2020, 9, 2316. [Google Scholar] [CrossRef]
- Duffy, K.A.; Hathaway, E.R.; Klein, S.D.; Ganguly, A.; Kalish, J.M. Epigenetic mosaicism and cell burden in Beckwith–Wiedemann syndrome due to loss of methylation at imprinting control region 2. Cold Spring Harb. Mol. Case Stud. 2021, 7, a006115. [Google Scholar] [CrossRef]
| This Report | The Literature | Total | |
|---|---|---|---|
| Total | 16 | 35 | 51 |
| Male | 5 (31%) | 16 (46%) | 21 (41%) |
| Female | 11 (69%) | 19 (54%) | 30 (59%) |
| Age of diagnosis min | Birth | in utero | in utero |
| Age of diagnosis max | 25 months | 22 years | 22 years |
| Epigenotype from Blood | 15 | 20 | 35 |
| upd(11)pat | 7 (47%) | 13 (65%) | 20 (57%) |
| IC2 LOM | 5 (33%) | 4 (20%) | 9 (26%) |
| Normal | 3 (20%) | n/a | 3 (9%) |
| Other | 0 | 3 (15%) | 3 (9%) |
| Pathology | 14 | 16 | 30 |
| Mixed epithelial | 14 (100%) | 16 (100%) | 30 (100%) |
| +Mesenchymal | 4 (29%) | 1 (6%) | 5 (17%) |
| +Cholangioblastic | 1 (7%) | 1 (6%) | 2 (7%) |
| Clinical Features | # of Patients (n) | % |
|---|---|---|
| Lateralized overgrowth | 14 (16) | 88% |
| Nevus simplex | 8 (10) | 80% |
| Macroglossia | 11 (15) | 80% |
| Ear creases | 8 (10) | 73% |
| Umbilical hernia/diastasis recti | 5 (8) | 55% |
| Hypoglycemia | 8 (15) | 53% |
| Hyperinsulinism | 5 (13) | 38% |
| Placental mesenchymal dysplasia | 1 (3) | 33% |
| Omphalocele | 3 (13) | 23% |
| Nephromegaly | 3 (13) | 21% |
| Hepatomegaly | 2 (13) | 15% |
| ID | BWS Clinical Score | BWS Blood Results | BWS Liver Results | BWS HB Results | CTNNB1 Mutation |
|---|---|---|---|---|---|
| 1 | 9 | upd(11)pat | upd(11)pat | upd(11)pat | c.101G>T, c.98C>T |
| 2 | 10 | upd(11)pat | upd(11)pat | upd(11)pat | c.99_101dup |
| 3 | 8 | upd(11)pat | \ | upd(11)pat | c. 65_100del |
| 4 | 10 | upd(11)pat | \ | \ | c.100G>A |
| 5 | 10 | upd(11)pat | \ | \ | \ |
| 6 | 10 | upd(11)pat | \ | \ | \ |
| 7 | 3 | upd(11)pat | \ | \ | \ |
| 8 | 12 | IC2 LOM | IC2 LOM | IC2 LOM | \ |
| 9 | 6 | IC2 LOM | IC2 LOM | IC2 LOM | c.85_102del |
| 10 | 5 | IC2 LOM | IC2 LOM/IC1 GOM | IC2 LOM/IC1 GOM | c.122C>T |
| 11 | 6 | IC2 LOM | \ | \ | \ |
| 12 | 10 | IC2 LOM | \ | \ | \ |
| 13 | 3 | Normal | \ | IC2 LOM | c.14-28_125del |
| 14 | 2 | \ | IC1GOM | upd(11)pat | c.87_241+71del |
| 15 | 4 | Normal | IC1 GOM | upd(11)pat | c.95A>T |
| 16 | 6 | Normal | IC1 GOM | upd(11)pat | \ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klein, S.D.; DeMarchis, M.; Linn, R.L.; MacFarland, S.P.; Kalish, J.M. Occurrence of Hepatoblastomas in Patients with Beckwith–Wiedemann Spectrum (BWSp). Cancers 2023, 15, 2548. https://doi.org/10.3390/cancers15092548
Klein SD, DeMarchis M, Linn RL, MacFarland SP, Kalish JM. Occurrence of Hepatoblastomas in Patients with Beckwith–Wiedemann Spectrum (BWSp). Cancers. 2023; 15(9):2548. https://doi.org/10.3390/cancers15092548
Chicago/Turabian StyleKlein, Steven D., Madison DeMarchis, Rebecca L. Linn, Suzanne P. MacFarland, and Jennifer M. Kalish. 2023. "Occurrence of Hepatoblastomas in Patients with Beckwith–Wiedemann Spectrum (BWSp)" Cancers 15, no. 9: 2548. https://doi.org/10.3390/cancers15092548
APA StyleKlein, S. D., DeMarchis, M., Linn, R. L., MacFarland, S. P., & Kalish, J. M. (2023). Occurrence of Hepatoblastomas in Patients with Beckwith–Wiedemann Spectrum (BWSp). Cancers, 15(9), 2548. https://doi.org/10.3390/cancers15092548






