Antiproliferative Activity of Krukovine by Regulating Transmembrane Protein 139 (TMEM139) in Oxaliplatin-Resistant Pancreatic Cancer Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultures and Reagents
2.2. MTT Assay (Cell Viability Assay)
2.3. RNA Preparation, Library Preparation, and RNA-Seq
2.3.1. Preprocessing of the RNA-Seq Data
2.3.2. Differential Transcriptome and Functional Analysis
2.3.3. Molecular Pathway and Functional Analysis
2.4. Kaplan–Meier Plotter Analysis
2.5. Protein–Protein Interaction (PPI) Network Analysis
2.6. Western Blot
2.7. Wound Healing Assay (Cell Migration Assay)
2.8. Transwell Cell Invasion Assay
2.9. Organoid Medium
2.10. Organoid Viability Assay
2.11. Mutation Profile of PDPCOs
2.12. Data Analysis
3. Results
3.1. Krukovine Shows an Antiproliferative Effect toward KRAS-Mutated Pancreatic Cancer Cells and Oxaliplatin-Resistant Pancreatic Cancer Cells
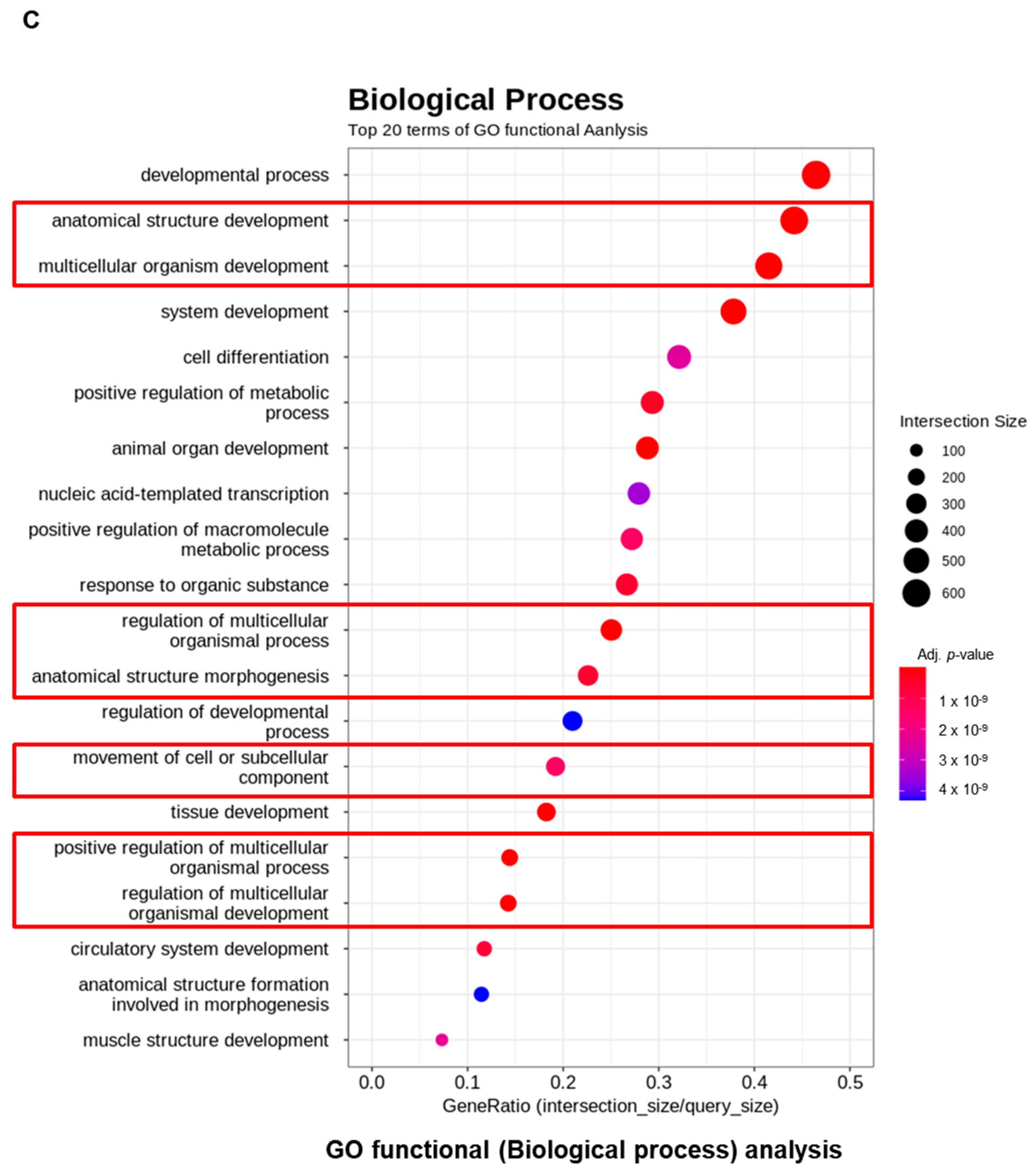
3.2. RNA Levels of KV-Treated Pancreatic Cancer Cells Show Major Metabolic Pathway
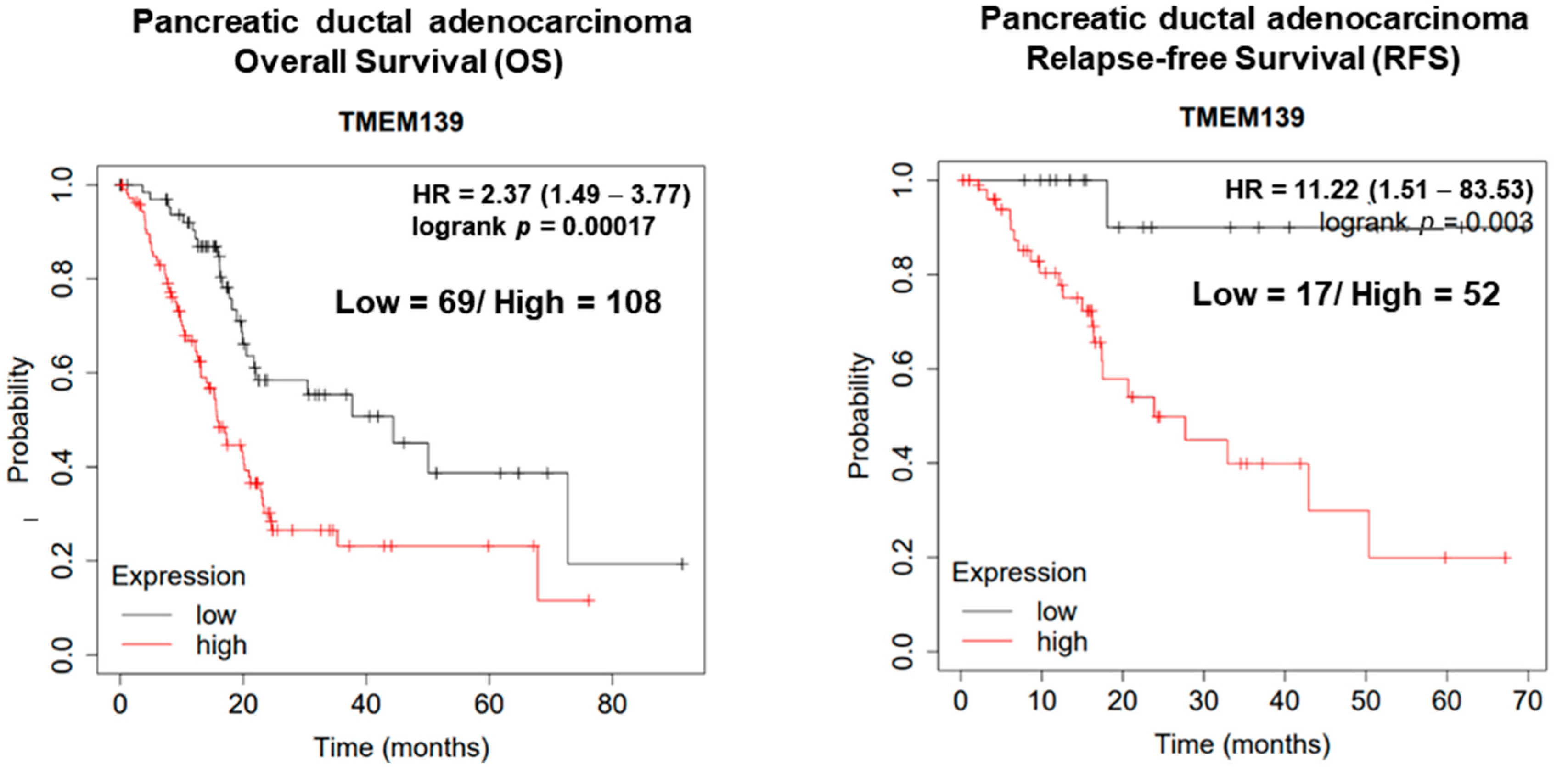
3.3. The Clinical Significance of TMEM139 Expression in Pancreatic Cancer Patients
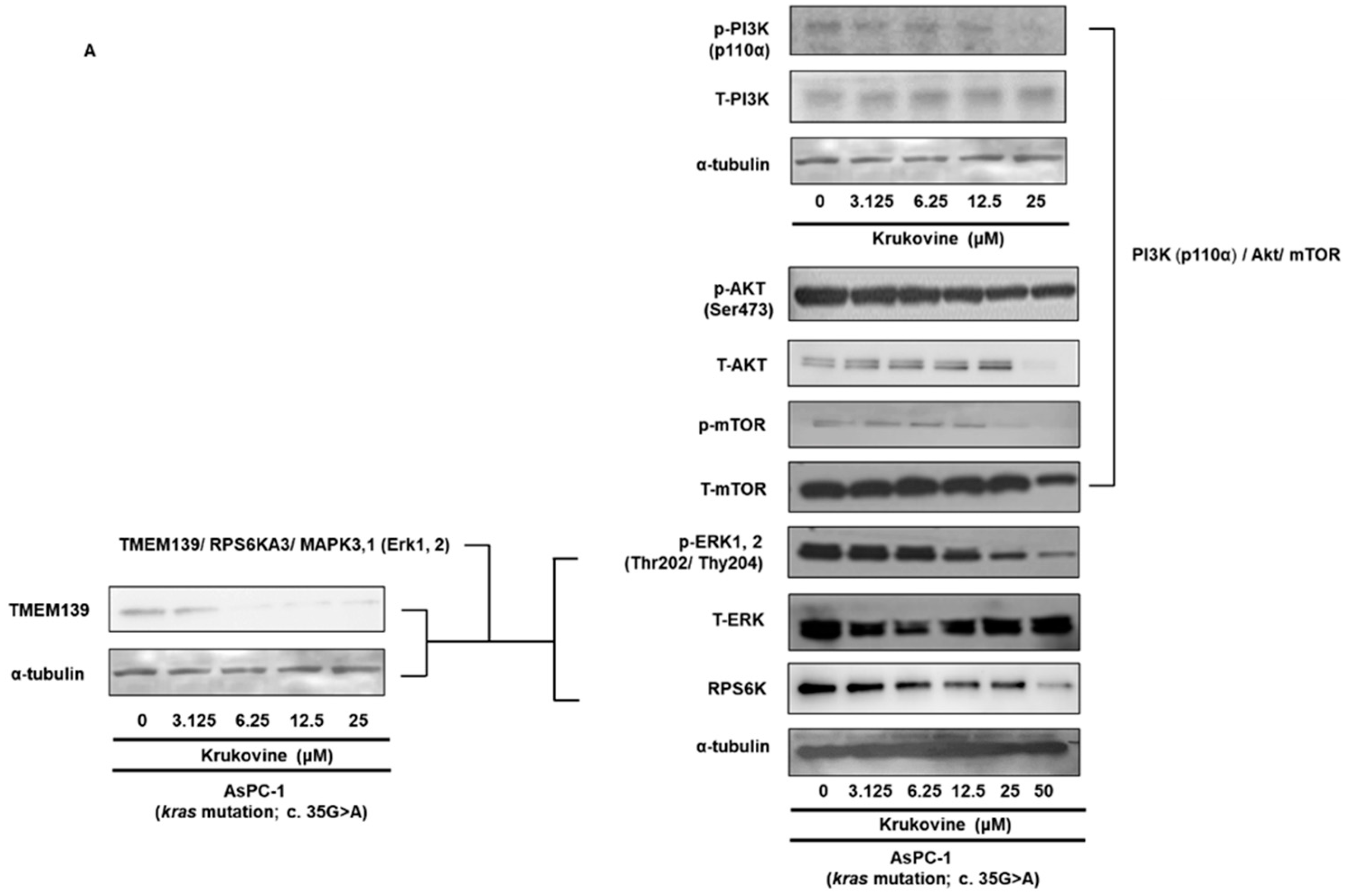
3.4. Effects of KV on the TMEM 139 Expression and TMEM139-Associated Signaling Pathway in Oxaliplatin-Resistant AsPC-1 Cells
3.5. KV Inhibits the PI3K-Akt-mTOR Pathway in Oxaliplatin-Resistant AsPC-1 Cells
3.6. KV Inhibits Migration and Invasion in Oxaliplatin-Resistant AsPC-1 Cells
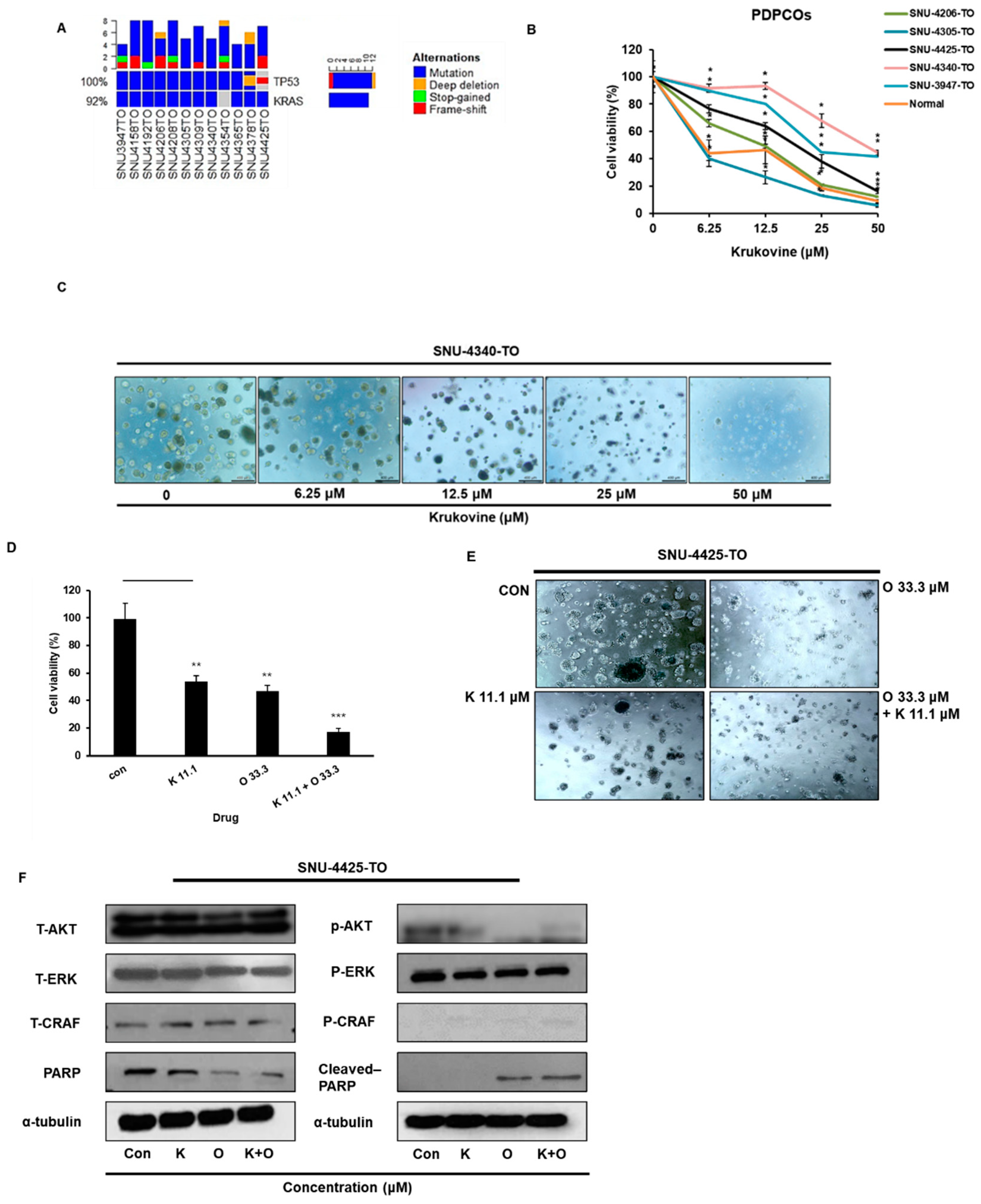
3.7. KV Showed an Antiproliferative Effect and Enhanced the Anticancer Effects of Oxaliplatin in KRAS-Mutated Patient-Derived Pancreatic Cancer Organoids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Iovanna, J.; Mallmann, M.C.; Gonçalves, A.; Turrini, O.; Dagorn, J.-C. Current knowledge on pancreatic cancer. Front. Oncol. 2012, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Nogueira, L.; Mariotto, A.B.; Rowland, J.H.; Yabroff, K.R.; Alfano, C.M.; Ahmedin Jemal, D.V.M.; Kramer, J.L.; Siege, R.L. Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 2019, 69, 363–385. [Google Scholar] [CrossRef]
- Rawla, P.; Sunkara, T.; Gaduputi, V. Epidemiology of pancreatic cancer: Global trends, etiology and risk factors. World J. Oncol. 2019, 10, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Feng, Z.; Miao, R.; Liu, X.; Liu, C.; Liu, Z. Prognosis and survival analysis of patients with pancreatic cancer: Retrospective experience of a single institution. World J. Surg. Oncol. 2022, 20, 11. [Google Scholar] [CrossRef]
- Otsu, T.; Inokawa, Y.; Takami, H.; Hayashi, M.; Kurimoto, K.; Tanaka, N.; Shimizu, D.; Hattori, N.; Kanda, M.; Tanaka, C.; et al. Comparison between FOLFIRINOX and nal-IRI/FL as Second-line Treatment After Gemcitabine Plus Nab-paclitaxel for Pancreatic Cancer. Anticancer Res. 2022, 42, 3889–3894. [Google Scholar] [CrossRef]
- Santucci, J.; Tacey, M.; Thomson, B.; Michael, M.; Wong, R.; Shapiro, J.; Jennens, R.; Clarke, K.; Pattison, S.; Burge, M.; et al. Impact of first-line FOLFIRINOX versus Gemcitabine/Nab-Paclitaxel chemotherapy on survival in advanced pancreatic cancer: Evidence from the prospective international multicentre PURPLE pancreatic cancer registry. Eur. J. Cancer 2022, 174, 102–112. [Google Scholar] [CrossRef]
- Mizrahi, J.D.; Surana, R.; Valle, J.W.; Shroff, R.T. Pancreatic cancer. Lancet 2020, 395, 2008–2020. [Google Scholar] [CrossRef]
- Micalizzi, D.S.; Che, D.; Nicholson, B.T.; Edd, J.F.; Desai, N.; Lang, E.R.; Toner, M.; Maheswaran, S.; Ting, D.T.; Haber, D.A. Targeting breast and pancreatic cancer metastasis using a dual-cadherin antibody. Proc. Natl. Acad. Sci. USA 2022, 119, e2209563119. [Google Scholar] [CrossRef] [PubMed]
- Marx, S.; Dal Maso, T.; Chen, J.-W.; Bury, M.; Wouters, J.; Michiels, C.; Le Calve, B. (Eds.) Transmembrane (TMEM) protein family members: Poorly characterized even if essential for the metastatic process. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Schmit, K.; Michiels, C. TMEM proteins in cancer: A review. Front. Pharmacol. 2018, 9, 1345. [Google Scholar] [CrossRef]
- Chen, K.; Zhang, Y.; Qian, L.; Wang, P. Emerging strategies to target RAS signaling in human cancer therapy. J. Hematol. Oncol. 2021, 14, 116. [Google Scholar] [CrossRef]
- Lai, H.; Wang, Y.; Duan, F.; Li, Y.; Jiang, Z.; Luo, L.; Liu, L.; Leung, E.L.H.; Yao, X. Krukovine suppresses KRAS-mutated lung Cancer cell growth and proliferation by inhibiting the RAF-ERK pathway and inactivating AKT pathway. Front. Pharmacol. 2018, 9, 958. [Google Scholar] [CrossRef] [PubMed]
- Zhiyao, F.; Kun, F.; Chao, Y.; Qiuyi, H.; Yitao, G.; He, C.; Kaizhou, J.; Chen, L.; Quanxing, N.; Xianjun, Y.; et al. Critical role of KRAS mutation in pancreatic ductal adenocarcinom. Transl. Cancer Res. 2018, 7, 1728–1736. [Google Scholar]
- Waters, A.M.; Der, C.J. KRAS: The critical driver and therapeutic target for pancreatic cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a031435. [Google Scholar] [CrossRef] [PubMed]
- Cragg, G.M.; Pezzuto, J.M. Natural products as a vital source for the discovery of cancer chemotherapeutic and chemopreventive agents. Med. Princ. Pract. 2016, 25 (Suppl. S2), 41–59. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Chavda, V.P.; Ertas, Y.N.; Walhekar, V.; Modh, D.; Doshi, A.; Shah, N.; Anand, K.; Chhabria, M. Advanced Computational Methodologies Used in the Discovery of New Natural Anticancer Compounds. Front. Pharmacol. 2021, 12, 702611. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Tilaoui, M.; Zyad, A. Update and New Insights on Future Cancer Drug Candidates from Plant-Based Alkaloids. Front. Pharmacol. 2021, 12, 719694. [Google Scholar] [CrossRef]
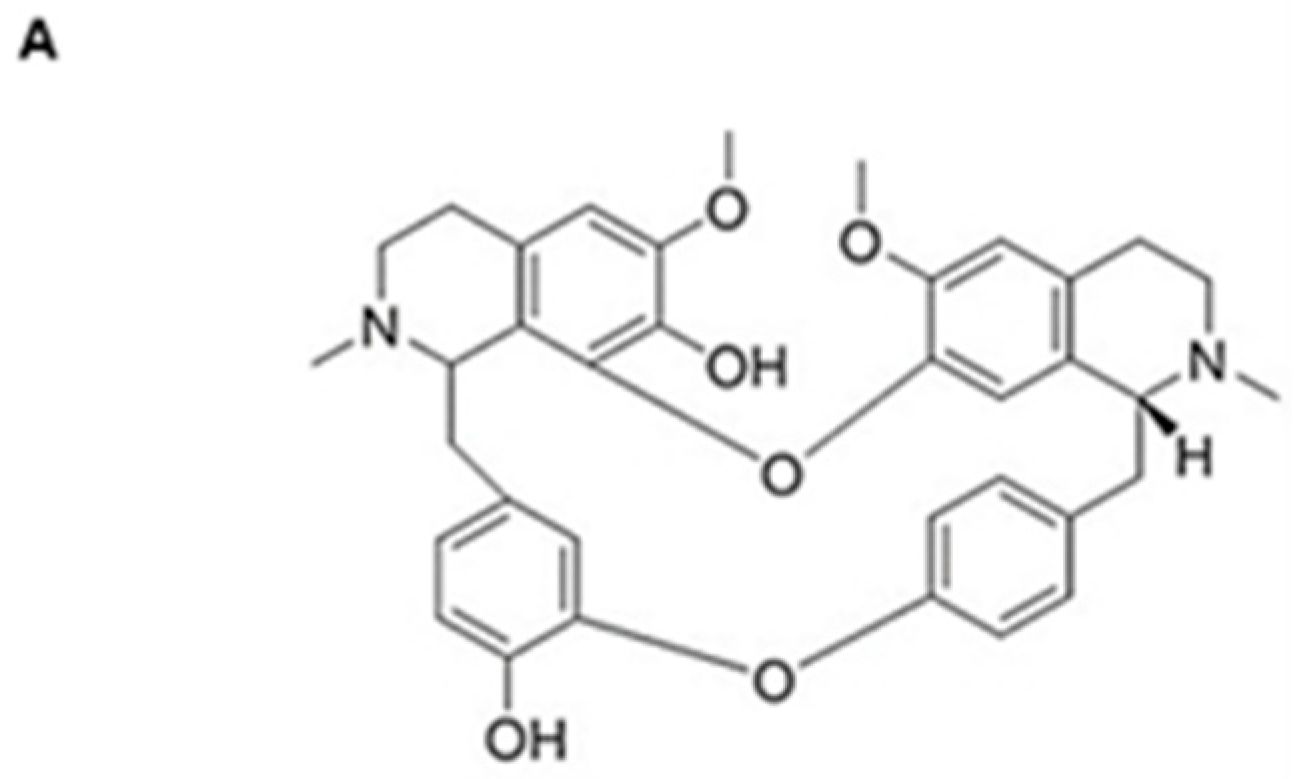
- Saa, J.M.; Lakshmikantham, M.; Mitchell, M.J.; Cava, M.P. Krukovine, a new bis (benzylisoquinoline) alkaloid from Abuta splendida. J. Org. Chem. 1976, 41, 317–319. [Google Scholar] [CrossRef]
- Weber, C.; Opatz, T. Bisbenzylisoquinoline alkaloids. Alkaloids Chem. Biol. 2019, 81, 1–114. [Google Scholar]
- Nagle, P.W.; Plukker, J.T.M.; Muijs, C.T.; van Luijk, P.; Coppes, R.P. (Eds.) Patient-derived tumor organoids for prediction of cancer treatment response. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Kim, J.; Koo, B.-K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Vivarelli, S.; Candido, S.; Caruso, G.; Falzone, L.; Libra, M. Patient-derived tumor organoids for drug repositioning in cancer care: A promising approach in the era of tailored treatment. Cancers 2020, 12, 3636. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yu, L.; Li, Z.; Li, W.; Huang, W. Patient-derived organoid (PDO) platforms to facilitate clinical decision making. J. Transl. Med. 2021, 19, 40. [Google Scholar] [CrossRef] [PubMed]
- Kondo, J.; Inoue, M. Application of cancer organoid model for drug screening and personalized therapy. Cells 2019, 8, 470. [Google Scholar] [CrossRef] [PubMed]
- Kevin, D.; Dimakatso, A. Senthebane and Collet Dandara. The Tumor Microenvironment in Tumorigenesis and Therapy Resistance Revisited. Cancers 2023, 15, 376. [Google Scholar]
- Haque, M.R.; Wessel, C.R.; Leary, D.D.; Wang, C.; Bhushan, A.; Bishehsari, F. Patient-derived pancreatic cancer-on-a-chip recapitulates the tumor microenvironment. Microsyst. Nanoeng. 2022, 8, 36. [Google Scholar] [CrossRef]
- Tan, C.; Wang, X.; Wang, X.; Weng, W.; Ni, S.-J.; Zhang, M.; Jiang, H.; Wang, L.; Huang, D.; Sheng, W.; et al. Molecular signatures of tumor progression in pancreatic adenocarcinoma identified by energy metabolism characteristics. BMC Cancer 2022, 22, 404. [Google Scholar] [CrossRef]
- Wang, S.; Zheng, Y.; Yang, F.; Zhu, L.; Zhu, X.-Q.; Wang, Z.-F.; Wu, X.-L.; Zhou, C.-H.; Yan, J.-Y.; Hu, B.-Y.; et al. The molecular biology of pancreatic adenocarcinoma: Translational challenges and clinical perspectives. Signal Transduct. Target. Ther. 2021, 6, 249. [Google Scholar] [CrossRef]
- He, X.; Wang, N.; Zhang, Y.; Huang, X.; Wang, Y. The therapeuticpotential of natural products for treatingpancreatic cancer. Front. Pharmacol. 2022, 13, 1051952. [Google Scholar] [CrossRef]
- Ruochen, D.; Ping, C.; Qi, C. Extract of the Medicinal Plant Pao Pereira Inhibits Pancreatic Cancer Stem-Like Cell In Vitro and In Vivo. Integr. Cancer Ther. 2018, 17, 1204–1215. [Google Scholar]
- Hu, H.-F.; Ye, Z.; Qin, Y.; Xu, X.-W.; Yu, X.-J.; Zhuo, Q.-F.; Ji, S.-R. Mutations in key driver genes of pancreatic cancer: Molecularly targeted therapies and other clinical implications. Acta Pharmacol. Sin. 2021, 42, 1725–1741. [Google Scholar] [CrossRef]
- McCubrey, J.A.; Steelman, L.S.; Chappell, W.H.; Abrams, S.L.; Montalto, G.; Cervello, M.; Nicoletti, F.; Fagone, P.; Malaponte, G.; Mazzarino, M.C.; et al. Mutations and deregulation of Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR cascades which alter therapy response. Oncotarget 2012, 3, 954. [Google Scholar] [CrossRef] [PubMed]
- Ning, W.; Yang, Z.; Kocher, G.J.; Dorn, P.; Peng, R.-W. A Breakthrough Brought about by Targeting KRASG12C: Nonconformity Is Punished. Cancers 2022, 14, 390. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Sun, M.M.; Zhang, G.G.; Yang, J.; Chen, K.S.; Xu, W.W.; Li, B. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct. Target. Ther. 2021, 6, 425. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.; Zheng, W.; Dong, J.; Wang, Y.; Lai, J.; Liu, X.; Yin, F. Tumor microenvironment in pancreatic ductal adenocarcinoma: Implications in immunotherapy. World J. Gastroenterol. 2022, 28, 3297–3313S. [Google Scholar] [CrossRef]
- Jin, Y.; Xiaoyang, L.; Shengji, Y. Cancer organoid co-culture model system: Novel approach to guide precision medicine. Front. Immunol. 2023, 13, 1061388. [Google Scholar]
- Heinemann, V. Gemcitabine-based combination treatment of pancreatic cancer. Semin. Oncol. 2002, 29, 25–35. [Google Scholar] [CrossRef]
- Lei, F.; Xi, X.; Batra, S.K.; Bronich, T.K. Combination therapies and drug delivery platforms in combating pancreatic cancer. J. Pharmacol. Exp. Ther. 2019, 370, 682–694. [Google Scholar] [CrossRef]
- Kroep, J.; Pinedo, H.; van Groeningen, C.; Peters, G. Experimental drugs and drug combinations in pancreatic cancer. Ann. Oncol. 1999, 10, S234–S238. [Google Scholar] [CrossRef]



| Gene Symbol | Description | kruko/con.fc | kruko/con.raw.pval |
|---|---|---|---|
| TMEM139 | transmembrane protein 139 | −20.08857481 | 0.000184391 |
| TMEM160 | transmembrane protein 160 | −10.07644718 | 0.0002668 |
| TMEM102 | transmembrane protein 102 | −9.08294669 | 0.000505557 |
| TMEM187 | transmembrane protein 187 | −7.717754905 | 0.001085602 |
| TMEM221 | transmembrane protein 221 | −7.22073532 | 0.019434951 |
| TMEM249 | transmembrane protein 249 | −6.628590914 | 0.006072763 |
| TMEM177 | transmembrane protein 177 | −5.850793145 | 0.004004917 |
| TMEM92 | transmembrane protein 92 | −5.793088235 | 0.005077808 |
| TMEM121 | transmembrane protein 121 | −5.624075595 | 0.005226063 |
| TMEM203 | transmembrane protein 203 | −4.929095646 | 0.008670705 |
| TMEM223 | transmembrane protein 223 | −4.591863583 | 0.011703338 |
| TMEM238 | transmembrane protein 238 | −4.352286379 | 0.028704807 |
| TMEM191B | transmembrane protein 191B | −3.783402356 | 0.034476139 |
| TMEM191A | transmembrane protein 191A | −3.7829524 | 0.031869069 |
| TMEM53 | transmembrane protein 53 | −3.782392019 | 0.027051 |
| TMEM191C | transmembrane protein 191C | −3.556386263 | 0.043266828 |
| TMEM141 | transmembrane protein 141 | −3.384076799 | 0.040373276 |
| TMEM115 | transmembrane protein 115 | −3.287999353 | 0.045333404 |
| TMEM186 | transmembrane protein 186 | −3.242921449 | 0.049370419 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.-H.; Lee, S.-H.; Lee, S.-K.; Choi, J.-H.; Lim, S.; Kim, M.-S.; Lee, K.-M.; Lee, M.-W.; Ku, J.-L.; Kim, D.-H.; et al. Antiproliferative Activity of Krukovine by Regulating Transmembrane Protein 139 (TMEM139) in Oxaliplatin-Resistant Pancreatic Cancer Cells. Cancers 2023, 15, 2642. https://doi.org/10.3390/cancers15092642
Lee J-H, Lee S-H, Lee S-K, Choi J-H, Lim S, Kim M-S, Lee K-M, Lee M-W, Ku J-L, Kim D-H, et al. Antiproliferative Activity of Krukovine by Regulating Transmembrane Protein 139 (TMEM139) in Oxaliplatin-Resistant Pancreatic Cancer Cells. Cancers. 2023; 15(9):2642. https://doi.org/10.3390/cancers15092642
Chicago/Turabian StyleLee, Jee-Hyung, Sang-Hyub Lee, Sang-Kook Lee, Jin-Ho Choi, Seohyun Lim, Min-Song Kim, Kyung-Min Lee, Min-Woo Lee, Ja-Lok Ku, Dae-Hyun Kim, and et al. 2023. "Antiproliferative Activity of Krukovine by Regulating Transmembrane Protein 139 (TMEM139) in Oxaliplatin-Resistant Pancreatic Cancer Cells" Cancers 15, no. 9: 2642. https://doi.org/10.3390/cancers15092642
APA StyleLee, J.-H., Lee, S.-H., Lee, S.-K., Choi, J.-H., Lim, S., Kim, M.-S., Lee, K.-M., Lee, M.-W., Ku, J.-L., Kim, D.-H., Cho, I.-R., Paik, W.-H., Ryu, J.-K., & Kim, Y.-T. (2023). Antiproliferative Activity of Krukovine by Regulating Transmembrane Protein 139 (TMEM139) in Oxaliplatin-Resistant Pancreatic Cancer Cells. Cancers, 15(9), 2642. https://doi.org/10.3390/cancers15092642








