Regulation of RORα Stability through PRMT5-Dependent Symmetric Dimethylation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Two-Dimensional and Three-Dimensional Cell Culture
2.2. Virus Preparation and Stable Cell Lines
2.3. Coimmunoprecipitation
2.4. GST Pull-Down Assay
2.5. Cycloheximide Chase Assay
2.6. Western Blotting
2.7. Real-Time PCR
2.8. Invasion Assay
2.9. Single-Cell Migration Assay
2.10. Mammosphere Assay
2.11. Statistical Analysis
3. Results
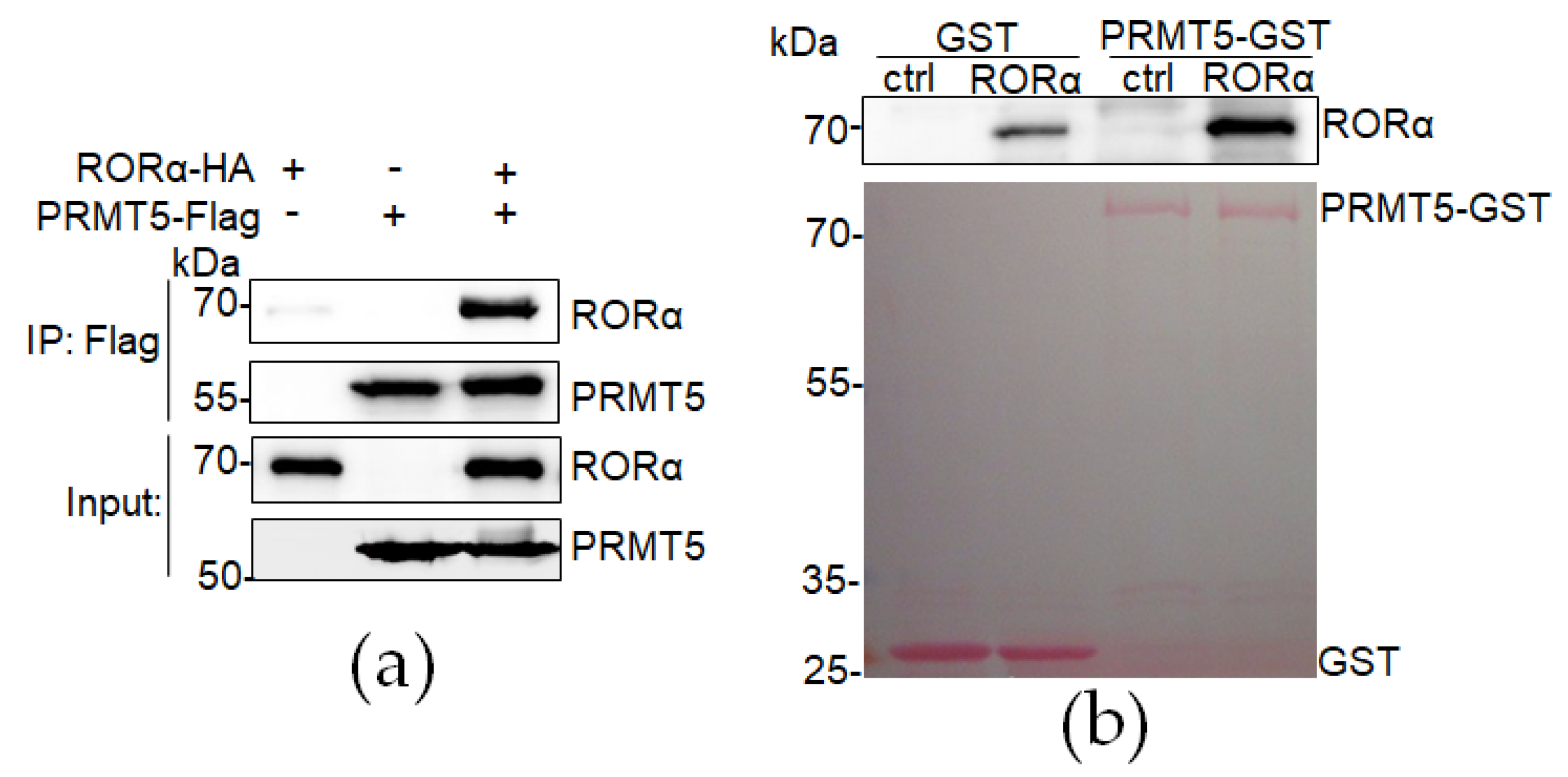
3.1. RORα Directly Interacts with PRMT5
3.2. RORα Is Symmetrically Dimethylated and Stablized by PRMT5
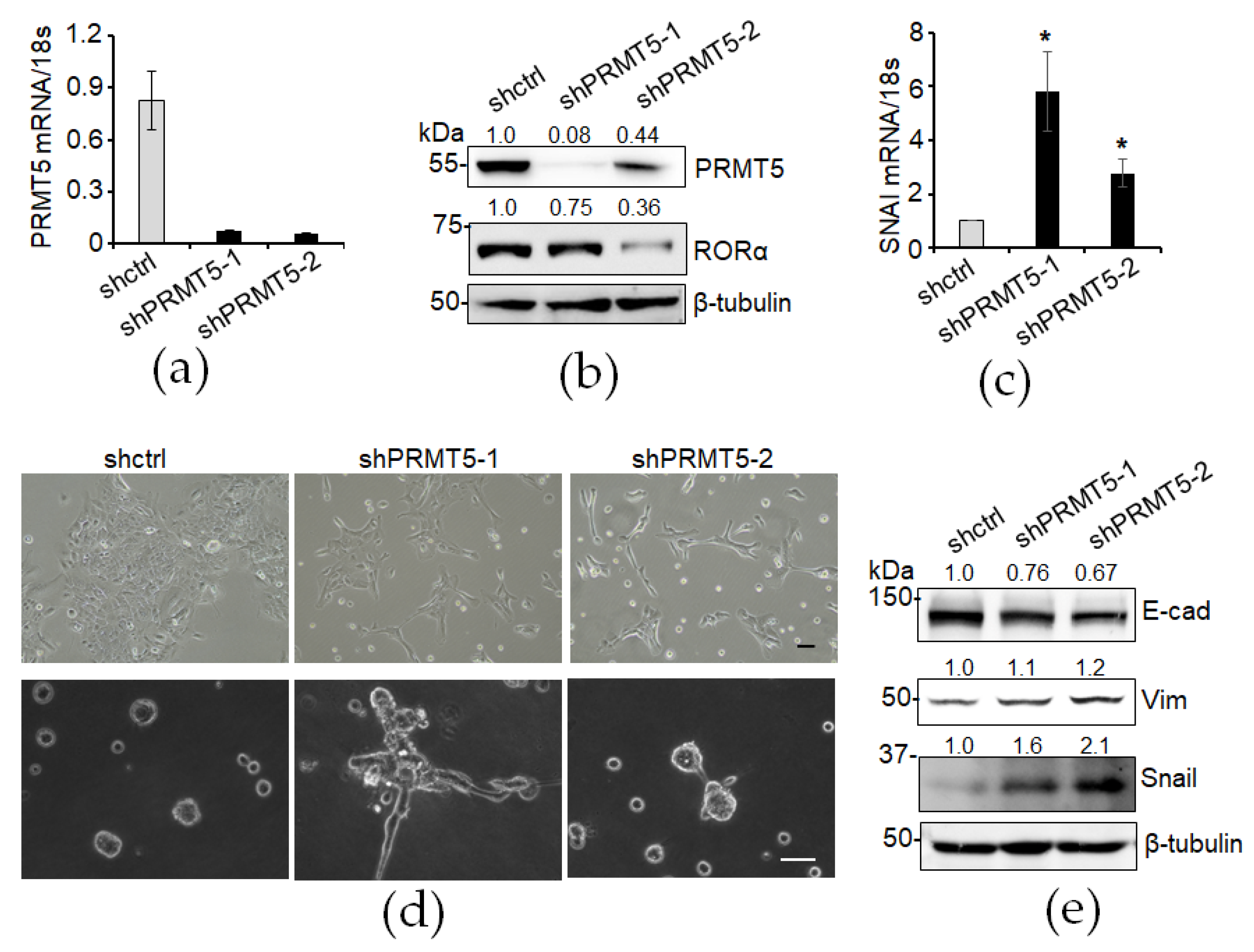
3.3. Knockdown of PRMT5 Decreases RORα Expression and Enhances Epithelial–Mesenchymal Transition (EMT)
3.4. Knockdown of PRMT5 Enhances Cell Invasion and Migration
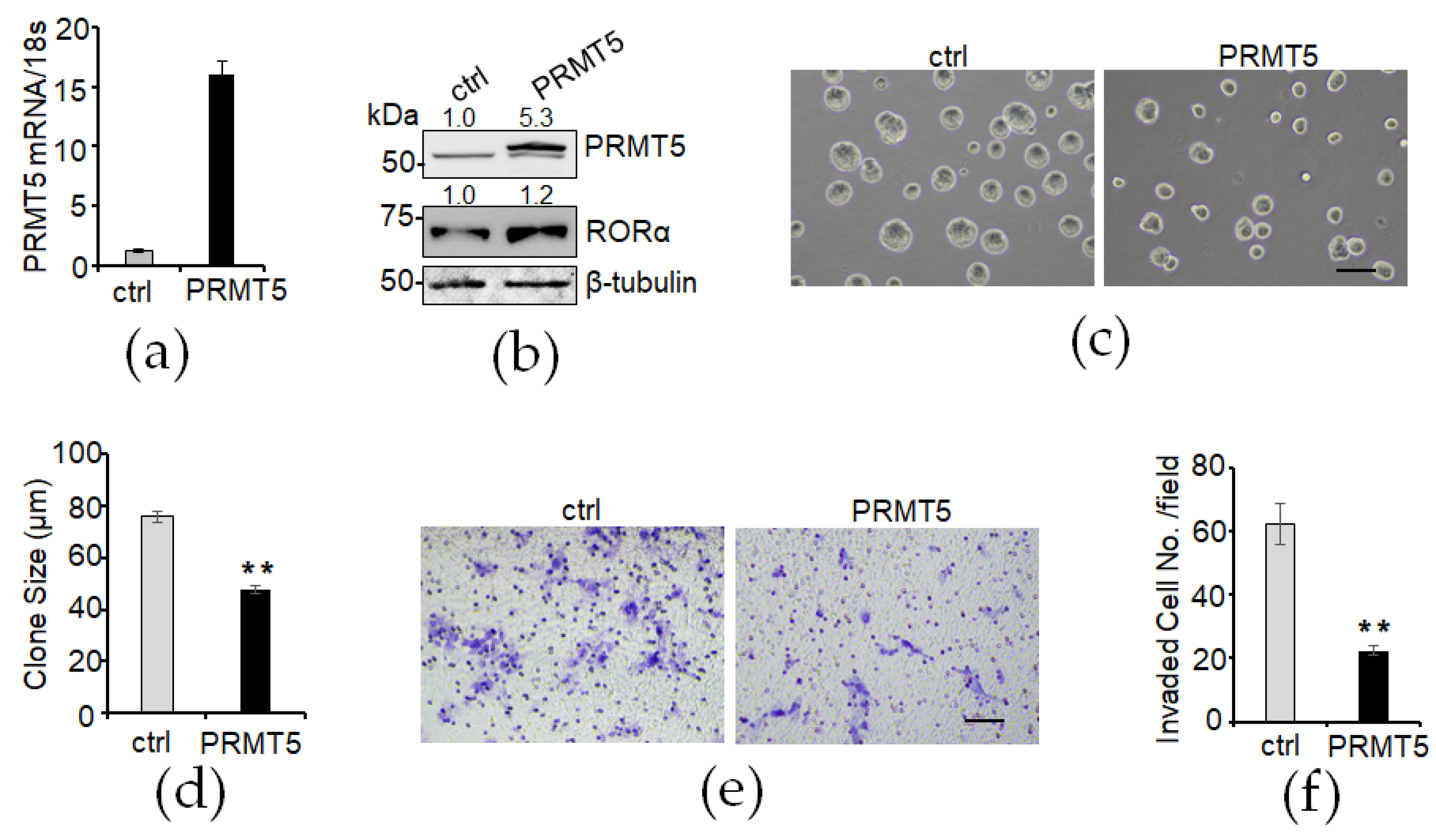
3.5. Overexpression of PRMT5 Elevates RORα Expression and Suppresses Cell Growth and Invasion
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jetten, A.M. Retinoid-related orphan receptors (RORs): Critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl. Recept. Signal. 2009, 7, e003. [Google Scholar] [CrossRef]
- Jetten, A.M.; Kurebayashi, S.; Ueda, E. The ROR nuclear orphan receptor subfamily: Critical regulators of multiple biological processes. Prog. Nucleic Acid Res. Mol. Biol. 2001, 69, 205–247. [Google Scholar] [CrossRef] [PubMed]
- Dzhagalov, I.; Giguere, V.; He, Y.W. Lymphocyte development and function in the absence of retinoic acid-related orphan receptor alpha. J. Immunol. 2004, 173, 2952–2959. [Google Scholar] [CrossRef]
- Sato, T.K.; Panda, S.; Miraglia, L.J.; Reyes, T.M.; Rudic, R.D.; McNamara, P.; Naik, K.A.; FitzGerald, G.A.; Kay, S.A.; Hogenesch, J.B. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 2004, 43, 527–537. [Google Scholar] [CrossRef]
- Lau, P.; Nixon, S.J.; Parton, R.G.; Muscat, G.E. RORalpha regulates the expression of genes involved in lipid homeostasis in skeletal muscle cells: Caveolin-3 and CPT-1 are direct targets of ROR. J. Biol. Chem. 2004, 279, 36828–36840. [Google Scholar] [CrossRef]
- Lee, J.M.; Kim, I.S.; Kim, H.; Lee, J.S.; Kim, K.; Yim, H.Y.; Jeong, J.; Kim, J.H.; Kim, J.Y.; Lee, H.; et al. RORalpha attenuates Wnt/beta-catenin signaling by PKCalpha-dependent phosphorylation in colon cancer. Mol. Cell. 2010, 37, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, H.; Katayama, M.; Ohishi, A.; Miya, K.; Tokunaga, R.; Kobayashi, S.; Nishimoto, Y.; Hirooka, K.; Shima, A.; Michihara, A. Orphan Nuclear Receptor RORalpha Regulates Enzymatic Metabolism of Cerebral 24S-Hydroxycholesterol through CYP39A1 Intronic Response Element Activation. Int. J. Mol. Sci. 2020, 21, 3309. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, S.; Sherrard, R.M. Cerebellum and neurodevelopmental disorders: RORalpha is a unifying force. Front. Cell. Neurosci. 2023, 17, 1108339. [Google Scholar] [CrossRef]
- Xiong, G.; Wang, C.; Evers, B.M.; Zhou, B.P.; Xu, R. RORalpha suppresses breast tumor invasion by inducing SEMA3F expression. Cancer Res. 2012, 72, 1728–1739. [Google Scholar] [CrossRef]
- Xiong, G.; Xu, R. Retinoid orphan nuclear receptor alpha (RORalpha) suppresses the epithelial-mesenchymal transition (EMT) by directly repressing Snail transcription. J. Biol. Chem. 2022, 298, 102059. [Google Scholar] [CrossRef]
- Fu, R.D.; Qiu, C.H.; Chen, H.A.; Zhang, Z.G.; Lu, M.Q. Retinoic acid receptor-related receptor alpha (RORalpha) is a prognostic marker for hepatocellular carcinoma. Tumour Biol. 2014, 35, 7603–7610. [Google Scholar] [CrossRef] [PubMed]
- Moretti, R.M.; Montagnani Marelli, M.; Sala, A.; Motta, M.; Limonta, P. Activation of the orphan nuclear receptor RORalpha counteracts the proliferative effect of fatty acids on prostate cancer cells: Crucial role of 5-lipoxygenase. Int. J. Cancer 2004, 112, 87–93. [Google Scholar] [CrossRef]
- Zhao, J.; Jiang, Y.; Chen, L.; Ma, Y.; Zhang, H.; Zhou, J.; Li, H.; Jing, Z. The EIF4A3/CASC2/RORA Feedback Loop Regulates the Aggressive Phenotype in Glioblastomas. Front. Oncol. 2021, 11, 699933. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Geng, W.; Zhou, M.; Xu, J.; Wang, S.; Huang, Q.; Sun, Y.; Li, Y.; Yang, G.; Jin, Y. POU6F1 cooperates with RORA to suppress the proliferation of lung adenocarcinoma by downregulation HIF1A signaling pathway. Cell Death Dis. 2022, 13, 427. [Google Scholar] [CrossRef]
- Brozyna, A.A.; Jozwicki, W.; Skobowiat, C.; Jetten, A.; Slominski, A.T. RORalpha and RORgamma expression inversely correlates with human melanoma progression. Oncotarget 2016, 7, 63261–63282. [Google Scholar] [CrossRef] [PubMed]
- Mao, W.; Xiong, G.; Wu, Y.; Wang, C.; St Clair, D.; Li, J.D.; Xu, R. RORalpha Suppresses Cancer-Associated Inflammation by Repressing Respiratory Complex I-Dependent ROS Generation. Int. J. Mol. Sci. 2021, 22, 665. [Google Scholar] [CrossRef] [PubMed]
- Xiong, G.; Xu, R. RORalpha binds to E2F1 to inhibit cell proliferation and regulate mammary gland branching morphogenesis. Mol. Cell. Biol. 2014, 34, 3066–3075. [Google Scholar] [CrossRef] [PubMed]
- Bedford, M.T.; Clarke, S.G. Protein arginine methylation in mammals: Who, what, and why. Mol. Cell 2009, 33, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Stopa, N.; Krebs, J.E.; Shechter, D. The PRMT5 arginine methyltransferase: Many roles in development, cancer and beyond. Cell. Mol. Life Sci. 2015, 72, 2041–2059. [Google Scholar] [CrossRef]
- Sun, L.; Wang, M.; Lv, Z.; Yang, N.; Liu, Y.; Bao, S.; Gong, W.; Xu, R.M. Structural insights into protein arginine symmetric dimethylation by PRMT5. Proc. Natl. Acad. Sci. USA 2011, 108, 20538–20543. [Google Scholar] [CrossRef]
- Ye, Q.; Zhang, J.; Zhang, C.; Yi, B.; Kazama, K.; Liu, W.; Sun, X.; Liu, Y.; Sun, J. Endothelial PRMT5 plays a crucial role in angiogenesis after acute ischemic injury. JCI Insight 2022, 7, e152481. [Google Scholar] [CrossRef] [PubMed]
- Favia, A.; Salvatori, L.; Nanni, S.; Iwamoto-Stohl, L.K.; Valente, S.; Mai, A.; Scagnoli, F.; Fontanella, R.A.; Totta, P.; Nasi, S.; et al. The Protein Arginine Methyltransferases 1 and 5 affect Myc properties in glioblastoma stem cells. Sci. Rep. 2019, 9, 15925. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wu, J.; Wu, C.; Chen, W.; Lin, R.; Zhou, Y.; Huang, X. The LINC01138 interacts with PRMT5 to promote SREBP1-mediated lipid desaturation and cell growth in clear cell renal cell carcinoma. Biochem. Biophys. Res. Commun. 2018, 507, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Moehlenbrink, J.; Lu, Y.C.; Zalmas, L.P.; Sagum, C.A.; Carr, S.; McGouran, J.F.; Alexander, L.; Fedorov, O.; Munro, S.; et al. Arginine methylation-dependent reader-writer interplay governs growth control by E2F-1. Mol. Cell. 2013, 52, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Gur, M.; Zhou, Z.; Gamper, A.; Hung, M.C.; Fujita, N.; Lan, L.; Bahar, I.; Wan, Y. Interplay between arginine methylation and ubiquitylation regulates KLF4-mediated genome stability and carcinogenesis. Nat. Commun. 2015, 6, 8419. [Google Scholar] [CrossRef] [PubMed]
- Vinet, M.; Suresh, S.; Maire, V.; Monchecourt, C.; Nemati, F.; Lesage, L.; Pierre, F.; Ye, M.; Lescure, A.; Brisson, A.; et al. Protein arginine methyltransferase 5: A novel therapeutic target for triple-negative breast cancers. Cancer Med. 2019, 8, 2414–2428. [Google Scholar] [CrossRef] [PubMed]
- Lattouf, H.; Kassem, L.; Jacquemetton, J.; Choucair, A.; Poulard, C.; Tredan, O.; Corbo, L.; Diab-Assaf, M.; Hussein, N.; Treilleux, I.; et al. LKB1 regulates PRMT5 activity in breast cancer. Int. J. Cancer 2019, 144, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; McAvoy, S.; Kuhn, R.; Smith, D.I. RORA, a large common fragile site gene, is involved in cellular stress response. Oncogene 2006, 25, 2901–2908. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Chen, H.; Li, L.; Yang, F.; Wu, C.; Tao, K. CircGSK3B promotes RORA expression and suppresses gastric cancer progression through the prevention of EZH2 trans-inhibition. J. Exp. Clin. Cancer Res. 2021, 40, 330. [Google Scholar] [CrossRef]
- Sarachana, T.; Hu, V.W. Differential recruitment of coregulators to the RORA promoter adds another layer of complexity to gene (dys) regulation by sex hormones in autism. Mol. Autism 2013, 4, 39. [Google Scholar] [CrossRef]
- Duplus, E.; Gras, C.; Soubeyre, V.; Vodjdani, G.; Lemaigre-Dubreuil, Y.; Brugg, B. Phosphorylation and transcriptional activity regulation of retinoid-related orphan receptor alpha 1 by protein kinases C. J. Neurochem. 2008, 104, 1321–1332. [Google Scholar] [CrossRef] [PubMed]
- Hwang, E.J.; Lee, J.M.; Jeong, J.; Park, J.H.; Yang, Y.; Lim, J.S.; Kim, J.H.; Baek, S.H.; Kim, K.I. SUMOylation of RORalpha potentiates transcriptional activation function. Biochem. Biophys. Res. Commun. 2009, 378, 513–517. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Lee, J.S.; Kim, H.; Kim, K.; Park, H.; Kim, J.Y.; Lee, S.H.; Kim, I.S.; Kim, J.; Lee, M.; et al. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol. Cell. 2012, 48, 572–586. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Qiu, T.; Wu, Y.; Yang, C.; Li, Y.; Du, G.; He, Y.; Liu, W.; Liu, R.; Chen, C.H.; et al. Arginine methyltransferase PRMT5 methylates and stabilizes KLF5 via decreasing its phosphorylation and ubiquitination to promote basal-like breast cancer. Cell. Death Differ. 2021, 28, 2931–2945. [Google Scholar] [CrossRef]
- Gu, Z.; Gao, S.; Zhang, F.; Wang, Z.; Ma, W.; Davis, R.E.; Wang, Z. Protein arginine methyltransferase 5 is essential for growth of lung cancer cells. Biochem. J. 2012, 446, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, C.; Yang, J.; Peters, S.B.; Bill, M.A.; Baiocchi, R.A.; Yan, F.; Sif, S.; Tae, S.; Gaudio, E.; Wu, X.; et al. PRMT5 is upregulated in malignant and metastatic melanoma and regulates expression of MITF and p27(Kip1.). PLoS ONE 2013, 8, e74710. [Google Scholar] [CrossRef]
- Yan, F.; Alinari, L.; Lustberg, M.E.; Martin, L.K.; Cordero-Nieves, H.M.; Banasavadi-Siddegowda, Y.; Virk, S.; Barnholtz-Sloan, J.; Bell, E.H.; Wojton, J.; et al. Genetic validation of the protein arginine methyltransferase PRMT5 as a candidate therapeutic target in glioblastoma. Cancer Res. 2014, 74, 1752–1765. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Li, Y.; Lee, P.; Liu, T.; Wan, C.; Wang, Z. Protein arginine methyltransferase 5 functions in opposite ways in the cytoplasm and nucleus of prostate cancer cells. PLoS ONE 2012, 7, e44033. [Google Scholar] [CrossRef]
- Pal, S.; Baiocchi, R.A.; Byrd, J.C.; Grever, M.R.; Jacob, S.T.; Sif, S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007, 26, 3558–3569. [Google Scholar] [CrossRef]
- Liang, Z.; Liu, L.; Wen, C.; Jiang, H.; Ye, T.; Ma, S.; Liu, X. Clinicopathological and Prognostic Significance of PRMT5 in Cancers: A System Review and Meta-Analysis. Cancer Control 2021, 28, 10732748211050583. [Google Scholar] [CrossRef]
- Feustel, K.; Falchook, G.S. Protein Arginine Methyltransferase 5 (PRMT5) Inhibitors in Oncology Clinical Trials: A review. J. Immunother. Precis. Oncol. 2022, 5, 58–67. [Google Scholar] [CrossRef]
- Yuan, Y.; Nie, H. Protein arginine methyltransferase 5: A potential cancer therapeutic target. Cell. Oncol. 2021, 44, 33–44. [Google Scholar] [CrossRef]
- Kim, H.; Ronai, Z.A. PRMT5 function and targeting in cancer. Cell. Stress 2020, 4, 199–215. [Google Scholar] [CrossRef]
- Rengasamy, M.; Zhang, F.; Vashisht, A.; Song, W.M.; Aguilo, F.; Sun, Y.; Li, S.; Zhang, W.; Zhang, B.; Wohlschlegel, J.A.; et al. The PRMT5/WDR77 complex regulates alternative splicing through ZNF326 in breast cancer. Nucleic Acids Res. 2017, 45, 11106–11120. [Google Scholar] [CrossRef]
- Han, X.; Wei, L.; Wu, B. PRMT5 Promotes Aerobic Glycolysis and Invasion of Breast Cancer Cells by Regulating the LXRalpha/NF-kappaBp65 Pathway. Onco Targets Ther. 2020, 13, 3347–3357. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Z.; Zhang, J.; Ling, R. Elevated expression of protein arginine methyltransferase 5 predicts the poor prognosis of breast cancer. Tumour Biol. 2017, 39, 1010428317695917. [Google Scholar] [CrossRef]
- Poulard, C.; Ha Pham, T.; Drouet, Y.; Jacquemetton, J.; Surmielova, A.; Kassem, L.; Mery, B.; Lasset, C.; Reboulet, J.; Treilleux, I.; et al. Nuclear PRMT5 is a biomarker of sensitivity to tamoxifen in ERalpha(+) breast cancer. EMBO Mol. Med. 2023, 15, e17248. [Google Scholar] [CrossRef]
- Shilo, K.; Wu, X.; Sharma, S.; Welliver, M.; Duan, W.; Villalona-Calero, M.; Fukuoka, J.; Sif, S.; Baiocchi, R.; Hitchcock, C.L.; et al. Cellular localization of protein arginine methyltransferase-5 correlates with grade of lung tumors. Diagn. Pathol. 2013, 8, 201. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, G.; Obringer, B.; Jones, A.; Horton, E.; Xu, R. Regulation of RORα Stability through PRMT5-Dependent Symmetric Dimethylation. Cancers 2024, 16, 1914. https://doi.org/10.3390/cancers16101914
Xiong G, Obringer B, Jones A, Horton E, Xu R. Regulation of RORα Stability through PRMT5-Dependent Symmetric Dimethylation. Cancers. 2024; 16(10):1914. https://doi.org/10.3390/cancers16101914
Chicago/Turabian StyleXiong, Gaofeng, Brynne Obringer, Austen Jones, Elise Horton, and Ren Xu. 2024. "Regulation of RORα Stability through PRMT5-Dependent Symmetric Dimethylation" Cancers 16, no. 10: 1914. https://doi.org/10.3390/cancers16101914
APA StyleXiong, G., Obringer, B., Jones, A., Horton, E., & Xu, R. (2024). Regulation of RORα Stability through PRMT5-Dependent Symmetric Dimethylation. Cancers, 16(10), 1914. https://doi.org/10.3390/cancers16101914





