Molecular and Histopathological Characterization of Metastatic Cutaneous Squamous Cell Carcinomas: A Case–Control Study
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Selection
2.2. Clinical and Histopathological Data Collection
2.3. Nanostring Analysis
2.4. Network Analysis
2.5. Statistical Analysis
3. Results
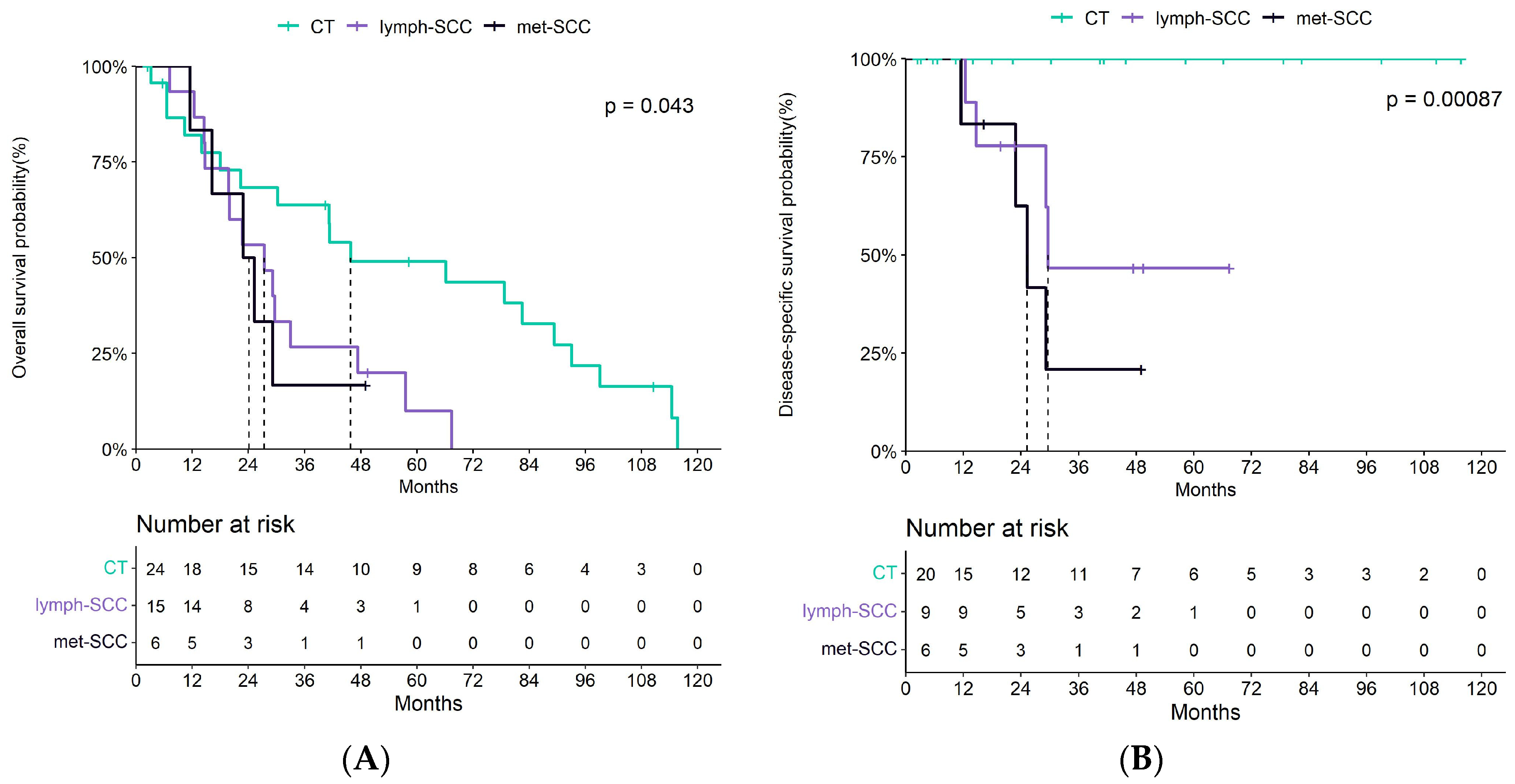
3.1. Patient and Tumor Features
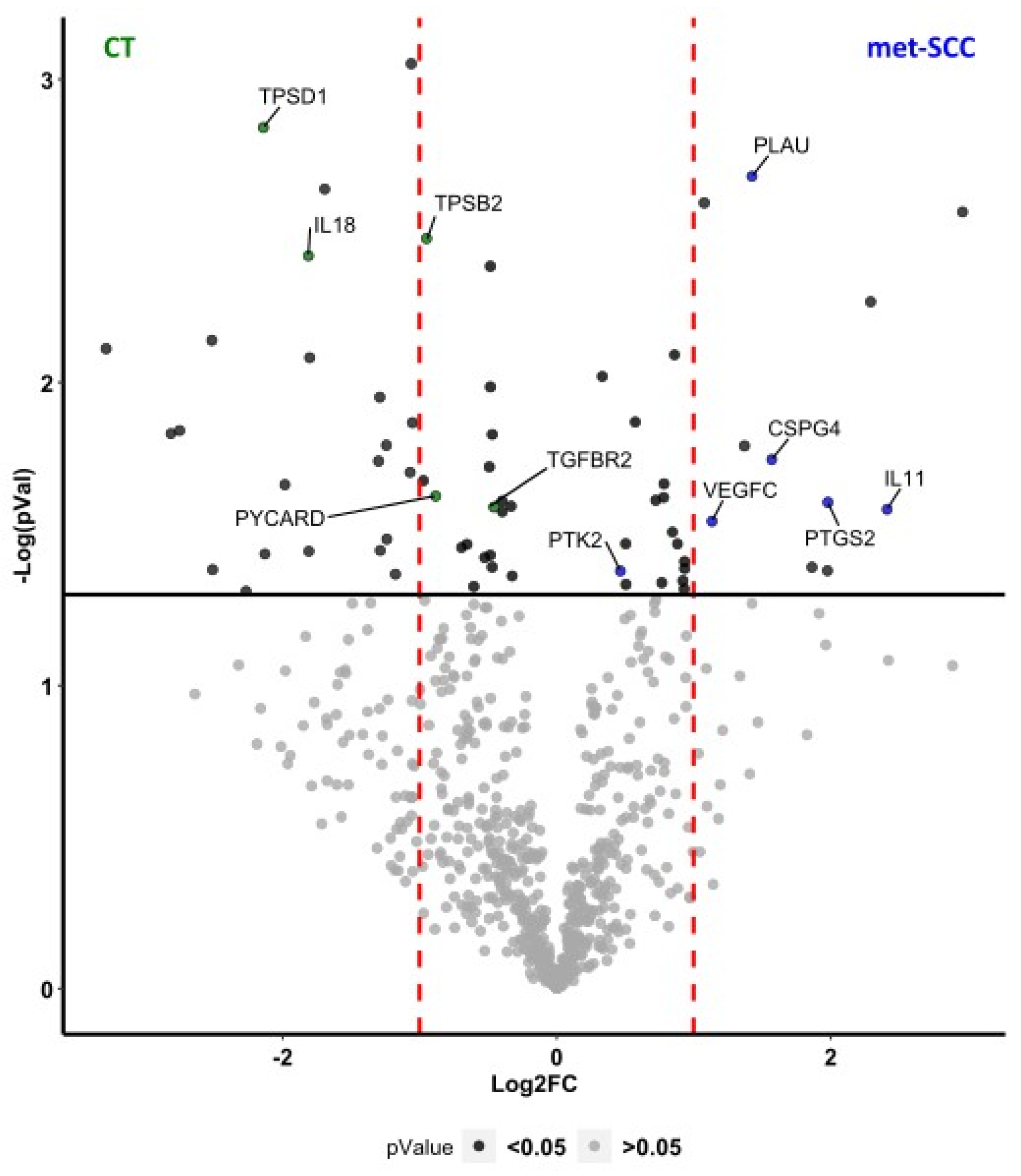
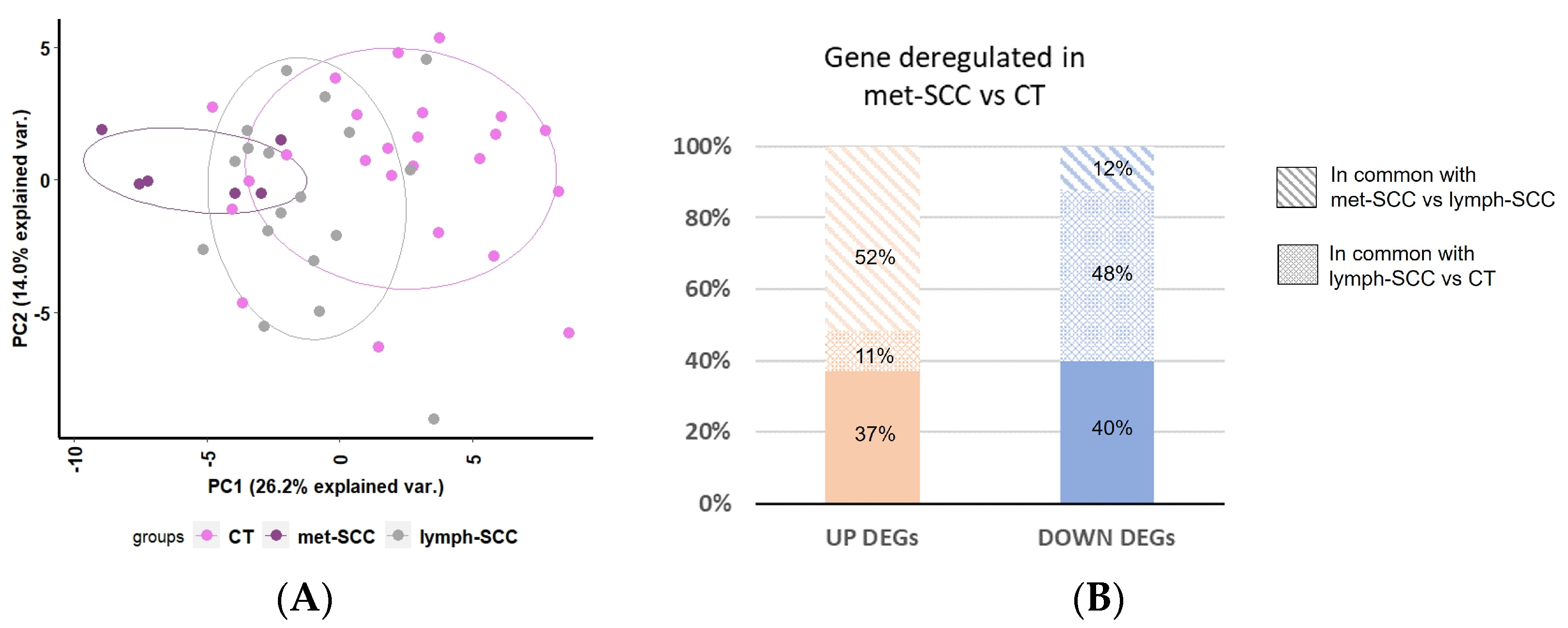
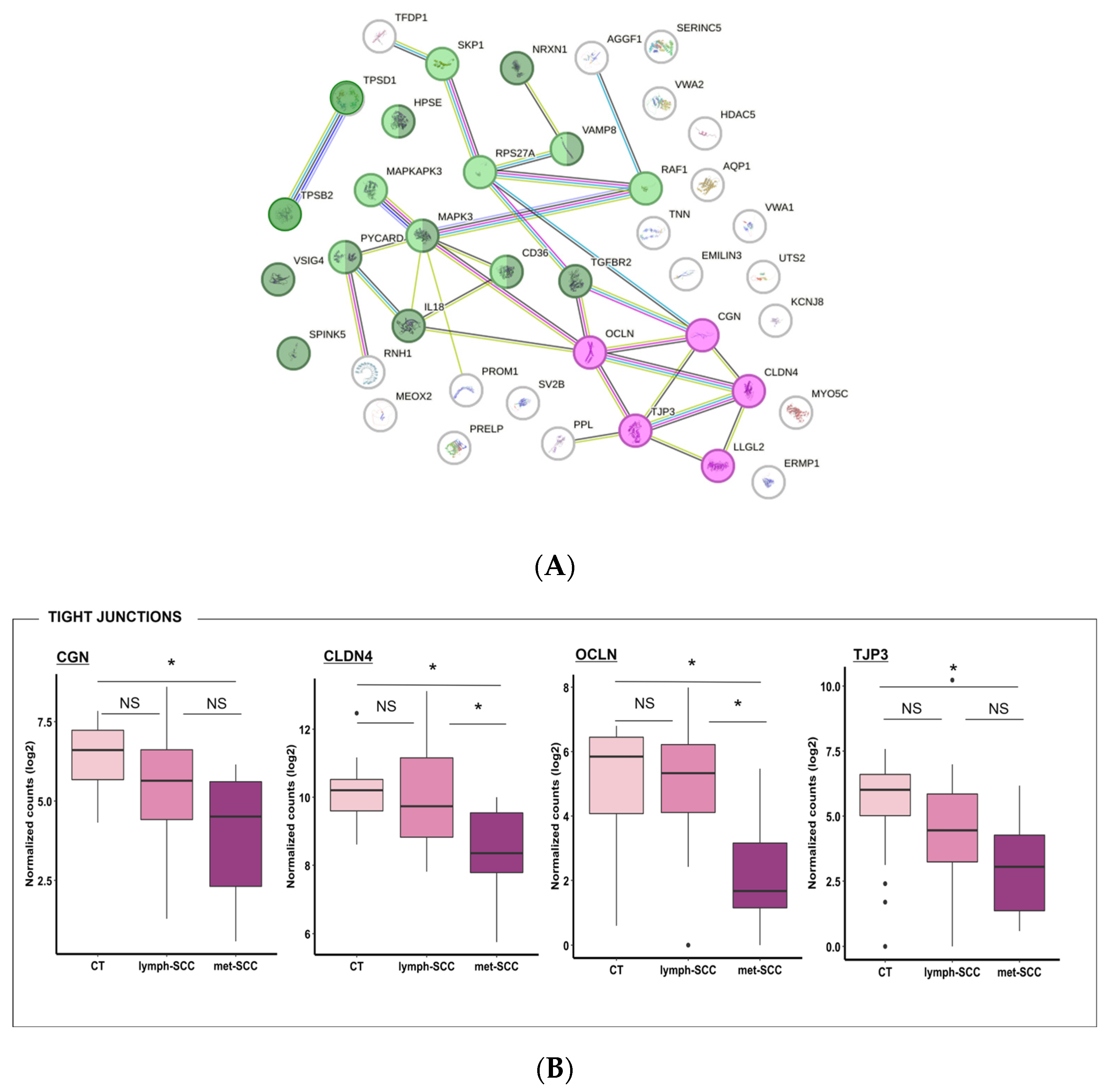
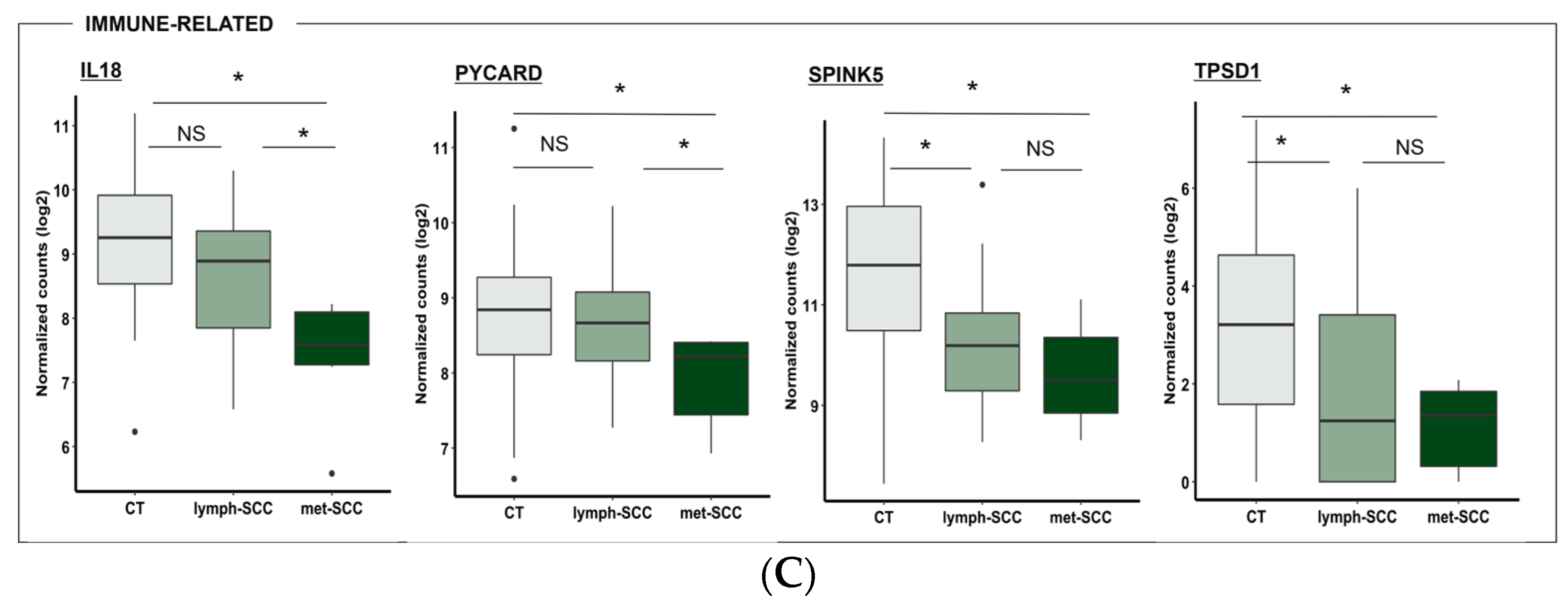
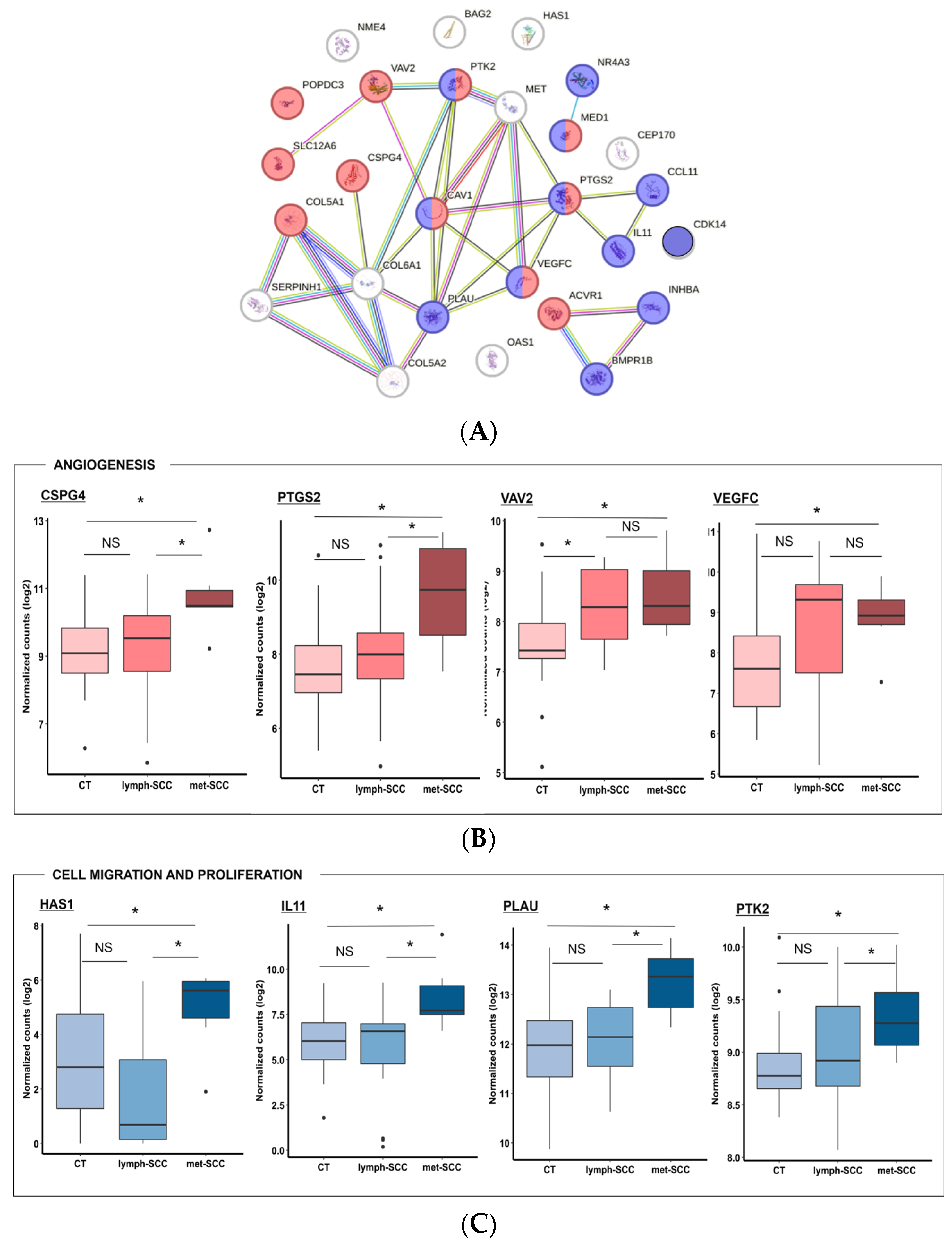
3.2. Gene Expression Profile Associated with Metastatic Behavior of cSCC
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Que, S.K.T.; Zwald, F.O.; Schmults, C.D. Cutaneous Squamous Cell Carcinoma: Incidence, Risk Factors, Diagnosis, and Staging. J. Am. Acad. Dermatol. 2018, 78, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Work Group; Invited Reviewers; Kim, J.Y.S.; Kozlow, J.H.; Mittal, B.; Moyer, J.; Olenecki, T.; Rodgers, P. Guidelines of Care for the Management of Cutaneous Squamous Cell Carcinoma. J. Am. Acad. Dermatol. 2018, 78, 560–578. [Google Scholar] [CrossRef] [PubMed]
- Corchado-Cobos, R.; García-Sancha, N.; González-Sarmiento, R.; Pérez-Losada, J.; Cañueto, J. Cutaneous Squamous Cell Carcinoma: From Biology to Therapy. Int. J. Mol. Sci. 2020, 21, 2956. [Google Scholar] [CrossRef] [PubMed]
- Karia, P.S.; Han, J.; Schmults, C.D. Cutaneous Squamous Cell Carcinoma: Estimated Incidence of Disease, Nodal Metastasis, and Deaths from Disease in the United States, 2012. J. Am. Acad. Dermatol. 2013, 68, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.K.; Kelley, B.F.; Prokop, L.J.; Murad, M.H.; Baum, C.L. Risk Factors for Cutaneous Squamous Cell Carcinoma Recurrence, Metastasis, and Disease-Specific Death: A Systematic Review and Meta-Analysis. JAMA Dermatol. 2016, 152, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Xiang, F.; Lucas, R.; Hales, S.; Neale, R. Incidence of Nonmelanoma Skin Cancer in Relation to Ambient UV Radiation in White Populations, 1978–2012: Empirical Relationships. JAMA Dermatol. 2014, 150, 1063–1071. [Google Scholar] [CrossRef] [PubMed]
- Urso, B.; Kelsey, A.; Bordelon, J.; Sheiner, P.; Finch, J.; Cohen, J.L. Risk Factors and Prevention Strategies for Cutaneous Squamous Cell Carcinoma in Transplant Recipients. Int. J. Dermatol. 2022, 61, 1218–1224. [Google Scholar] [CrossRef] [PubMed]
- Fania, L.; Abeni, D.; Esposito, I.; Spagnoletti, G.; Citterio, F.; Romagnoli, J.; Castriota, M.; Ricci, F.; Moro, F.; Perino, F.; et al. Behavioral and Demographic Factors Associated with Occurrence of Non-Melanoma Skin Cancer in Organ Transplant Recipients. G. Ital. Dermatol. Venereol. 2020, 155, 669–675. [Google Scholar] [CrossRef]
- Stevens, J.S.; Murad, F.; Smile, T.D.; O’Connor, D.M.; Ilori, E.; Koyfman, S.; Vidimos, A.; Waldman, A.B.; Ruiz, E.S. Validation of the 2022 National Comprehensive Cancer Network Risk Stratification for Cutaneous Squamous Cell Carcinoma. JAMA Dermatol. 2023, 159, 728–735. [Google Scholar] [CrossRef]
- Karia, P.S.; Morgan, F.C.; Califano, J.A.; Schmults, C.D. Comparison of Tumor Classifications for Cutaneous Squamous Cell Carcinoma of the Head and Neck in the 7th vs. 8th Edition of the AJCC Cancer Staging Manual. JAMA Dermatol. 2018, 154, 175–181. [Google Scholar] [CrossRef]
- Stratigos, A.J.; Garbe, C.; Dessinioti, C.; Lebbe, C.; van Akkooi, A.; Bataille, V.; Bastholt, L.; Dreno, B.; Dummer, R.; Fargnoli, M.C.; et al. European Consensus-Based Interdisciplinary Guideline for Invasive Cutaneous Squamous Cell Carcinoma: Part 2. Treatment-Update 2023. Eur. J. Cancer 2023, 193, 113252. [Google Scholar] [CrossRef] [PubMed]
- Puebla-Tornero, L.; Corchete-Sánchez, L.A.; Conde-Ferreirós, A.; García-Sancha, N.; Corchado-Cobos, R.; Román-Curto, C.; Cañueto, J. Performance of Salamanca Refinement of the T3-AJCC8 versus the Brigham and Women’s Hospital and Tübingen Alternative Staging Systems for High-Risk Cutaneous Squamous Cell Carcinoma. J. Am. Acad. Dermatol. 2021, 84, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Brancaccio, G.; Briatico, G.; Pellegrini, C.; Rocco, T.; Moscarella, E.; Fargnoli, M.C. Risk Factors and Diagnosis of Advanced Cutaneous Squamous Cell Carcinoma. Dermatol. Pract. Concept. 2021, 11, e2021166S. [Google Scholar] [CrossRef] [PubMed]
- Stratigos, A.J.; Garbe, C.; Dessinioti, C.; Lebbe, C.; van Akkooi, A.; Bataille, V.; Bastholt, L.; Dreno, B.; Dummer, R.; Fargnoli, M.C.; et al. European Consensus-Based Interdisciplinary Guideline for Invasive Cutaneous Squamous Cell Carcinoma. Part 1: Diagnostics and Prevention-Update 2023. Eur. J. Cancer 2023, 193, 113251. [Google Scholar] [CrossRef] [PubMed]
- de Jong, E.; Lammerts, M.U.P.A.; Genders, R.E.; Bouwes Bavinck, J.N. Update of Advanced Cutaneous Squamous Cell Carcinoma. J. Eur. Acad. Dermatol. Venereol. 2022, 36 (Suppl. S1), 6–10. [Google Scholar] [CrossRef] [PubMed]
- Winge, M.C.G.; Kellman, L.N.; Guo, K.; Tang, J.Y.; Swetter, S.M.; Aasi, S.Z.; Sarin, K.Y.; Chang, A.L.S.; Khavari, P.A. Advances in Cutaneous Squamous Cell Carcinoma. Nat. Rev. Cancer 2023, 23, 430–449. [Google Scholar] [CrossRef] [PubMed]
- Mulvaney, P.M.; Schmults, C.D. Molecular Prediction of Metastasis in Cutaneous Squamous Cell Carcinoma. Curr. Opin. Oncol. 2020, 32, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Piipponen, M.; Riihilä, P.; Nissinen, L.; Kähäri, V.-M. The Role of P53 in Progression of Cutaneous Squamous Cell Carcinoma. Cancers 2021, 13, 4507. [Google Scholar] [CrossRef]
- Di Nardo, L.; Pellegrini, C.; Di Stefani, A.; Del Regno, L.; Sollena, P.; Piccerillo, A.; Longo, C.; Garbe, C.; Fargnoli, M.C.; Peris, K. Molecular Genetics of Cutaneous Squamous Cell Carcinoma: Perspective for Treatment Strategies. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 932–941. [Google Scholar] [CrossRef]
- Hedberg, M.L.; Berry, C.T.; Moshiri, A.S.; Xiang, Y.; Yeh, C.J.; Attilasoy, C.; Capell, B.C.; Seykora, J.T. Molecular Mechanisms of Cutaneous Squamous Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 3478. [Google Scholar] [CrossRef]
- Tsang, D.A.; Tam, S.Y.C.; Oh, C.C. Molecular Alterations in Cutaneous Squamous Cell Carcinoma in Immunocompetent and Immunosuppressed Hosts—A Systematic Review. Cancers 2023, 15, 1832. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.; Collie, C.J. What’s New in Dermatopathology 2023: WHO 5th Edition Updates. J. Pathol. Transl. Med. 2023, 57, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Drexler, K.; Drexler, H.; Karrer, S.; Landthaler, M.; Haferkamp, S.; Zeman, F.; Berneburg, M.; Niebel, D. Degree of Actinic Elastosis Is a Surrogate of Exposure to Chronic Ultraviolet Radiation and Correlates More Strongly with Cutaneous Squamous Cell Carcinoma than Basal Cell Carcinoma. Life 2023, 13, 811. [Google Scholar] [CrossRef] [PubMed]
- Claveau, J.; Archambault, J.; Ernst, D.S.; Giacomantonio, C.; Limacher, J.J.; Murray, C.; Parent, F.; Zloty, D. Multidisciplinary Management of Locally Advanced and Metastatic Cutaneous Squamous Cell Carcinoma. Curr. Oncol. 2020, 27, e399–e407. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Tillman, B.N.; Nijhawan, R.I.; Srivastava, D.; Sher, D.J.; Avkshtol, V.; Homsi, J.; Bishop, J.A.; Wynings, E.M.; Lee, R.; et al. High-Risk Cutaneous Squamous Cell Carcinoma of the Head and Neck: A Clinical Review. Ann. Surg. Oncol. 2021, 28, 9009–9030. [Google Scholar] [CrossRef] [PubMed]
- Tokez, S.; Venables, Z.C.; Hollestein, L.M.; Qi, H.; Bramer, E.M.; Rentroia-Pacheco, B.; Van Den Bos, R.R.; Rous, B.; Leigh, I.M.; Nijsten, T.; et al. Risk Factors for Metastatic Cutaneous Squamous Cell Carcinoma: Refinement and Replication Based on 2 Nationwide Nested Case-Control Studies. J. Am. Acad. Dermatol. 2022, 87, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Wohltmann, W.E. JAAD Game Changer: Risk Factors for Metastatic Cutaneous Squamous Cell Carcinoma: Refinement and Replication Based on 2 Nationwide Nested Case-Control Studies. J. Am. Acad. Dermatol. 2023, 90, 1120. [Google Scholar] [CrossRef]
- Zakhem, G.A.; Pulavarty, A.N.; Carucci, J.; Stevenson, M.L. Association of Patient Risk Factors, Tumor Characteristics, and Treatment Modality With Poor Outcomes in Primary Cutaneous Squamous Cell Carcinoma: A Systematic Review and Meta-Analysis. JAMA Dermatol. 2023, 159, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Roccuzzo, G.; Orlando, G.; Rumore, M.R.; Morrone, A.; Fruttero, E.; Caliendo, V.; Picciotto, F.; Sciarrillo, A.; Quaglino, P.; Cassoni, P.; et al. Predictors of Recurrence and Progression in Poorly Differentiated Cutaneous Squamous Cell Carcinomas: Insights from a Real-Life Experience. Dermatology 2024, 240, 329–336. [Google Scholar] [CrossRef]
- Marsidi, N.; Ottevanger, R.; Bouwes Bavinck, J.N.; Krekel-Taminiau, N.M.A.; Goeman, J.J.; Genders, R.E. Risk Factors for Incomplete Excision of Cutaneous Squamous Cell Carcinoma: A Large Cohort Study. J. Eur. Acad. Dermatol. Venereol. 2022, 36, 1229–1234. [Google Scholar] [CrossRef]
- Griffith, C.F. Skin Cancer in Immunosuppressed Patients. JAAPA 2022, 35, 19–27. [Google Scholar] [CrossRef] [PubMed]
- US Preventive Services Task Force; Mangione, C.M.; Barry, M.J.; Nicholson, W.K.; Chelmow, D.; Coker, T.R.; Davis, E.M.; Donahue, K.E.; Jaén, C.R.; Kubik, M.; et al. Screening for Skin Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2023, 329, 1290–1295. [Google Scholar] [CrossRef] [PubMed]
- Banchereau, R.; Leng, N.; Zill, O.; Sokol, E.; Liu, G.; Pavlick, D.; Maund, S.; Liu, L.-F.; Kadel, E.; Baldwin, N.; et al. Molecular Determinants of Response to PD-L1 Blockade across Tumor Types. Nat. Commun. 2021, 12, 3969. [Google Scholar] [CrossRef] [PubMed]
- Rentroia-Pacheco, B.; Tokez, S.; Bramer, E.M.; Venables, Z.C.; van de Werken, H.J.G.; Bellomo, D.; van Klaveren, D.; Mooyaart, A.L.; Hollestein, L.M.; Wakkee, M. Personalised Decision Making to Predict Absolute Metastatic Risk in Cutaneous Squamous Cell Carcinoma: Development and Validation of a Clinico-Pathological Model. eClinicalMedicine 2023, 63, 102150. [Google Scholar] [CrossRef] [PubMed]
- Wysong, A.; Newman, J.G.; Covington, K.R.; Kurley, S.J.; Ibrahim, S.F.; Farberg, A.S.; Bar, A.; Cleaver, N.J.; Somani, A.-K.; Panther, D.; et al. Validation of a 40-Gene Expression Profile Test to Predict Metastatic Risk in Localized High-Risk Cutaneous Squamous Cell Carcinoma. J. Am. Acad. Dermatol. 2021, 84, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.A.; Jiang, W.G. Loss of Tight Junction Barrier Function and Its Role in Cancer Metastasis. Biochim. Biophys. Acta 2009, 1788, 872–891. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, S.; Moon, E.-H.; Kim, G.J.; Choi, J. Hypoxia Disrupt Tight Junctions and Promote Metastasis of Oral Squamous Cell Carcinoma via Loss of Par3. Cancer Cell Int. 2023, 23, 79. [Google Scholar] [CrossRef] [PubMed]
- Ashrafzadeh, S.; Foreman, R.K.; Kalra, S.; Mandinova, A.; Asgari, M.M. Patterns of Tumor-Infiltrating Lymphocytes Used to Distinguish Primary Cutaneous Squamous Cell Carcinomas That Metastasize. JAMA Dermatol. 2023, 159, 788. [Google Scholar] [CrossRef] [PubMed]
- Briso, E.M.; Guinea-Viniegra, J.; Bakiri, L.; Rogon, Z.; Petzelbauer, P.; Eils, R.; Wolf, R.; Rincón, M.; Angel, P.; Wagner, E.F. Inflammation-Mediated Skin Tumorigenesis Induced by Epidermal c-Fos. Genes Dev. 2013, 27, 1959–1973. [Google Scholar] [CrossRef]
- Lichterman, J.N.; Reddy, S.M. Mast Cells: A New Frontier for Cancer Immunotherapy. Cells 2021, 10, 1270. [Google Scholar] [CrossRef]
- Brockmeyer, P.; Kling, A.; Schulz, X.; Perske, C.; Schliephake, H.; Hemmerlein, B. High Mast Cell Density Indicates a Longer Overall Survival in Oral Squamous Cell Carcinoma. Sci. Rep. 2017, 7, 14677. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, K.; Yoshimoto, T.; Torigoe, K.; Kurimoto, M.; Matsui, K.; Hada, T.; Okamura, H.; Nakanishi, K. IL-12 Synergizes with IL-18 or IL-1beta for IFN-Gamma Production from Human T Cells. Int. Immunol. 2000, 12, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Busch, S.; Acar, A.; Magnusson, Y.; Gregersson, P.; Rydén, L.; Landberg, G. TGF-Beta Receptor Type-2 Expression in Cancer-Associated Fibroblasts Regulates Breast Cancer Cell Growth and Survival and Is a Prognostic Marker in Pre-Menopausal Breast Cancer. Oncogene 2015, 34, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Drexler, S.K.; Bonsignore, L.; Masin, M.; Tardivel, A.; Jackstadt, R.; Hermeking, H.; Schneider, P.; Gross, O.; Tschopp, J.; Yazdi, A.S. Tissue-Specific Opposing Functions of the Inflammasome Adaptor ASC in the Regulation of Epithelial Skin Carcinogenesis. Proc. Natl. Acad. Sci. USA 2012, 109, 18384–18389. [Google Scholar] [CrossRef] [PubMed]
- Sand, J.; Fenini, G.; Grossi, S.; Hennig, P.; Di Filippo, M.; Levesque, M.; Werner, S.; French, L.E.; Beer, H.-D. The NLRP1 Inflammasome Pathway Is Silenced in Cutaneous Squamous Cell Carcinoma. J. Investig. Dermatol. 2019, 139, 1788–1797.e6. [Google Scholar] [CrossRef] [PubMed]
- Minaei, E.; Mueller, S.A.; Ashford, B.; Thind, A.S.; Mitchell, J.; Perry, J.R.; Genenger, B.; Clark, J.R.; Gupta, R.; Ranson, M. Cancer Progression Gene Expression Profiling Identifies the Urokinase Plasminogen Activator Receptor as a Biomarker of Metastasis in Cutaneous Squamous Cell Carcinoma. Front. Oncol. 2022, 12, 835929. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhang, W.; Liu, C. FAK Downregulation Suppresses Stem-like Properties and Migration of Human Colorectal Cancer Cells. PLoS ONE 2023, 18, e0284871. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-Y.; Liu, J.-F.; Tsai, H.-C.; Tzeng, H.-E.; Hsieh, T.-H.; Wang, M.; Lin, Y.-F.; Lu, C.-C.; Lien, M.-Y.; Tang, C.-H. Interleukin-11/Gp130 Upregulates MMP-13 Expression and Cell Migration in OSCC by Activating PI3K/Akt and AP-1 Signaling. J. Cell Physiol. 2022, 237, 4551–4562. [Google Scholar] [CrossRef] [PubMed]
- Kamal, M.V.; Damerla, R.R.; Dikhit, P.S.; Kumar, N.A. Prostaglandin-Endoperoxide Synthase 2 (PTGS2) Gene Expression and Its Association with Genes Regulating the VEGF Signaling Pathway in Head and Neck Squamous Cell Carcinoma. J. Oral. Biol. Craniofac. Res. 2023, 13, 567–574. [Google Scholar] [CrossRef]
- Chen, K.; Yong, J.; Zauner, R.; Wally, V.; Whitelock, J.; Sajinovic, M.; Kopecki, Z.; Liang, K.; Scott, K.F.; Mellick, A.S. Chondroitin Sulfate Proteoglycan 4 as a Marker for Aggressive Squamous Cell Carcinoma. Cancers 2022, 14, 5564. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Yu, L.; Sakakura, K.; Visus, C.; Schwab, J.H.; Ferrone, C.R.; Favoino, E.; Koya, Y.; Campoli, M.R.; et al. CSPG4 in Cancer: Multiple Roles. Curr. Mol. Med. 2010, 10, 419–429. [Google Scholar] [CrossRef] [PubMed]
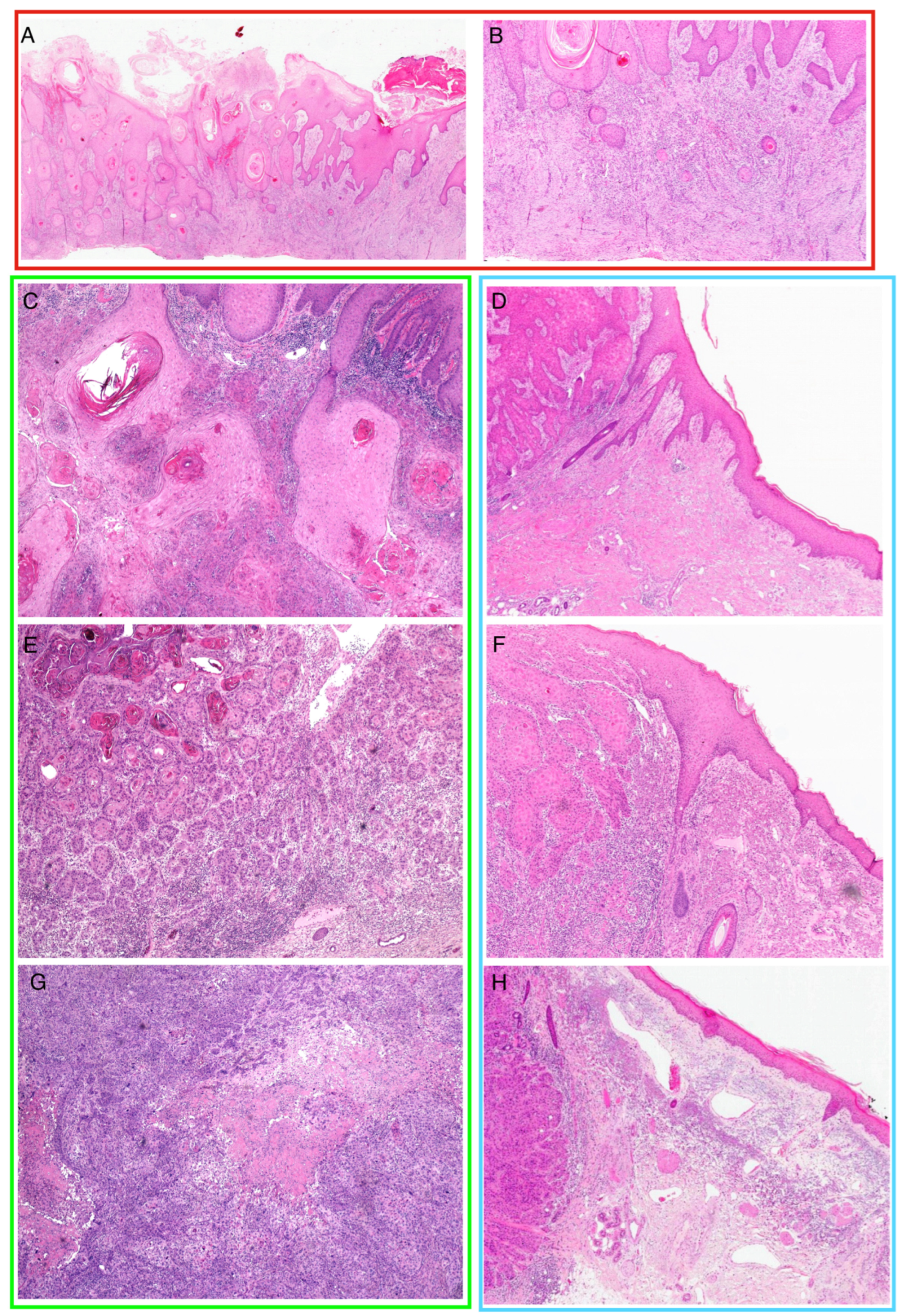
| CT (N = 24) | Lymph-SCC (N = 18) | Met-SCC (N = 6) | Total (N = 48) | p Value | |
|---|---|---|---|---|---|
| Sex | 0.293 | ||||
| F | 8 (33.3%) | 5 (27.8%) | 0 (0.0%) | 13 (27.1%) | |
| M | 16 (66.7%) | 13 (72.2%) | 6 (100.0%) | 35 (72.9%) | |
| Age | 0.183 | ||||
| Mean (SD) | 81.583 (7.454) | 81.389 (7.655) | 76.833 (4.956) | 80.917 (7.310) | |
| Site | 0.980 | ||||
| Lower limbs | 2 (8.3%) | 2 (11.1%) | 0 (0.0%) | 4 (8.3%) | |
| Upper limbs | 5 (20.8%) | 3 (16.7%) | 2 (33.3%) | 10 (20.8%) | |
| Head/neck | 14 (58.3%) | 10 (55.6%) | 4 (66.7%) | 28 (58.3%) | |
| torso | 3 (12.5%) | 3 (16.7%) | 0 (0.0%) | 6 (12.5%) | |
| Dimension | 0.899 | ||||
| Mean (SD) | 2.733 (2.311) | 2.917 (2.036) | 2.167 (0.489) | 2.731 (2.047) | |
| Dimension (grouped) | 1.000 | ||||
| >2 cm | 11 (45.8%) | 9 (50.0%) | 3 (50.0%) | 23 (47.9%) | |
| ≤2cm | 13 (54.2%) | 9 (50.0%) | 3 (50.0%) | 25 (52.1%) | |
| Breslow | 0.058 | ||||
| Mean (SD) | 6.348 (4.519) | 9.500 (4.885) | 7.500 (1.761) | 7.622 (4.564) | |
| NA | 1 | 2 | 0 | 3 | |
| Clark | 0.223 | ||||
| II | 4 (17.4%) | 0 (0.0%) | 0 (0.0%) | 4 (8.9%) | |
| III | 3 (13.0%) | 3 (18.8%) | 0 (0.0%) | 6 (13.3%) | |
| IV | 9 (39.1%) | 3 (18.8%) | 3 (50.0%) | 15 (33.3%) | |
| V | 7 (30.4%) | 10 (62.5%) | 3 (50.0%) | 20 (44.4%) | |
| NA | 1 | 2 | 0 | 3 | |
| Differentiation | 0.006 | ||||
| Well/moderately differentiated | 17 (70.8%) | 5 (27.8%) | 1 (16.7%) | 23 (47.9%) | |
| Poorly differentiated | 7 (29.2%) | 13 (72.2%) | 5 (83.3%) | 25 (52.1%) | |
| Desmoplasia | |||||
| 0 | 17 (70.8%) | 12 (70.6%) | 4 (66.7%) | 33 (70.2%) | 0.999 |
| I/II/III | 7 (29.2%) | 5 (29.4%) | 2 (33.3%) | 14 (29.8%) | |
| NA | 0 | 1 | 0 | 1 | |
| Elastosis | 0.702 | ||||
| + | 10 (43.5%) | 5 (31.2%) | 3 (50.0%) | 18 (40.0%) | |
| ++/+++ | 13 (56.5%) | 11 (68.8%) | 3 (50.0%) | 27 (60.0%) | |
| N-Miss | 1 | 2 | 0 | 3 | |
| Margins | 0.122 | ||||
| Free | 21 (87.5%) | 13 (76.5%) | 3 (50.0%) | 37 (78.7%) | |
| Affected | 3 (12.5%) | 4 (23.5%) | 3 (50.0%) | 10 (21.3%) | |
| NA | 0 | 1 | 0 | 1 | |
| T, N (AJCC/UIC 8th ed) | 0.245 | ||||
| T1N0 | 12 (50.0%) | 7 (41.2%) | 2 (33.3%) | 21 (44.7%) | |
| T1N1 | 0 (0.0%) | 1 (5.9%) | 1 (16.7%) | 2 (4.3%) | |
| T2N0 | 9 (37.5%) | 4 (23.5%) | 3 (50.0%) | 16 (34.0%) | |
| T2N1 | 0 (0.0%) | 2 (11.8%) | 0 (0.0%) | 2 (4.3%) | |
| T3N0 | 3 (12.5%) | 1 (5.9%) | 0 (0.0%) | 4 (8.5%) | |
| T3N1 | 0 (0.0%) | 2 (11.8%) | 0 (0.0%) | 2 (4.3%) | |
| NA | 0 | 1 | 0 | 1 | |
| Stage (AJCC/UIC 8th ed) | <0.001 | ||||
| I | 12 (50.0%) | 0 (0.0%) | 0 (0.0%) | 12 (25.5%) | |
| II | 9 (37.5%) | 0 (0.0%) | 0 (0.0%) | 9 (19.1%) | |
| III | 3 (12.5%) | 12 (70.6%) | 0 (0.0%) | 15 (31.9%) | |
| IV | 0 (0.0%) | 5 (29.4%) | 6 (100.0%) | 11 (23.4%) | |
| NA | 0 | 1 | 0 | 1 | |
| Disease-specific mortality | <0.001 | ||||
| No | 20 (100.0%) | 5 (45.5%) | 2 (33.3%) | 27 (73.0%) | |
| Yes | 0 (0.0%) | 6 (54.5%) | 4 (66.7%) | 10 (27.0%) | |
| NA | 4 | 7 | 0 | 11 | |
| Overall mortality | 0.449 | ||||
| No | 5 (20.8%) | 1 (5.9%) | 1 (16.7%) | 7 (14.9%) | |
| Yes | 19 (79.2%) | 16 (94.1%) | 5 (83.3%) | 40 (85.1%) | |
| NA | 0 | 1 | 0 | 1 |
| UP Regulated Genes | DOWN Regulated Genes | ||||
|---|---|---|---|---|---|
| Gene | Absolute Fold Change | p Value | Gene | Absolute Fold Change | p Value |
| ACVR1 | 1.49 | 0.013 | AGGF1 | 0.71 | 0.019 |
| BAG2 | 1.82 | 0.008 | AQP1 | 0.62 | 0.035 |
| BMPR1B | 3.94 | 0.042 | CD36 | 0.41 | 0.011 |
| CAV1 | 1.65 | 0.024 | CGN | 0.18 | 0.041 |
| CCL11 | 4.90 | 0.005 | CLDN4 | 0.29 | 0.036 |
| CDK14 | 1.84 | 0.034 | EMILIN3 | 0.41 | 0.018 |
| CEP170 | 1.42 | 0.034 | ERMP1 | 0.48 | 0.001 |
| COL5A1 | 1.90 | 0.045 | HDAC5 | 0.72 | 0.010 |
| COL5A2 | 1.70 | 0.046 | HPSE | 0.72 | 0.041 |
| COL6A1 | 1.91 | 0.041 | IL18 | 0.29 | 0.004 |
| CSPG4 | 2.97 | 0.018 | KCNJ8 | 0.70 | 0.038 |
| HAS1 | 3.64 | 0.041 | LLGL2 | 0.42 | 0.033 |
| IL11 | 5.32 | 0.026 | MAPK3 | 0.79 | 0.026 |
| INHBA | 2.59 | 0.016 | MAPKAPK3 | 0.48 | 0.014 |
| MED1 | 1.26 | 0.010 | MEOX2 | 0.51 | 0.021 |
| MET | 1.72 | 0.022 | MYO5C | 0.64 | 0.034 |
| NME4 | 2.11 | 0.003 | NRXN1 | 0.42 | 0.016 |
| NR4A3 | 1.91 | 0.048 | OCLN | 0.14 | 0.015 |
| OAS1 | 1.80 | 0.031 | PPL | 0.48 | 0.020 |
| PLAU | 2.69 | 0.002 | PRELP | 0.10 | 0.008 |
| POPDC3 | 7.80 | 0.003 | PROM1 | 0.25 | 0.022 |
| PTGS2 | 3.94 | 0.025 | PYCARD | 0.54 | 0.024 |
| PTK2 | 1.38 | 0.042 | RAF1 | 0.76 | 0.025 |
| SERPINH1 | 1.72 | 0.024 | RNH1 | 0.72 | 0.015 |
| SLC12A6 | 1.42 | 0.046 | RPS27A | 0.71 | 0.004 |
| VAV2 | 1.91 | 0.039 | SERINC5 | 0.71 | 0.037 |
| VEGFC | 2.19 | 0.029 | SKP1 | 0.80 | 0.043 |
| SPINK5 | 0.29 | 0.008 | |||
| SV2B | 0.23 | 0.037 | |||
| TFDP1 | 0.76 | 0.027 | |||
| TGFBR2 | 0.73 | 0.026 | |||
| TJP3 | 0.21 | 0.049 | |||
| TNN | 0.31 | 0.002 | |||
| TPSB2 | 0.52 | 0.003 | |||
| TPSD1 | 0.23 | 0.001 | |||
| UTS2 | 0.41 | 0.036 | |||
| VAMP8 | 0.66 | 0.047 | |||
| VSIG4 | 0.44 | 0.043 | |||
| VWA1 | 0.17 | 0.007 | |||
| VWA2 | 0.15 | 0.014 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paganelli, A.; Zaffonato, M.; Donati, B.; Torricelli, F.; Manicardi, V.; Lai, M.; Spadafora, M.; Piana, S.; Ciarrocchi, A.; Longo, C. Molecular and Histopathological Characterization of Metastatic Cutaneous Squamous Cell Carcinomas: A Case–Control Study. Cancers 2024, 16, 2233. https://doi.org/10.3390/cancers16122233
Paganelli A, Zaffonato M, Donati B, Torricelli F, Manicardi V, Lai M, Spadafora M, Piana S, Ciarrocchi A, Longo C. Molecular and Histopathological Characterization of Metastatic Cutaneous Squamous Cell Carcinomas: A Case–Control Study. Cancers. 2024; 16(12):2233. https://doi.org/10.3390/cancers16122233
Chicago/Turabian StylePaganelli, Alessia, Marco Zaffonato, Benedetta Donati, Federica Torricelli, Veronica Manicardi, Michela Lai, Marco Spadafora, Simonetta Piana, Alessia Ciarrocchi, and Caterina Longo. 2024. "Molecular and Histopathological Characterization of Metastatic Cutaneous Squamous Cell Carcinomas: A Case–Control Study" Cancers 16, no. 12: 2233. https://doi.org/10.3390/cancers16122233
APA StylePaganelli, A., Zaffonato, M., Donati, B., Torricelli, F., Manicardi, V., Lai, M., Spadafora, M., Piana, S., Ciarrocchi, A., & Longo, C. (2024). Molecular and Histopathological Characterization of Metastatic Cutaneous Squamous Cell Carcinomas: A Case–Control Study. Cancers, 16(12), 2233. https://doi.org/10.3390/cancers16122233







