The Spectrum of CAR Cellular Effectors: Modes of Action in Anti-Tumor Immunity
Abstract
:Simple Summary
Abstract
1. Introduction
2. T-Lymphocytes
3. Natural Killer Cells
3.1. Activation by the Chimeric Antigen Receptor
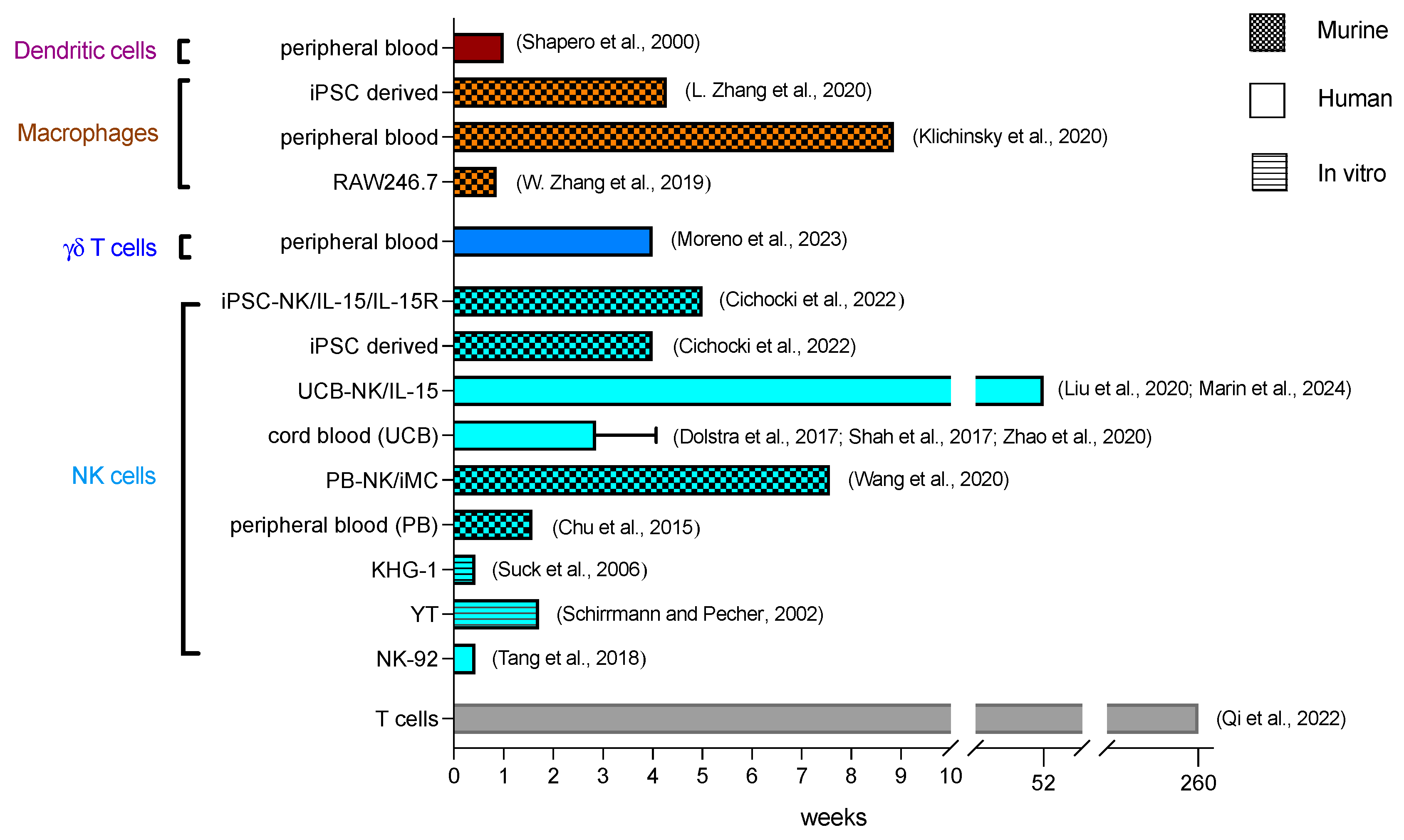
3.2. Kinetics
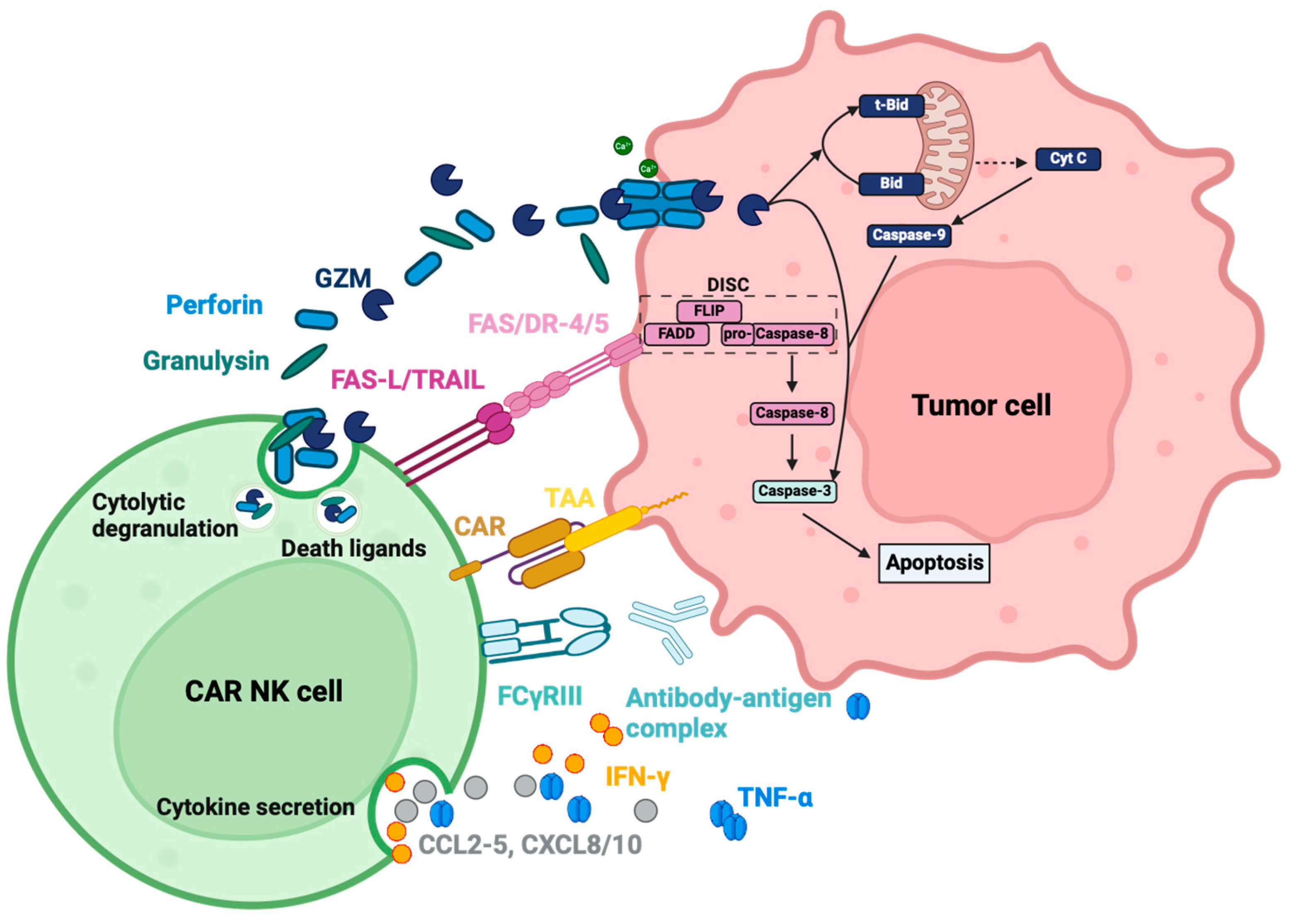
3.3. Functions
3.3.1. Soluble Effector Molecules
Cytolytic Granules
Cytokines and Chemokines
3.3.2. Membrane-Bound Effector Molecules
3.3.3. Interaction with Other Cells
4. Macrophages
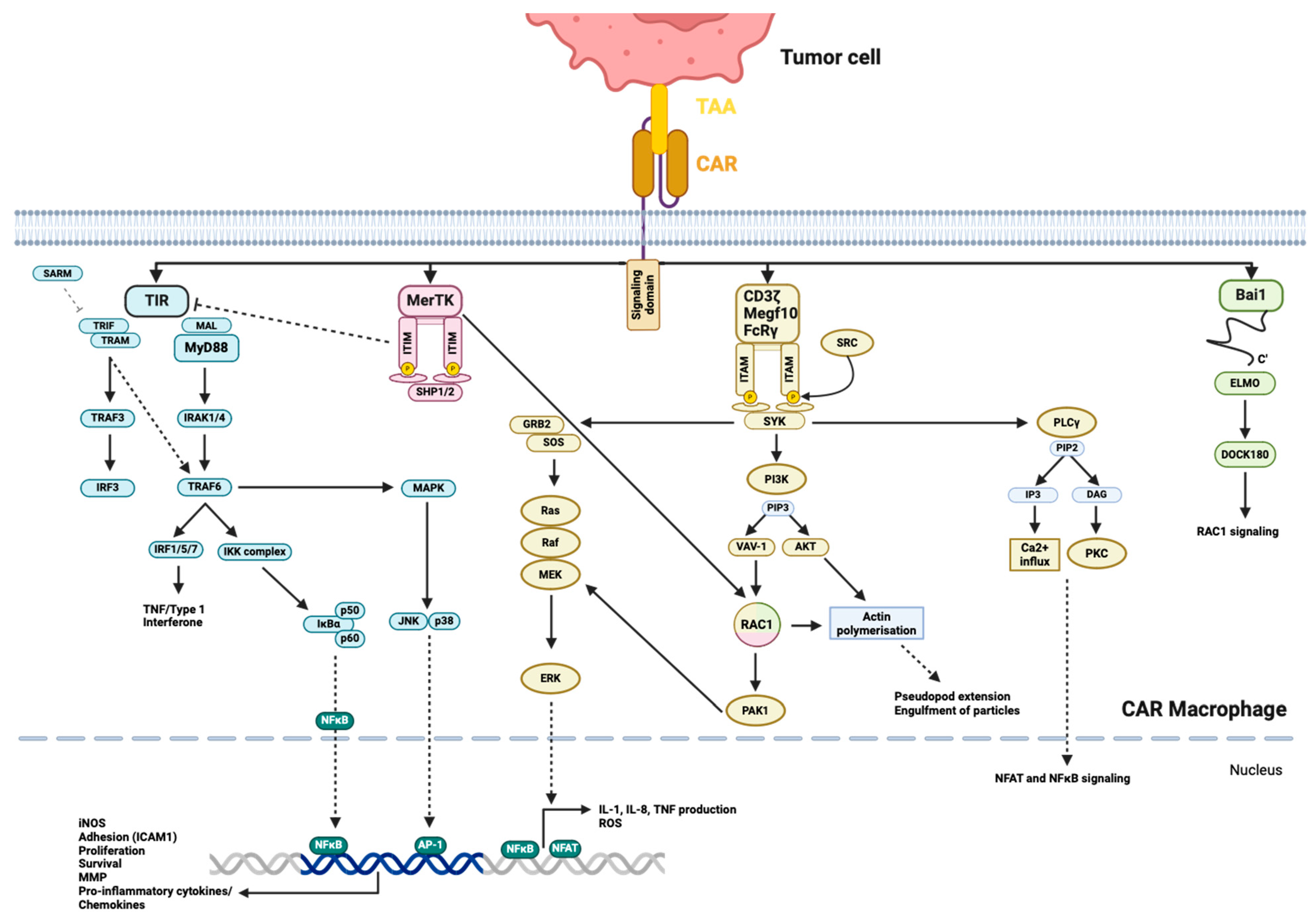
4.1. Activation by the CAR
4.2. Kinetics
4.3. Functions
4.3.1. Soluble Effector Molecules
Cytokines and Chemokines
Matrix Metalloprotease (MMP)
ROS, RNS, NO and Other Soluble Molecules
Apoptotic, Necroptotic and Pyroptotic Effects of Soluble Effector Molecules
4.3.2. Membrane-Bound Effectors
4.3.3. Phagocytosis and Antigen Presentation
4.3.4. TME Remodeling
5. γδ T Cells
5.1. Activation by the CAR
5.2. Kinetics
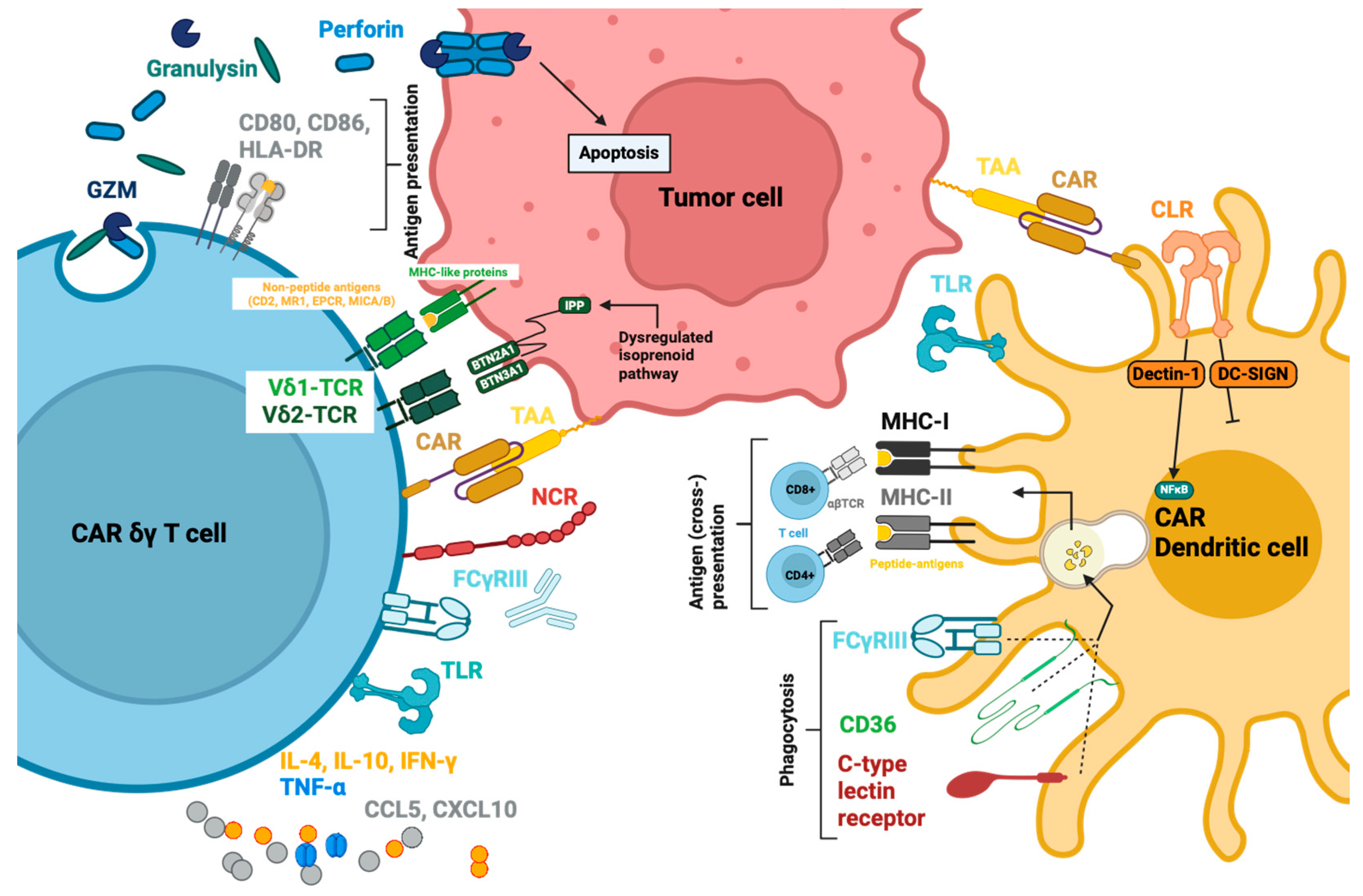
5.3. Functions
5.3.1. Soluble Effector Molecules
5.3.2. Membrane-bound effectors.
5.3.3. Antigen-Presenting Function
6. Dendritic Cells
6.1. Activation by the CAR
6.2. Kinetics
6.3. Functions
6.3.1. Endocytosis, Antigen Presentation and Immune Modulation
6.3.2. Soluble Effector Molecules
Cytokines, Chemokines, Exosomes
6.3.3. Membrane-Bound Effectors
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Prehn, R.T.; Main, J.M. Immunity to Methylcholanthrene-Induced Sarcomas. J. Natl. Cancer Inst. 1957, 18, 769–778. [Google Scholar] [PubMed]
- Klein, G.; Sjogren, H.O.; Klein, E.; Hellstrom, K.E. Demonstration of Resistance against Methylcholanthrene-Induced Sarcomas in the Primary Autochthonous Host. Cancer Res. 1960, 20, 1561–1572. [Google Scholar] [PubMed]
- Abken, H. Building on Synthetic Immunology and T Cell Engineering: A Brief Journey through the History of Chimeric Antigen Receptors. Hum. Gene Ther. 2021, 32, 1011–1028. [Google Scholar] [CrossRef]
- Rosenberg, S.A.; Spiess, P.; Lafreniere, R. A New Approach to the Adoptive Immunotherapy of Cancer with Tumor-Infiltrating Lymphocytes. Science 1986, 233, 1318–1321. [Google Scholar] [CrossRef]
- Rosenberg, S.A.; Packard, B.S.; Aebersold, P.M.; Solomon, D.; Topalian, S.L.; Toy, S.T.; Simon, P.; Lotze, M.T.; Yang, J.C.; Seipp, C.A.; et al. Use of Tumor-Infiltrating Lymphocytes and Interleukin-2 in the Immunotherapy of Patients with Metastatic Melanoma. N. Engl. J. Med. 1988, 319, 1676–1680. [Google Scholar] [CrossRef]
- Morgan, R.A.; Dudley, M.E.; Wunderlich, J.R.; Hughes, M.S.; Yang, J.C.; Sherry, R.M.; Royal, R.E.; Topalian, S.L.; Kammula, U.S.; Restifo, N.P.; et al. Cancer Regression in Patients After Transfer of Genetically Engineered Lymphocytes. Science 2006, 314, 126–129. [Google Scholar] [CrossRef]
- Eshhar, Z.; Waks, T.; Gross, G.; Schindler, D.G. Specific Activation and Targeting of Cytotoxic Lymphocytes through Chimeric Single Chains Consisting of Antibody-Binding Domains and the Gamma or Zeta Subunits of the Immunoglobulin and T-Cell Receptors. Proc. Natl. Acad. Sci. USA 1993, 90, 720–724. [Google Scholar] [CrossRef]
- Brocker, T.; Peter, A.; Traunecker, A.; Karjalainen, K. New Simplified Molecular Design for Functional T Cell Receptor. Eur. J. Immunol. 1993, 23, 1435–1439. [Google Scholar] [CrossRef] [PubMed]
- Guedan, S.; Calderon, H.; Posey, A.D.; Maus, M.V. Engineering and Design of Chimeric Antigen Receptors. Mol. Ther. Methods Clin. Dev. 2019, 12, 145–156. [Google Scholar] [CrossRef]
- Rafiq, S.; Hackett, C.S.; Brentjens, R.J. Engineering Strategies to Overcome the Current Roadblocks in CAR T Cell Therapy. Nat. Rev. Clin. Oncol. 2020, 17, 147–167. [Google Scholar] [CrossRef]
- Porter, D.L.; Levine, B.L.; Kalos, M.; Bagg, A.; June, C.H. Chimeric Antigen Receptor–Modified T Cells in Chronic Lymphoid Leukemia. N. Engl. J. Med. 2011, 365, 725–733. [Google Scholar] [CrossRef]
- Johnson, P.C.; Abramson, J.S. Patient Selection for Chimeric Antigen Receptor (CAR) T-Cell Therapy for Aggressive B-Cell Non-Hodgkin Lymphomas. Leuk. Lymphoma 2020, 61, 2561–2567. [Google Scholar] [CrossRef] [PubMed]
- Schuster, S.J.; Bishop, M.R.; Tam, C.S.; Waller, E.K.; Borchmann, P.; McGuirk, J.P.; Jäger, U.; Jaglowski, S.; Andreadis, C.; Westin, J.R.; et al. Tisagenlecleucel in Adult Relapsed or Refractory Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2019, 380, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Neelapu, S.S.; Jacobson, C.A.; Ghobadi, A.; Miklos, D.B.; Lekakis, L.J.; Oluwole, O.O.; Lin, Y.; Braunschweig, I.; Hill, B.T.; Timmerman, J.M.; et al. 5-Year Follow-Up Supports Curative Potential of Axicabtagene Ciloleucel in Refractory Large B-Cell Lymphoma (ZUMA-1). Blood 2023, 141, 2307–2315. [Google Scholar] [CrossRef] [PubMed]
- Sterner, R.C.; Sterner, R.M. CAR-T Cell Therapy: Current Limitations and Potential Strategies. Blood Cancer J. 2021, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Quintero, J.; Díaz, M.P.; Palmar, J.; Galan-Freyle, N.J.; Morillo, V.; Escalona, D.; González-Torres, H.J.; Torres, W.; Navarro-Quiroz, E.; Rivera-Porras, D.; et al. Car T Cells in Solid Tumors: Overcoming Obstacles. Int. J. Mol. Sci. 2024, 25, 4170. [Google Scholar] [CrossRef]
- Daei Sorkhabi, A.; Mohamed Khosroshahi, L.; Sarkesh, A.; Mardi, A.; Aghebati-Maleki, A.; Aghebati-Maleki, L.; Baradaran, B. The Current Landscape of CAR T-Cell Therapy for Solid Tumors: Mechanisms, Research Progress, Challenges, and Counterstrategies. Front. Immunol. 2023, 14, 1113882. [Google Scholar] [CrossRef] [PubMed]
- Benmebarek, M.-R.; Karches, C.; Cadilha, B.; Lesch, S.; Endres, S.; Kobold, S. Killing Mechanisms of Chimeric Antigen Receptor (CAR) T Cells. Int. J. Mol. Sci. 2019, 20, 1283. [Google Scholar] [CrossRef]
- Zheng, Z.; Li, S.; Liu, M.; Chen, C.; Zhang, L.; Zhou, D. Fine-Tuning through Generations: Advances in Structure and Production of CAR-T Therapy. Cancers 2023, 15, 3476. [Google Scholar] [CrossRef]
- Asmamaw Dejenie, T.; Tiruneh, G.; Medhin, M.; Dessie Terefe, G.; Tadele Admasu, F.; Wale Tesega, W.; Chekol Abebe, E. Current Updates on Generations, Approvals, and Clinical Trials of CAR T-Cell Therapy. Hum. Vaccin. Immunother. 2022, 18, 2114254. [Google Scholar] [CrossRef]
- Wu, L.; Wei, Q.; Brzostek, J.; Gascoigne, N.R.J. Signaling from T Cell Receptors (TCRs) and Chimeric Antigen Receptors (CARs) on T Cells. Cell Mol. Immunol. 2020, 17, 600–612. [Google Scholar] [CrossRef]
- Xiong, Y.; Libby, K.A.; Su, X. The Physical Landscape of CAR-T Synapse. Biophys. J. 2023, 123, 1–12. [Google Scholar] [CrossRef]
- Hwang, J.-R.; Byeon, Y.; Kim, D.; Park, S.-G. Recent Insights of T Cell Receptor-Mediated Signaling Pathways for T Cell Activation and Development. Exp. Mol. Med. 2020, 52, 750–761. [Google Scholar] [CrossRef]
- Qi, T.; McGrath, K.; Ranganathan, R.; Dotti, G.; Cao, Y. Cellular Kinetics: A Clinical and Computational Review of CAR-T Cell Pharmacology. Adv. Drug Deliv. Rev. 2022, 188, 114421. [Google Scholar] [CrossRef]
- Wittibschlager, V.; Bacher, U.; Seipel, K.; Porret, N.; Wiedemann, G.; Haslebacher, C.; Hoffmann, M.; Daskalakis, M.; Akhoundova, D.; Pabst, T. CAR T-Cell Persistence Correlates with Improved Outcome in Patients with B-Cell Lymphoma. Int. J. Mol. Sci. 2023, 24, 5688. [Google Scholar] [CrossRef]
- Fraietta, J.A.; Lacey, S.F.; Orlando, E.J.; Pruteanu-Malinici, I.; Gohil, M.; Lundh, S.; Boesteanu, A.C.; Wang, Y.; O’Connor, R.S.; Hwang, W.-T.; et al. Determinants of Response and Resistance to CD19 Chimeric Antigen Receptor (CAR) T Cell Therapy of Chronic Lymphocytic Leukemia. Nat. Med. 2018, 24, 563–571. [Google Scholar] [CrossRef]
- Sommermeyer, D.; Hudecek, M.; Kosasih, P.L.; Gogishvili, T.; Maloney, D.G.; Turtle, C.J.; Riddell, S.R. Chimeric Antigen Receptor-Modified T Cells Derived from Defined CD8+ and CD4+ Subsets Confer Superior Antitumor Reactivity in Vivo. Leukemia 2016, 30, 492–500. [Google Scholar] [CrossRef]
- López-Cantillo, G.; Urueña, C.; Camacho, B.A.; Ramírez-Segura, C. CAR-T Cell Performance: How to Improve Their Persistence? Front. Immunol. 2022, 13, 878209. [Google Scholar] [CrossRef]
- Wang, Y.; Tong, C.; Lu, Y.; Wu, Z.; Guo, Y.; Liu, Y.; Wei, J.; Wang, C.; Yang, Q.; Han, W. Characteristics of Premanufacture CD8+ T Cells Determine CAR-T Efficacy in Patients with Diffuse Large B-Cell Lymphoma. Signal Transduct. Target. Ther. 2023, 8, 409. [Google Scholar] [CrossRef]
- Turtle, C.J.; Hanafi, L.-A.; Berger, C.; Gooley, T.A.; Cherian, S.; Hudecek, M.; Sommermeyer, D.; Melville, K.; Pender, B.; Budiarto, T.M.; et al. CD19 CAR–T Cells of Defined CD4+:CD8+ Composition in Adult B Cell ALL Patients. J. Clin. Investig. 2016, 126, 2123–2138. [Google Scholar] [CrossRef]
- Garfall, A.L.; Dancy, E.K.; Cohen, A.D.; Hwang, W.-T.; Fraietta, J.A.; Davis, M.M.; Levine, B.L.; Siegel, D.L.; Stadtmauer, E.A.; Vogl, D.T.; et al. T-Cell Phenotypes Associated with Effective CAR T-Cell Therapy in Postinduction vs Relapsed Multiple Myeloma. Blood Adv. 2019, 3, 2812–2815. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.M.; Weaver, C. Janeway’s Immunobiology, 9th ed.; Garland Science: New York, NY, USA, 2017; ISBN 9780815345053. [Google Scholar]
- Greenberg, A.H.; Hudson, L.; Shen, L.; Roitt, I.M. Antibody-Dependent Cell-Mediated Cytotoxicity Due to a “Null” Lymphoid Cell. Nat. New Biol. 1973, 242, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, A.H.; Playfair, J.H. Spontaneously Arising Cytotoxicity to the P-815-Y Mastocytoma in NZB Mice. Clin. Exp. Immunol. 1974, 16, 99–109. [Google Scholar] [PubMed]
- Greenberg, A.H. The Origins of the NK Cell, or a Canadian in King Ivan’s Court. Clin. Invest. Med. 1994, 17, 626–631. [Google Scholar] [PubMed]
- Kiessling, R.; Klein, E.; Wigzell, H. „Natural” Killer Cells in the Mouse. I. Cytotoxic Cells with Specificity for Mouse Moloney Leukemia Cells. Specificity and Distribution According to Genotype. Eur. J. Immunol. 1975, 5, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Vojdani, A.; Koksoy, S.; Vojdani, E.; Engelman, M.; Benzvi, C.; Lerner, A. Natural Killer Cells and Cytotoxic T Cells: Complementary Partners against Microorganisms and Cancer. Microorganisms 2024, 12, 230. [Google Scholar] [CrossRef] [PubMed]
- Khawar, M.B.; Sun, H. CAR-NK Cells: From Natural Basis to Design for Kill. Front. Immunol. 2021, 12, 707542. [Google Scholar] [CrossRef] [PubMed]
- Tran, A.C.; Zhang, D.; Byrn, R.; Roberts, M.R. Chimeric Zeta-Receptors Direct Human Natural Killer (NK) Effector Function to Permit Killing of NK-Resistant Tumor Cells and HIV-Infected T Lymphocytes. J. Immunol. 1995, 155, 1000–1009. [Google Scholar] [CrossRef] [PubMed]
- Vivier, E.; Nunès, J.A.; Vély, F. Natural Killer Cell Signaling Pathways. Science 2004, 306, 1517–1519. [Google Scholar] [CrossRef]
- Chen, Y.; Lu, D.; Churov, A.; Fu, R. Research Progress on NK Cell Receptors and Their Signaling Pathways. Mediat. Inflamm. 2020, 2020, 1–14. [Google Scholar] [CrossRef]
- Lanier, L.L. DAP10- and DAP12-associated Receptors in Innate Immunity. Immunol. Rev. 2009, 227, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Oberschmidt, O.; Kloess, S.; Koehl, U. Redirected Primary Human Chimeric Antigen Receptor Natural Killer Cells As an “Off-the-Shelf Immunotherapy” for Improvement in Cancer Treatment. Front. Immunol. 2017, 8, 654. [Google Scholar] [CrossRef] [PubMed]
- Imai, C.; Iwamoto, S.; Campana, D. Genetic Modification of Primary Natural Killer Cells Overcomes Inhibitory Signals and Induces Specific Killing of Leukemic Cells. Blood 2005, 106, 376–383. [Google Scholar] [CrossRef] [PubMed]
- Müller, N.; Michen, S.; Tietze, S.; Töpfer, K.; Schulte, A.; Lamszus, K.; Schmitz, M.; Schackert, G.; Pastan, I.; Temme, A. Engineering NK Cells Modified With an EGFRvIII-Specific Chimeric Antigen Receptor to Overexpress CXCR4 Improves Immunotherapy of CXCL12/SDF-1α-Secreting Glioblastoma. J. Immunother. 2015, 38, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Clémenceau, B.; Valsesia-Wittmann, S.; Jallas, A.-C.; Vivien, R.; Rousseau, R.; Marabelle, A.; Caux, C.; Vié, H. In Vitro and In Vivo Comparison of Lymphocytes Transduced with a Human CD16 or with a Chimeric Antigen Receptor Reveals Potential Off-Target Interactions Due to the IgG2 CH2-CH3 CAR-Spacer. J. Immunol. Res. 2015, 2015, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Töpfer, K.; Cartellieri, M.; Michen, S.; Wiedemuth, R.; Müller, N.; Lindemann, D.; Bachmann, M.; Füssel, M.; Schackert, G.; Temme, A. DAP12-Based Activating Chimeric Antigen Receptor for NK Cell Tumor Immunotherapy. J. Immunol. 2015, 194, 3201–3212. [Google Scholar] [CrossRef] [PubMed]
- Siegler, E.L.; Kim, Y.J.; Chen, X.; Siriwon, N.; Mac, J.; Rohrs, J.A.; Bryson, P.D.; Wang, P. Combination Cancer Therapy Using Chimeric Antigen Receptor-Engineered Natural Killer Cells as Drug Carriers. Mol. Ther. 2017, 25, 2607–2619. [Google Scholar] [CrossRef] [PubMed]
- Oelsner, S.; Friede, M.E.; Zhang, C.; Wagner, J.; Badura, S.; Bader, P.; Ullrich, E.; Ottmann, O.G.; Klingemann, H.; Tonn, T.; et al. Continuously Expanding CAR NK-92 Cells Display Selective Cytotoxicity against B-Cell Leukemia and Lymphoma. Cytotherapy 2017, 19, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hermanson, D.L.; Moriarity, B.S.; Kaufman, D.S. Human IPSC-Derived Natural Killer Cells Engineered with Chimeric Antigen Receptors Enhance Anti-Tumor Activity. Cell Stem Cell 2018, 23, 181–192. [Google Scholar] [CrossRef]
- Altvater, B.; Landmeier, S.; Pscherer, S.; Temme, J.; Schweer, K.; Kailayangiri, S.; Campana, D.; Juergens, H.; Pule, M.; Rossig, C. 2B4 (CD244) Signaling by Recombinant Antigen-Specific Chimeric Receptors Costimulates Natural Killer Cell Activation to Leukemia and Neuroblastoma Cells. Clin. Cancer Res. 2009, 15, 4857–4866. [Google Scholar] [CrossRef]
- Kailayangiri, S.; Altvater, B.; Spurny, C.; Jamitzky, S.; Schelhaas, S.; Jacobs, A.H.; Wiek, C.; Roellecke, K.; Hanenberg, H.; Hartmann, W.; et al. Targeting Ewing Sarcoma with Activated and GD2-Specific Chimeric Antigen Receptor-Engineered Human NK Cells Induces Upregulation of Immune-Inhibitory HLA-G. Oncoimmunology 2017, 6, e1250050. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Klein Wolterink, R.G.J.; Wang, J.; Bos, G.M.J.; Germeraad, W.T. V. Chimeric Antigen Receptor Natural Killer (CAR-NK) Cell Design and Engineering for Cancer Therapy. J. Hematol. Oncol. 2021, 14, 73. [Google Scholar] [CrossRef] [PubMed]
- Schirrmann, T.; Pecher, G. Human Natural Killer Cell Line Modified with a Chimeric Immunoglobulin T-Cell Receptor Gene Leads to Tumor Growth Inhibition in Vivo. Cancer Gene Ther. 2002, 9, 390–398. [Google Scholar] [CrossRef]
- Tang, X.; Yang, L.; Li, Z.; Nalin, A.P.; Dai, H.; Xu, T.; Yin, J.; You, F.; Zhu, M.; Shen, W.; et al. First-in-Man Clinical Trial of CAR NK-92 Cells: Safety Test of CD33-CAR NK-92 Cells in Patients with Relapsed and Refractory Acute Myeloid Leukemia. Am. J. Cancer Res. 2018, 8, 1083–1089. [Google Scholar] [PubMed]
- Suck, G.; Branch, D.R.; Keating, A. Irradiated KHYG-1 Retains Cytotoxicity: Potential for Adoptive Immunotherapy with a Natural Killer Cell Line. Int. J. Radiat. Biol. 2006, 82, 355–361. [Google Scholar] [CrossRef]
- Chu, Y.; Hochberg, J.; Yahr, A.; Ayello, J.; van de Ven, C.; Barth, M.; Czuczman, M.; Cairo, M.S. Targeting CD20+ Aggressive B-Cell Non–Hodgkin Lymphoma by Anti-CD20 CAR MRNA-Modified Expanded Natural Killer Cells In Vitro and in NSG Mice. Cancer Immunol. Res. 2015, 3, 333–344. [Google Scholar] [CrossRef]
- Wang, X.; Jasinski, D.L.; Medina, J.L.; Spencer, D.M.; Foster, A.E.; Bayle, J.H. Inducible MyD88/CD40 Synergizes with IL-15 to Enhance Antitumor Efficacy of CAR-NK Cells. Blood Adv. 2020, 4, 1950–1964. [Google Scholar] [CrossRef] [PubMed]
- Dolstra, H.; Roeven, M.W.H.; Spanholtz, J.; Hangalapura, B.N.; Tordoir, M.; Maas, F.; Leenders, M.; Bohme, F.; Kok, N.; Trilsbeek, C.; et al. Successful Transfer of Umbilical Cord Blood CD34+ Hematopoietic Stem and Progenitor-Derived NK Cells in Older Acute Myeloid Leukemia Patients. Clin. Cancer Res. 2017, 23, 4107–4118. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Li, L.; McCarty, J.; Kaur, I.; Yvon, E.; Shaim, H.; Muftuoglu, M.; Liu, E.; Orlowski, R.Z.; Cooper, L.; et al. Phase I Study of Cord Blood-Derived Natural Killer Cells Combined with Autologous Stem Cell Transplantation in Multiple Myeloma. Br. J. Haematol. 2017, 177, 457–466. [Google Scholar] [CrossRef]
- Zhao, X.; Cai, L.; Hu, Y.; Wang, H. Cord-Blood Natural Killer Cell-Based Immunotherapy for Cancer. Front. Immunol. 2020, 11, 584099. [Google Scholar] [CrossRef]
- Liu, E.; Marin, D.; Banerjee, P.; Macapinlac, H.A.; Thompson, P.; Basar, R.; Nassif Kerbauy, L.; Overman, B.; Thall, P.; Kaplan, M.; et al. Use of CAR-Transduced Natural Killer Cells in CD19-Positive Lymphoid Tumors. N. Engl. J. Med. 2020, 382, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Marin, D.; Li, Y.; Basar, R.; Rafei, H.; Daher, M.; Dou, J.; Mohanty, V.; Dede, M.; Nieto, Y.; Uprety, N.; et al. Safety, Efficacy and Determinants of Response of Allogeneic CD19-Specific CAR-NK Cells in CD19+ B Cell Tumors: A Phase 1/2 Trial. Nat. Med. 2024, 30, 772–784. [Google Scholar] [CrossRef] [PubMed]
- Cichocki, F.; Goodridge, J.P.; Bjordahl, R.; Mahmood, S.; Davis, Z.B.; Gaidarova, S.; Abujarour, R.; Groff, B.; Witty, A.; Wang, H.; et al. Dual Antigen–Targeted off-the-Shelf NK Cells Show Durable Response and Prevent Antigen Escape in Lymphoma and Leukemia. Blood 2022, 140, 2451–2462. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, L.; Su, H.; Liu, Q.; Shen, J.; Dai, H.; Zheng, W.; Lu, Y.; Zhang, W.; Bei, Y.; et al. Chimeric Antigen Receptor Macrophage Therapy for Breast Tumours Mediated by Targeting the Tumour Extracellular Matrix. Br. J. Cancer 2019, 121, 837–845. [Google Scholar] [CrossRef]
- Zhang, L.; Tian, L.; Dai, X.; Yu, H.; Wang, J.; Lei, A.; Zhu, M.; Xu, J.; Zhao, W.; Zhu, Y.; et al. Pluripotent Stem Cell-Derived CAR-Macrophage Cells with Antigen-Dependent Anti-Cancer Cell Functions. J. Hematol. Oncol. 2020, 13, 153. [Google Scholar] [CrossRef] [PubMed]
- Klichinsky, M.; Ruella, M.; Shestova, O.; Lu, X.M.; Best, A.; Zeeman, M.; Schmierer, M.; Gabrusiewicz, K.; Anderson, N.R.; Petty, N.E.; et al. Human Chimeric Antigen Receptor Macrophages for Cancer Immunotherapy. Nat. Biotechnol. 2020, 38, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.A.; Kennedy-Wilde, J.; Wrong, A.D.; Jahchan, N.; Galimi, F.; Ye, Y.; Lai, R.; Aftab, B.T. Expansion, Persistence and Pharmacodynamic Profile of ADI-001, a First-in-Class Allogeneic CD20-Targeted CAR Gamma Delta T Cell Therapy, in Patients with Relapsed/Refractory Aggressive B-Cell Non-Hodgkin’s Lymphoma. Blood 2023, 142, 3478–3478. [Google Scholar] [CrossRef]
- Shapero, M.H.; Kundu, S.K.; Engleman, E.; Laus, R.; Van Schooten, W.C.A.; Merigan, T.C. In Vivo Persistence of Donor Cells Following Adoptive Transfer of Allogeneic Dendritic Cells in HIV-Infected Patients. Cell Transplant. 2000, 9, 307–317. [Google Scholar] [CrossRef]
- Tonn, T.; Schwabe, D.; Klingemann, H.G.; Becker, S.; Esser, R.; Koehl, U.; Suttorp, M.; Seifried, E.; Ottmann, O.G.; Bug, G. Treatment of Patients with Advanced Cancer with the Natural Killer Cell Line NK-92. Cytotherapy 2013, 15, 1563–1570. [Google Scholar] [CrossRef]
- Arai, S.; Meagher, R.; Swearingen, M.; Myint, H.; Rich, E.; Martinson, J.; Klingemann, H. Infusion of the Allogeneic Cell Line NK-92 in Patients with Advanced Renal Cell Cancer or Melanoma: A Phase I Trial. Cytotherapy 2008, 10, 625–632. [Google Scholar] [CrossRef]
- Schönfeld, K.; Sahm, C.; Zhang, C.; Naundorf, S.; Brendel, C.; Odendahl, M.; Nowakowska, P.; Bönig, H.; Köhl, U.; Kloess, S.; et al. Selective Inhibition of Tumor Growth by Clonal NK Cells Expressing an ErbB2/HER2-Specific Chimeric Antigen Receptor. Mol. Ther. 2015, 23, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Tonn, T.; Becker, S.; Esser, R.; Schwabe, D.; Seifried, E. Cellular Immunotherapy of Malignancies Using the Clonal Natural Killer Cell Line NK-92. J. Hematother Stem Cell Res. 2001, 10, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Lapteva, N.; Szmania, S.M.; van Rhee, F.; Rooney, C.M. Clinical Grade Purification and Expansion of Natural Killer Cells. Crit. Rev. Oncog. 2014, 19, 121–132. [Google Scholar] [CrossRef]
- Becker, P.S.A.; Suck, G.; Nowakowska, P.; Ullrich, E.; Seifried, E.; Bader, P.; Tonn, T.; Seidl, C. Selection and Expansion of Natural Killer Cells for NK Cell-Based Immunotherapy. Cancer Immunol. Immunother. 2016, 65, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Ahn, Y.-O.; Kim, S.; Kim, T.M.; Song, E.Y.; Park, M.H.; Heo, D.S. Irradiated and Activated Autologous PBMCs Induce Expansion of Highly Cytotoxic Human NK Cells In Vitro. J. Immunother. 2013, 36, 373–381. [Google Scholar] [CrossRef]
- Koehl, U.; Brehm, C.; Huenecke, S.; Zimmermann, S.-Y.; Kloess, S.; Bremm, M.; Ullrich, E.; Soerensen, J.; Quaiser, A.; Erben, S.; et al. Clinical Grade Purification and Expansion of NK Cell Products for an Optimized Manufacturing Protocol. Front. Oncol. 2013, 3, 118. [Google Scholar] [CrossRef]
- Suerth, J.D.; Morgan, M.A.; Kloess, S.; Heckl, D.; Neudörfl, C.; Falk, C.S.; Koehl, U.; Schambach, A. Efficient Generation of Gene-Modified Human Natural Killer Cells via Alpharetroviral Vectors. J. Mol. Med. 2016, 94, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Lapteva, N.; Durett, A.G.; Sun, J.; Rollins, L.A.; Huye, L.L.; Fang, J.; Dandekar, V.; Mei, Z.; Jackson, K.; Vera, J.; et al. Large-Scale Ex Vivo Expansion and Characterization of Natural Killer Cells for Clinical Applications. Cytotherapy 2012, 14, 1131–1143. [Google Scholar] [CrossRef]
- Herrera, L.; Santos, S.; Vesga, M.A.; Anguita, J.; Martin-Ruiz, I.; Carrascosa, T.; Juan, M.; Eguizabal, C. Adult Peripheral Blood and Umbilical Cord Blood NK Cells Are Good Sources for Effective CAR Therapy against CD19 Positive Leukemic Cells. Sci. Rep. 2019, 9, 18729. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Martin-Antonio, B.; Yang, H.; Ku, S.; Lee, D.A.; Cooper, L.J.N.; Decker, W.K.; Li, S.; Robinson, S.N.; Sekine, T.; et al. Antigen Presenting Cell-Mediated Expansion of Human Umbilical Cord Blood Yields Log-Scale Expansion of Natural Killer Cells with Anti-Myeloma Activity. PLoS ONE 2013, 8, e76781. [Google Scholar] [CrossRef]
- Wen, W.; Chen, X.; Shen, X.-Y.; Li, H.-Y.; Zhang, F.; Fang, F.-Q.; Zhang, X.-B. Enhancing Cord Blood Stem Cell-Derived NK Cell Growth and Differentiation through Hyperosmosis. Stem Cell Res. Ther. 2023, 14, 295. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.; Tong, Y.; Dotti, G.; Shaim, H.; Savoldo, B.; Mukherjee, M.; Orange, J.; Wan, X.; Lu, X.; Reynolds, A.; et al. Cord Blood NK Cells Engineered to Express IL-15 and a CD19-Targeted CAR Show Long-Term Persistence and Potent Antitumor Activity. Leukemia 2018, 32, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Knorr, D.A.; Ni, Z.; Hermanson, D.; Hexum, M.K.; Bendzick, L.; Cooper, L.J.N.; Lee, D.A.; Kaufman, D.S. Clinical-Scale Derivation of Natural Killer Cells From Human Pluripotent Stem Cells for Cancer Therapy. Stem Cells Transl. Med. 2013, 2, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Ueda, T.; Kumagai, A.; Iriguchi, S.; Yasui, Y.; Miyasaka, T.; Nakagoshi, K.; Nakane, K.; Saito, K.; Takahashi, M.; Sasaki, A.; et al. Non–Clinical Efficacy, Safety and Stable Clinical Cell Processing of Induced Pluripotent Stem Cell-derived Anti–Glypican-3 Chimeric Antigen Receptor-expressing Natural Killer/Innate Lymphoid Cells. Cancer Sci. 2020, 111, 1478–1490. [Google Scholar] [CrossRef] [PubMed]
- Cichocki, F.; Bjordahl, R.; Goodridge, J.P.; Mahmood, S.; Gaidarova, S.; Abujarour, R.; Davis, Z.B.; Merino, A.; Tuininga, K.; Wang, H.; et al. Quadruple Gene-Engineered Natural Killer Cells Enable Multi-Antigen Targeting for Durable Antitumor Activity against Multiple Myeloma. Nat. Commun. 2022, 13, 7341. [Google Scholar] [CrossRef] [PubMed]
- Bachanova, V.; Ghobadi, A.; Patel, K.; Park, J.H.; Flinn, I.W.; Shah, P.; Wong, C.; Bickers, C.; Szabo, P.; Wong, L.; et al. Safety and Efficacy of FT596, a First-in-Class, Multi-Antigen Targeted, Off-the-Shelf, IPSC-Derived CD19 CAR NK Cell Therapy in Relapsed/Refractory B-Cell Lymphoma. Blood 2021, 138, 823–823. [Google Scholar] [CrossRef]
- Uherek, C.; Tonn, T.; Uherek, B.; Becker, S.; Schnierle, B.; Klingemann, H.-G.; Wels, W. Retargeting of Natural Killer–Cell Cytolytic Activity to ErbB2-Expressing Cancer Cells Results in Efficient and Selective Tumor Cell Destruction. Blood 2002, 100, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Oberoi, P.; Oelsner, S.; Waldmann, A.; Lindner, A.; Tonn, T.; Wels, W.S. Chimeric Antigen Receptor-Engineered NK-92 Cells: An Off-the-Shelf Cellular Therapeutic for Targeted Elimination of Cancer Cells and Induction of Protective Antitumor Immunity. Front. Immunol. 2017, 8, 533. [Google Scholar] [CrossRef]
- Klingemann, H.; Boissel, L.; Toneguzzo, F. Natural Killer Cells for Immunotherapy—Advantages of the NK-92 Cell Line over Blood NK Cells. Front. Immunol. 2016, 7, 91. [Google Scholar] [CrossRef]
- Suck, G.; Branch, D.R.; Smyth, M.J.; Miller, R.G.; Vergidis, J.; Fahim, S.; Keating, A. KHYG-1, a Model for the Study of Enhanced Natural Killer Cell Cytotoxicity. Exp. Hematol. 2005, 33, 1160–1171. [Google Scholar] [CrossRef]
- Tagaya, Y.; Okada, M.; Sugie, K.; Kasahara, T.; Kondo, N.; Hamuro, J.; Matsushima, K.; Dinarello, C.A.; Yodoi, J. IL-2 Receptor(P55)/Tac-Inducing Factor. Purification and Characterization of Adult T Cell Leukemia-Derived Factor. J. Immunol. 1988, 140, 2614–2620. [Google Scholar] [CrossRef] [PubMed]
- Orange, J.S. Formation and Function of the Lytic NK-Cell Immunological Synapse. Nat. Rev. Immunol. 2008, 8, 713–725. [Google Scholar] [CrossRef] [PubMed]
- James, A.M.; Hsu, H.-T.; Dongre, P.; Uzel, G.; Mace, E.M.; Banerjee, P.P.; Orange, J.S. Rapid Activation Receptor– or IL-2–Induced Lytic Granule Convergence in Human Natural Killer Cells Requires Src, but Not Downstream Signaling. Blood 2013, 121, 2627–2637. [Google Scholar] [CrossRef] [PubMed]
- Voskoboinik, I.; Whisstock, J.C.; Trapani, J.A. Perforin and Granzymes: Function, Dysfunction and Human Pathology. Nat. Rev. Immunol. 2015, 15, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Gwalani, L.A.; Orange, J.S. Single Degranulations in NK Cells Can Mediate Target Cell Killing. J. Immunol. 2018, 200, 3231–3243. [Google Scholar] [CrossRef]
- Osińska, I.; Popko, K.; Demkow, U. Perforin: An Important Player in Immune Response. Cent. Eur. J. Immunol. 2014, 1, 109–115. [Google Scholar] [CrossRef]
- Kloess, S.; Oberschmidt, O.; Dahlke, J.; Vu, X.-K.; Neudoerfl, C.; Kloos, A.; Gardlowski, T.; Matthies, N.; Heuser, M.; Meyer, J.; et al. Preclinical Assessment of Suitable Natural Killer Cell Sources for Chimeric Antigen Receptor Natural Killer–Based “Off-the-Shelf” Acute Myeloid Leukemia Immunotherapies. Hum. Gene Ther. 2019, 30, 381–401. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, D.; Lieberman, J. Death by a Thousand Cuts: Granzyme Pathways of Programmed Cell Death. Annu. Rev. Immunol. 2008, 26, 389–420. [Google Scholar] [CrossRef] [PubMed]
- Grossman, W.J.; Revell, P.A.; Lu, Z.H.; Johnson, H.; Bredemeyer, A.J.; Ley, T.J. The Orphan Granzymes of Humans and Mice. Curr. Opin. Immunol. 2003, 15, 544–552. [Google Scholar] [CrossRef]
- Lieberman, J. Granzyme A Activates Another Way to Die. Immunol. Rev. 2010, 235, 93–104. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A Review of Programmed Cell Death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Esposti, M.D. The Roles of Bid. Apoptosis 2002, 7, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Zhao, T.; Zhang, H.; Lu, H.; Zhang, Q.; Sun, L.; Fan, Z. Granzyme H Induces Apoptosis of Target Tumor Cells Characterized by DNA Fragmentation and Bid-Dependent Mitochondrial Damage. Mol. Immunol. 2008, 45, 1044–1055. [Google Scholar] [CrossRef] [PubMed]
- Fellows, E.; Gil-Parrado, S.; Jenne, D.E.; Kurschus, F.C. Natural Killer Cell–Derived Human Granzyme H Induces an Alternative, Caspase-Independent Cell-Death Program. Blood 2007, 110, 544–552. [Google Scholar] [CrossRef]
- de Poot, S.A.H.; Bovenschen, N. Granzyme M: Behind Enemy Lines. Cell Death Differ. 2014, 21, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Bardine, C.; Lourenço, A.L.; Wang, Y.; Huang, Y.; Cleary, S.J.; Wilson, D.M.; Oh, D.Y.; Fong, L.; Looney, M.R.; et al. In Vivo Measurement of Granzyme Proteolysis from Activated Immune Cells with PET. ACS Cent. Sci. 2021, 7, 1638–1649. [Google Scholar] [CrossRef] [PubMed]
- Buzza, M.S.; Zamurs, L.; Sun, J.; Bird, C.H.; Smith, A.I.; Trapani, J.A.; Froelich, C.J.; Nice, E.C.; Bird, P.I. Extracellular Matrix Remodeling by Human Granzyme B via Cleavage of Vitronectin, Fibronectin, and Laminin. J. Biol. Chem. 2005, 280, 23549–23558. [Google Scholar] [CrossRef] [PubMed]
- Irmler, M.; Hertig, S.; MacDonald, H.R.; Sadoul, R.; Becherer, J.D.; Proudfoot, A.; Solari, R.; Tschopp, J. Granzyme A Is an Interleukin 1 Beta-Converting Enzyme. J. Exp. Med. 1995, 181, 1917–1922. [Google Scholar] [CrossRef] [PubMed]
- Bade, B.; Boettcher, H.E.; Lohrmann, J.; Hink-Schauer, C.; Bratke, K.; Jenne, D.E.; Virchow, J.C.; Luttmann, W. Differential Expression of the Granzymes A, K and M and Perforin in Human Peripheral Blood Lymphocytes. Int. Immunol. 2005, 17, 1419–1428. [Google Scholar] [CrossRef]
- Bratke, K.; Kuepper, M.; Bade, B.; Virchow, J.C.; Luttmann, W. Differential Expression of Human Granzymes A, B, and K in Natural Killer Cells and during CD8 + T Cell Differentiation in Peripheral Blood. Eur. J. Immunol. 2005, 35, 2608–2616. [Google Scholar] [CrossRef]
- Krensky, A.M.; Clayberger, C. Biology and Clinical Relevance of Granulysin. Tissue Antigens 2009, 73, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Aporta, A.; Catalán, E.; Galán-Malo, P.; Ramírez-Labrada, A.; Pérez, M.; Azaceta, G.; Palomera, L.; Naval, J.; Marzo, I.; Pardo, J.; et al. Granulysin Induces Apoptotic Cell Death and Cleavage of the Autophagy Regulator Atg5 in Human Hematological Tumors. Biochem. Pharmacol. 2014, 87, 410–423. [Google Scholar] [CrossRef] [PubMed]
- Clayberger, C.; Krensky, A.M. Granulysin. Curr. Opin. Immunol. 2003, 15, 560–565. [Google Scholar] [CrossRef]
- Deng, A.; Chen, S.; Li, Q.; Lyu, S.; Clayberger, C.; Krensky, A.M. Granulysin, a Cytolytic Molecule, Is Also a Chemoattractant and Proinflammatory Activator. J. Immunol. 2005, 174, 5243–5248. [Google Scholar] [CrossRef] [PubMed]
- Cohnen, A.; Chiang, S.C.; Stojanovic, A.; Schmidt, H.; Claus, M.; Saftig, P.; Janßen, O.; Cerwenka, A.; Bryceson, Y.T.; Watzl, C. Surface CD107a/LAMP-1 Protects Natural Killer Cells from Degranulation-Associated Damage. Blood 2013, 122, 1411–1418. [Google Scholar] [CrossRef] [PubMed]
- Jamali, A.; Hadjati, J.; Madjd, Z.; Mirzaei, H.R.; Thalheimer, F.B.; Agarwal, S.; Bonig, H.; Ullrich, E.; Hartmann, J. Highly Efficient Generation of Transgenically Augmented CAR NK Cells Overexpressing CXCR4. Front. Immunol. 2020, 11, 2028. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.A.; Fehniger, T.A.; Caligiuri, M.A. The Biology of Human Natural Killer-Cell Subsets. Trends Immunol. 2001, 22, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Fauriat, C.; Long, E.O.; Ljunggren, H.-G.; Bryceson, Y.T. Regulation of Human NK-Cell Cytokine and Chemokine Production by Target Cell Recognition. Blood 2010, 115, 2167–2176. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Lal, G. The Molecular Mechanism of Natural Killer Cells Function and Its Importance in Cancer Immunotherapy. Front. Immunol. 2017, 8, 1124. [Google Scholar] [CrossRef]
- Yu, M.; Luo, H.; Fan, M.; Wu, X.; Shi, B.; Di, S.; Liu, Y.; Pan, Z.; Jiang, H.; Li, Z. Development of GPC3-Specific Chimeric Antigen Receptor-Engineered Natural Killer Cells for the Treatment of Hepatocellular Carcinoma. Mol. Ther. 2018, 26, 366–378. [Google Scholar] [CrossRef]
- Müller, S.; Bexte, T.; Gebel, V.; Kalensee, F.; Stolzenberg, E.; Hartmann, J.; Koehl, U.; Schambach, A.; Wels, W.S.; Modlich, U.; et al. High Cytotoxic Efficiency of Lentivirally and Alpharetrovirally Engineered CD19-Specific Chimeric Antigen Receptor Natural Killer Cells Against Acute Lymphoblastic Leukemia. Front. Immunol. 2020, 10, 3123. [Google Scholar] [CrossRef]
- Chiossone, L.; Dumas, P.-Y.; Vienne, M.; Vivier, E. Natural Killer Cells and Other Innate Lymphoid Cells in Cancer. Nat. Rev. Immunol. 2018, 18, 671–688. [Google Scholar] [CrossRef] [PubMed]
- Guillerey, C.; Huntington, N.D.; Smyth, M.J. Targeting Natural Killer Cells in Cancer Immunotherapy. Nat. Immunol. 2016, 17, 1025–1036. [Google Scholar] [CrossRef]
- Braud, V.M.; Allan, D.S.J.; O’Callaghan, C.A.; Söderström, K.; D’Andrea, A.; Ogg, G.S.; Lazetic, S.; Young, N.T.; Bell, J.I.; Phillips, J.H.; et al. HLA-E Binds to Natural Killer Cell Receptors CD94/NKG2A, B and C. Nature 1998, 391, 795–799. [Google Scholar] [CrossRef]
- Zhuang, X.; Long, E.O. NK Cells Equipped With a Chimeric Antigen Receptor That Overcomes Inhibition by HLA Class I for Adoptive Transfer of CAR-NK Cells. Front. Immunol. 2022, 13, 840844. [Google Scholar] [CrossRef]
- Colamartino, A.B.L.; Lemieux, W.; Bifsha, P.; Nicoletti, S.; Chakravarti, N.; Sanz, J.; Roméro, H.; Selleri, S.; Béland, K.; Guiot, M.; et al. Efficient and Robust NK-Cell Transduction With Baboon Envelope Pseudotyped Lentivector. Front. Immunol. 2019, 10, 2873. [Google Scholar] [CrossRef] [PubMed]
- Wrona, E.; Borowiec, M.; Potemski, P. CAR-NK Cells in the Treatment of Solid Tumors. Int. J. Mol. Sci. 2021, 22, 5899. [Google Scholar] [CrossRef]
- Xie, G.; Dong, H.; Liang, Y.; Ham, J.D.; Rizwan, R.; Chen, J. CAR-NK Cells: A Promising Cellular Immunotherapy for Cancer. EBioMedicine 2020, 59, 102975. [Google Scholar] [CrossRef] [PubMed]
- Zamai, L.; Ahmad, M.; Bennett, I.M.; Azzoni, L.; Alnemri, E.S.; Perussia, B. Natural Killer (NK) Cell–Mediated Cytotoxicity: Differential Use of TRAIL and Fas Ligand by Immature and Mature Primary Human NK Cells. J. Exp. Med. 1998, 188, 2375–2380. [Google Scholar] [CrossRef]
- Strasser, A.; Jost, P.J.; Nagata, S. The Many Roles of FAS Receptor Signaling in the Immune System. Immunity 2009, 30, 180–192. [Google Scholar] [CrossRef]
- Prager, I.; Liesche, C.; van Ooijen, H.; Urlaub, D.; Verron, Q.; Sandström, N.; Fasbender, F.; Claus, M.; Eils, R.; Beaudouin, J.; et al. NK Cells Switch from Granzyme B to Death Receptor–Mediated Cytotoxicity during Serial Killing. J. Exp. Med. 2019, 216, 2113–2127. [Google Scholar] [CrossRef] [PubMed]
- Waring, P.; Müllbacher, A. Cell Death Induced by the Fas/Fas Ligand Pathway and Its Role in Pathology. Immunol. Cell Biol. 1999, 77, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, R.W.; Frew, A.J.; Smyth, M.J. The TRAIL Apoptotic Pathway in Cancer Onset, Progression and Therapy. Nat. Rev. Cancer 2008, 8, 782–798. [Google Scholar] [CrossRef] [PubMed]
- Bari, R.; Granzin, M.; Tsang, K.S.; Roy, A.; Krueger, W.; Orentas, R.; Schneider, D.; Pfeifer, R.; Moeker, N.; Verhoeyen, E.; et al. A Distinct Subset of Highly Proliferative and Lentiviral Vector (LV)-Transducible NK Cells Define a Readily Engineered Subset for Adoptive Cellular Therapy. Front. Immunol. 2019, 10, 2001. [Google Scholar] [CrossRef]
- Tang, S.Y.; Zha, S.; Du, Z.; Zeng, J.; Zhu, D.; Luo, Y.; Wang, S. Targeted Integration of EpCAM-Specific CAR in Human Induced Pluripotent Stem Cells and Their Differentiation into NK Cells. Stem Cell Res. Ther. 2021, 12, 580. [Google Scholar] [CrossRef] [PubMed]
- Peterson, E.E.; Barry, K.C. The Natural Killer–Dendritic Cell Immune Axis in Anti-Cancer Immunity and Immunotherapy. Front. Immunol. 2021, 11, 621254. [Google Scholar] [CrossRef] [PubMed]
- Moretta, A. Natural Killer Cells and Dendritic Cells: Rendezvous in Abused Tissues. Nat. Rev. Immunol. 2002, 2, 957–965. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Yang, M.; Zhang, W.; Liu, N.; Wang, D.; Jing, L.; Xu, N.; Yang, N.; Ren, T. Chimeric Antigen Receptor-Based Natural Killer Cell Immunotherapy in Cancer: From Bench to Bedside. Cell Death Dis. 2024, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- Land, W.G. The Role of Damage-Associated Molecular Patterns in Human Diseases: Part I—Promoting Inflammation and Immunity. Sultan Qaboos Univ. Med. J. 2015, 15, e9–e21. [Google Scholar]
- Zhou, J.; Zhang, S.; Guo, C. Crosstalk between Macrophages and Natural Killer Cells in the Tumor Microenvironment. Int. Immunopharmacol. 2021, 101, 108374. [Google Scholar] [CrossRef]
- Zhang, X.; Bai, W.; Hu, L.; Ha, H.; Du, Y.; Xiong, W.; Wang, H.; Shang, P. The Pleiotropic Mode and Molecular Mechanism of Macrophages in Promoting Tumor Progression and Metastasis. Clin. Transl. Oncol. 2022, 25, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Hadiloo, K.; Taremi, S.; Heidari, M.; Esmaeilzadeh, A. The CAR Macrophage Cells, a Novel Generation of Chimeric Antigen-Based Approach against Solid Tumors. Biomark. Res. 2023, 11, 103. [Google Scholar] [CrossRef]
- Li, M.; He, L.; Zhu, J.; Zhang, P.; Liang, S. Targeting Tumor-Associated Macrophages for Cancer Treatment. Cell Biosci. 2022, 12, 85. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Xu, J.; Lan, H. Tumor-Associated Macrophages in Tumor Metastasis: Biological Roles and Clinical Therapeutic Applications. J. Hematol. Oncol. 2019, 12, 76. [Google Scholar] [CrossRef] [PubMed]
- Strizova, Z.; Benesova, I.; Bartolini, R.; Novysedlak, R.; Cecrdlova, E.; Foley, L.K.; Striz, I. M1/M2 Macrophages and Their Overlaps—Myth or Reality? Clin. Sci. 2023, 137, 1067–1093. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xu, X.; Wei, S.; Jiang, P.; Xue, L.; Wang, J. Tumor-Associated Macrophages: Potential Therapeutic Strategies and Future Prospects in Cancer. J. Immunother. Cancer 2021, 9, e001341. [Google Scholar] [CrossRef]
- Yang, S.; Wang, Y.; Jia, J.; Fang, Y.; Yang, Y.; Yuan, W.; Hu, J. Advances in Engineered Macrophages: A New Frontier in Cancer Immunotherapy. Cell Death Dis. 2024, 15, 238. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Yu, Y.; Wang, X.; Zhang, T. Tumor-Associated Macrophages in Tumor Immunity. Front. Immunol. 2020, 11, 583084. [Google Scholar] [CrossRef]
- Bruni, D.; Angell, H.K.; Galon, J. The Immune Contexture and Immunoscore in Cancer Prognosis and Therapeutic Efficacy. Nat. Rev. Cancer 2020, 20, 662–680. [Google Scholar] [CrossRef]
- Biglari, A.; Southgate, T.D.; Fairbairn, L.J.; Gilham, D.E. Human Monocytes Expressing a CEA-Specific Chimeric CD64 Receptor Specifically Target CEA-Expressing Tumour Cells in Vitro and in Vivo. Gene Ther. 2006, 13, 602–610. [Google Scholar] [CrossRef]
- Morrissey, M.A.; Williamson, A.P.; Steinbach, A.M.; Roberts, E.W.; Kern, N.; Headley, M.B.; Vale, R.D. Chimeric Antigen Receptors That Trigger Phagocytosis. eLife 2018, 7, e36688. [Google Scholar] [CrossRef] [PubMed]
- Tridandapani, S.; Anderson, C.L. Regulation of Phagocytosis by FcγRIIb and Phosphatases. In Molecular Mechanisms of Phagocytosis; Springer: Boston, MA, USA, 2005; pp. 85–96. [Google Scholar]
- Weng, Z.; Situ, C.; Lin, L.; Wu, Z.; Zhu, J.; Zhang, R. Structure of BAI1/ELMO2 Complex Reveals an Action Mechanism of Adhesion GPCRs via ELMO Family Scaffolds. Nat. Commun. 2019, 10, 51. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, J.R.; Purcell, R.H.; Hall, R.A. The BAI Subfamily of Adhesion GPCRs: Synaptic Regulation and Beyond. Trends Pharmacol. Sci. 2014, 35, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Wanke, F.; Gutbier, S.; Rümmelin, A.; Steinberg, M.; Hughes, L.D.; Koenen, M.; Komuczki, J.; Regan-Komito, D.; Wagage, S.; Hesselmann, J.; et al. Ligand-Dependent Kinase Activity of MERTK Drives Efferocytosis in Human IPSC-Derived Macrophages. Cell Death Dis. 2021, 12, 538. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-H.; Chun, T. Anti-Inflammatory Role of TAM Family of Receptor Tyrosine Kinases Via Modulating Macrophage Function. Mol. Cells 2019, 42, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Niu, Z.; Chen, G.; Chang, W.; Sun, P.; Luo, Z.; Zhang, H.; Zhi, L.; Guo, C.; Chen, H.; Yin, M.; et al. Chimeric Antigen Receptor-modified Macrophages Trigger Systemic Anti-tumour Immunity. J. Pathol. 2021, 253, 247–257. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.J.; Bowie, A.G. The Family of Five: TIR-Domain-Containing Adaptors in Toll-like Receptor Signalling. Nat. Rev. Immunol. 2007, 7, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Lendeckel, U.; Venz, S.; Wolke, C. Macrophages: Shapes and Functions. ChemTexts 2022, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Zarubin, T.; Han, J. Activation and Signaling of the P38 MAP Kinase Pathway. Cell Res. 2005, 15, 11–18. [Google Scholar] [CrossRef]
- Himes, S.R.; Sester, D.P.; Ravasi, T.; Cronau, S.L.; Sasmono, T.; Hume, D.A. The JNK Are Important for Development and Survival of Macrophages. J. Immunol. 2006, 176, 2219–2228. [Google Scholar] [CrossRef]
- Lei, A.; Yu, H.; Lu, S.; Lu, H.; Ding, X.; Tan, T.; Zhang, H.; Zhu, M.; Tian, L.; Wang, X.; et al. A Second-Generation M1-Polarized CAR Macrophage with Antitumor Efficacy. Nat. Immunol. 2024, 25, 102–116. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.A.; Pope, C.; Wojtacha, D.; Robson, A.J.; Gordon-Walker, T.T.; Hartland, S.; Ramachandran, P.; Van Deemter, M.; Hume, D.A.; Iredale, J.P.; et al. Macrophage Therapy for Murine Liver Fibrosis Recruits Host Effector Cells Improving Fibrosis, Regeneration, and Function. Hepatology 2011, 53, 2003–2015. [Google Scholar] [CrossRef] [PubMed]
- Reiss, K.A.; Yuan, Y.; Ueno, N.T.; Johnson, M.L.; Gill, S.; Dees, E.C.; Chao, J.; Angelos, M.; Shestova, O.; Serody, J.S.; et al. A Phase 1, First-in-Human (FIH) Study of the Anti-HER2 CAR Macrophage CT-0508 in Subjects with HER2 Overexpressing Solid Tumors. J. Clin. Oncol. 2022, 40, 2533–2533. [Google Scholar] [CrossRef]
- Andreesen, R.; Hennemann, B.; Krause, S.W. Adoptive Immunotherapy of Cancer Using Monocyte-Derived Macrophages: Rationale, Current Status, and Perspectives. J. Leukoc. Biol. 1998, 64, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.M.; Edwards, J.P. Exploring the Full Spectrum of Macrophage Activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, S.; Powers, A.; Lohmueller, J.; Luketich, J.; Dhupar, R.; Soloff, A. 112 Tumor-Specific Reactivity and Effector Function of Chimeric Antigen Receptor Engineered Macrophages Targeting MUC1. J. Immunother. Cancer 2021, 9, A122–A122. [Google Scholar] [CrossRef]
- Zhang, J.; Webster, S.; Duffin, B.; Bernstein, M.N.; Steill, J.; Swanson, S.; Forsberg, M.H.; Bolin, J.; Brown, M.E.; Majumder, A.; et al. Generation of Anti-GD2 CAR Macrophages from Human Pluripotent Stem Cells for Cancer Immunotherapies. Stem Cell Rep. 2023, 18, 585–596. [Google Scholar] [CrossRef]
- Newby, A.C. Metalloproteinase Production from Macrophages—A Perfect Storm Leading to Atherosclerotic Plaque Rupture and Myocardial Infarction. Exp. Physiol. 2016, 101, 1327–1337. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, K.; Tan, X.; Li, Z.; Wang, H. Immunomodulatory Role of Metalloproteases in Cancers: Current Progress and Future Trends. Front. Immunol. 2022, 13, 1064033. [Google Scholar] [CrossRef]
- Huang, W.-C.; Sala-Newby, G.B.; Susana, A.; Johnson, J.L.; Newby, A.C. Classical Macrophage Activation Up-Regulates Several Matrix Metalloproteinases through Mitogen Activated Protein Kinases and Nuclear Factor-ΚB. PLoS ONE 2012, 7, e42507. [Google Scholar] [CrossRef]
- Kim, J.-Y.; Kim, H.; Suk, K.; Lee, W.-H. Activation of CD147 with Cyclophilin A Induces the Expression of IFITM1 through ERK and PI3K in THP-1 Cells. Mediat. Inflamm. 2010, 2010, 821940. [Google Scholar] [CrossRef] [PubMed]
- Herb, M.; Schramm, M. Functions of ROS in Macrophages and Antimicrobial Immunity. Antioxidants 2021, 10, 313. [Google Scholar] [CrossRef] [PubMed]
- Canton, M.; Sánchez-Rodríguez, R.; Spera, I.; Venegas, F.C.; Favia, M.; Viola, A.; Castegna, A. Reactive Oxygen Species in Macrophages: Sources and Targets. Front. Immunol. 2021, 12, 734229. [Google Scholar] [CrossRef] [PubMed]
- Rahat, M.A.; Hemmerlein, B. Macrophage-Tumor Cell Interactions Regulate the Function of Nitric Oxide. Front. Physiol. 2013, 4, 144. [Google Scholar] [CrossRef] [PubMed]
- Cullen, S.P.; Brunet, M.; Martin, S.J. Granzymes in Cancer and Immunity. Cell Death Differ. 2010, 17, 616–623. [Google Scholar] [CrossRef]
- Kim, W.-J.; Kim, H.; Suk, K.; Lee, W.-H. Macrophages Express Granzyme B in the Lesion Areas of Atherosclerosis and Rheumatoid Arthritis. Immunol. Lett. 2007, 111, 57–65. [Google Scholar] [CrossRef]
- van Eck, J.A.; Shan, L.; Meeldijk, J.; Hack, C.E.; Bovenschen, N. A Novel Proinflammatory Role for Granzyme A. Cell Death Dis. 2017, 8, e2630. [Google Scholar] [CrossRef] [PubMed]
- Turner, C.T.; Zeglinski, M.R.; Richardson, K.C.; Zhao, H.; Shen, Y.; Papp, A.; Bird, P.I.; Granville, D.J. Granzyme K Expressed by Classically Activated Macrophages Contributes to Inflammation and Impaired Remodeling. J. Investig. Dermatol. 2019, 139, 930–939. [Google Scholar] [CrossRef] [PubMed]
- Wensink, A.C.; Hack, C.E.; Bovenschen, N. Granzymes Regulate Proinflammatory Cytokine Responses. J. Immunol. 2015, 194, 491–497. [Google Scholar] [CrossRef]
- McCormack, R.M.; de Armas, L.R.; Shiratsuchi, M.; Fiorentino, D.G.; Olsson, M.L.; Lichtenheld, M.G.; Morales, A.; Lyapichev, K.; Gonzalez, L.E.; Strbo, N.; et al. Perforin-2 Is Essential for Intracellular Defense of Parenchymal Cells and Phagocytes against Pathogenic Bacteria. eLife 2015, 4, e06508. [Google Scholar] [CrossRef]
- Dondelinger, Y.; Darding, M.; Bertrand, M.J.M.; Walczak, H. Poly-Ubiquitination in TNFR1-Mediated Necroptosis. Cell. Mol. Life Sci. 2016, 73, 2165–2176. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lin, X. Positive and Negative Signaling Components Involved in TNFα-Induced NF-ΚB Activation. Cytokine 2008, 41, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Dhuriya, Y.K.; Sharma, D. Necroptosis: A Regulated Inflammatory Mode of Cell Death. J. Neuroinflammation 2018, 15, 199. [Google Scholar] [CrossRef] [PubMed]
- Wajant, H.; Siegmund, D. TNFR1 and TNFR2 in the Control of the Life and Death Balance of Macrophages. Front. Cell Dev. Biol. 2019, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Reza, I.; Díaz, L.; García-Becerra, R. Preclinical and Clinical Aspects of TNF-α and Its Receptors TNFR1 and TNFR2 in Breast Cancer. J. Biomed. Sci. 2017, 24, 90. [Google Scholar] [CrossRef] [PubMed]
- Montfort, A.; Colacios, C.; Levade, T.; Andrieu-Abadie, N.; Meyer, N.; Ségui, B. The TNF Paradox in Cancer Progression and Immunotherapy. Front. Immunol. 2019, 10, 1818. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Huang, X.; Li, C.; Guan, N.; Pan, T.; Dong, J.; Li, L. Effect of Tumor-Associated Macrophages on the Pyroptosis of Breast Cancer Tumor Cells. Cell Commun. Signal. 2023, 21, 197. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ye, Q.; Wang, L.; Zhou, J.; Xiang, A.; Lin, X.; Guo, J.; Hu, S.; Rui, T.; Liu, J. Targeting Pyroptosis in Breast Cancer: Biological Functions and Therapeutic Potentials on It. Cell Death Discov. 2023, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Dockrell, D.H. The Multiple Roles of Fas Ligand in the Pathogenesis of Infectious Diseases. Clin. Microbiol. Infect. 2003, 9, 766–779. [Google Scholar] [CrossRef]
- Wang, L.; Qin, X.; Liang, J.; Ge, P. Induction of Pyroptosis: A Promising Strategy for Cancer Treatment. Front. Oncol. 2021, 11, 635774. [Google Scholar] [CrossRef]
- Kang, M.; Lee, S.H.; Kwon, M.; Byun, J.; Kim, D.; Kim, C.; Koo, S.; Kwon, S.P.; Moon, S.; Jung, M.; et al. Nanocomplex-Mediated In Vivo Programming to Chimeric Antigen Receptor-M1 Macrophages for Cancer Therapy. Adv. Mater. 2021, 33, 2103258. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, D.; Iida, T.; Nakase, H. The Phagocytic Function of Macrophage-Enforcing Innate Immunity and Tissue Homeostasis. Int. J. Mol. Sci. 2017, 19, 92. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wu, M. Pattern Recognition Receptors in Health and Diseases. Signal Transduct. Target. Ther. 2021, 6, 291. [Google Scholar] [CrossRef]
- Taylor, P.R.; Martinez-Pomares, L.; Stacey, M.; Lin, H.-H.; Brown, G.D.; Gordon, S. Macrophage Receptors and Immune Recognition. Annu. Rev. Immunol. 2005, 23, 901–944. [Google Scholar] [CrossRef] [PubMed]
- Law, S.K. C3 Receptors on Macrophages. J. Cell Sci. Suppl. 1988, 9, 67–97. [Google Scholar] [CrossRef] [PubMed]
- Nimmerjahn, F.; Ravetch, J.V. Fcγ Receptors as Regulators of Immune Responses. Nat. Rev. Immunol. 2008, 8, 34–47. [Google Scholar] [CrossRef]
- Lemke, G. Biology of the TAM Receptors. Cold Spring Harb. Perspect. Biol. 2013, 5, a009076. [Google Scholar] [CrossRef] [PubMed]
- Uribe-Querol, E.; Rosales, C. Phagocytosis: Our Current Understanding of a Universal Biological Process. Front. Immunol. 2020, 11, 1066. [Google Scholar] [CrossRef]
- Dockrell, D.H.; Badley, A.D.; Villacian, J.S.; Heppelmann, C.J.; Algeciras, A.; Ziesmer, S.; Yagita, H.; Lynch, D.H.; Roche, P.C.; Leibson, P.J.; et al. The Expression of Fas Ligand by Macrophages and Its Upregulation by Human Immunodeficiency Virus Infection. J. Clin. Investig. 1998, 101, 2394–2405. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, F.; Huang, Z.; Li, Y.; Shi, H.; Sun, Q.; Ma, Y.; Wang, Y.; Zhang, Y.; Yang, S.; et al. T Cells, NK Cells, and Tumor-Associated Macrophages in Cancer Immunotherapy and the Current State of the Art of Drug Delivery Systems. Front. Immunol. 2023, 14, 1199173. [Google Scholar] [CrossRef]
- Sugita, J.; Ohtani, H.; Mizoi, T.; Saito, K.; Shiiba, K.; Sasaki, I.; Matsuno, S.; Yagita, H.; Miyazawa, M.; Nagura, H. Close Association between Fas Ligand (FasL; CD95L)-positive Tumor-associated Macrophages and Apoptotic Cancer Cells along Invasive Margin of Colorectal Carcinoma: A Proposal on Tumor-Host Interactions. Jpn. J. Cancer Res. 2002, 93, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.B.; Savill, J. Phagocytosis Triggers Macrophage Release of Fas Ligand and Induces Apoptosis of Bystander Leukocytes. J. Immunol. 1999, 162, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Kiener, P.A.; Davis, P.M.; Starling, G.C.; Mehlin, C.; Klebanoff, S.J.; Ledbetter, J.A.; Liles, W.C. Differential Induction of Apoptosis by Fas–Fas Ligand Interactions in Human Monocytes and Macrophages. J. Exp. Med. 1997, 185, 1511–1516. [Google Scholar] [CrossRef] [PubMed]
- Herbeuval, J.-P.; Lambert, C.; Sabido, O.; Cottier, M.; Fournel, P.; Dy, M.; Genin, C. Macrophages From Cancer Patients: Analysis of TRAIL, TRAIL Receptors, and Colon Tumor Cell Apoptosis. JNCI J. Natl. Cancer Inst. 2003, 95, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Liguori, M.; Buracchi, C.; Pasqualini, F.; Bergomas, F.; Pesce, S.; Sironi, M.; Grizzi, F.; Mantovani, A.; Belgiovine, C.; Allavena, P. Functional TRAIL Receptors in Monocytes and Tumor-Associated Macrophages: A Possible Targeting Pathway in the Tumor Microenvironment. Oncotarget 2106, 7, 41662–41676. [Google Scholar] [CrossRef] [PubMed]
- Muntjewerff, E.M.; Meesters, L.D.; van den Bogaart, G. Antigen Cross-Presentation by Macrophages. Front. Immunol. 2020, 11, 1276. [Google Scholar] [CrossRef] [PubMed]
- Blohm, U.; Roth, E.; Brommer, K.; Dumrese, T.; Rosenthal, F.M.; Pircher, H. Lack of Effector Cell Function and Altered Tetramer Binding of Tumor-Infiltrating Lymphocytes. J. Immunol. 2002, 169, 5522–5530. [Google Scholar] [CrossRef]
- Lyadova, I.; Vasiliev, A. Macrophages Derived from Pluripotent Stem Cells: Prospective Applications and Research Gaps. Cell Biosci. 2022, 12, 96. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Hu, Q.; Li, Y.; Lu, L.; Xiang, Z.; Yin, Z.; Kabelitz, D.; Wu, Y. Γδ T Cells: Origin and Fate, Subsets, Diseases and Immunotherapy. Signal Transduct. Target. Ther. 2023, 8, 434. [Google Scholar] [CrossRef]
- Park, J.H.; Lee, H.K. Function of Γδ T Cells in Tumor Immunology and Their Application to Cancer Therapy. Exp. Mol. Med. 2021, 53, 318–327. [Google Scholar] [CrossRef]
- Ribot, J.C.; Lopes, N.; Silva-Santos, B. Γδ T Cells in Tissue Physiology and Surveillance. Nat. Rev. Immunol. 2021, 21, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Feng, Y.; Zhou, Z. A Close Look at Current Γδ T-Cell Immunotherapy. Front. Immunol. 2023, 14, 1140623. [Google Scholar] [CrossRef] [PubMed]
- Siegers, G.M.; Lamb, L.S. Cytotoxic and Regulatory Properties of Circulating Vδ1+ Γδ T Cells: A New Player on the Cell Therapy Field? Mol. Ther. 2014, 22, 1416–1422. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy, T.; Radhakrishnan, R.; Sakshi, S.; Martin, S. CAR Γδ T Cells for Cancer Immunotherapy. Is the Field More Yellow than Green? Cancer Immunol. Immunother. 2023, 72, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Fisher, J.; Abramowski, P.; Wisidagamage Don, N.D.; Flutter, B.; Capsomidis, A.; Cheung, G.W.-K.; Gustafsson, K.; Anderson, J. Avoidance of On-Target Off-Tumor Activation Using a Co-Stimulation-Only Chimeric Antigen Receptor. Mol. Ther. 2017, 25, 1234–1247. [Google Scholar] [CrossRef] [PubMed]
- Jhita, N.; Raikar, S.S. Allogeneic Gamma Delta T Cells as Adoptive Cellular Therapy for Hematologic Malignancies. Explor. Immunol. 2022, 2, 334–350. [Google Scholar] [CrossRef] [PubMed]
- Fleischer, L.C.; Becker, S.A.; Ryan, R.E.; Fedanov, A.; Doering, C.B.; Spencer, H.T. Non-Signaling Chimeric Antigen Receptors Enhance Antigen-Directed Killing by Γδ T Cells in Contrast to Aβ T Cells. Mol. Ther. Oncolytics 2020, 18, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Li, Y.; Li, J.; ter Haak, M.; Lamb, L.S. Gamma-Delta (Γδ) CAR-T Cells Lacking the CD3z Signaling Domain Enhance Targeted Killing of Tumor Cells and Preserve Healthy Tissues. Blood 2023, 142, 6835–6835. [Google Scholar] [CrossRef]
- Du, S.-H.; Li, Z.; Chen, C.; Tan, W.-K.; Chi, Z.; Kwang, T.W.; Xu, X.-H.; Wang, S. Co-Expansion of Cytokine-Induced Killer Cells and Vγ9Vδ2 T Cells for CAR T-Cell Therapy. PLoS ONE 2016, 11, e0161820. [Google Scholar] [CrossRef]
- Moser, B.; Brandes, M. Γδ T Cells: An Alternative Type of Professional APC. Trends Immunol. 2006, 27, 112–118. [Google Scholar] [CrossRef]
- Morath, A.; Schamel, W.W. Aβ and Γδ T Cell Receptors: Similar but Different. J. Leukoc. Biol. 2020, 107, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Muro, R.; Takayanagi, H.; Nitta, T. T Cell Receptor Signaling for ΓδT Cell Development. Inflamm. Regen. 2019, 39, 6. [Google Scholar] [CrossRef] [PubMed]
- Shah, K.; Al-Haidari, A.; Sun, J.; Kazi, J.U. T Cell Receptor (TCR) Signaling in Health and Disease. Signal Transduct. Target. Ther. 2021, 6, 412. [Google Scholar] [CrossRef] [PubMed]
- Ohno, H.; Ono, S.; Hirayama, N.; Shimada, S.; Saito, T. Preferential Usage of the Fc Receptor Gamma Chain in the T Cell Antigen Receptor Complex by Gamma/Delta T Cells Localized in Epithelia. J. Exp. Med. 1994, 179, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Khattri, R.; Sperling, A.I.; Qian, D.; Fitch, F.W.; Shores, E.W.; Love, P.E.; Bluestone, J.A. TCR-Gamma Delta Cells in CD3 Zeta-Deficient Mice Contain Fc Epsilon RI Gamma in the Receptor Complex but Are Specifically Unresponsive to Antigen. J. Immunol. 1996, 157, 2320–2327. [Google Scholar] [CrossRef]
- Cipriani, B.; Knowles, H.; Chen, L.; Battistini, L.; Brosnan, C.F. Involvement of Classical and Novel Protein Kinase C Isoforms in the Response of Human Vγ9Vδ2 T Cells to Phosphate Antigens. J. Immunol. 2002, 169, 5761–5770. [Google Scholar] [CrossRef] [PubMed]
- Correia, D.V.; d’Orey, F.; Cardoso, B.A.; Lança, T.; Grosso, A.R.; deBarros, A.; Martins, L.R.; Barata, J.T.; Silva-Santos, B. Highly Active Microbial Phosphoantigen Induces Rapid yet Sustained MEK/Erk- and PI-3K/Akt-Mediated Signal Transduction in Anti-Tumor Human Γδ T-Cells. PLoS ONE 2009, 4, e5657. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, S.T.; Ribot, J.C.; Silva-Santos, B. Five Layers of Receptor Signaling in Î3Î’ T-Cell Differentiation and Activation. Front. Immunol. 2015, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Fisher, J.; Sharma, R.; Don, D.W.; Barisa, M.; Hurtado, M.O.; Abramowski, P.; Porter, L.; Day, W.; Borea, R.; Inglott, S.; et al. Engineering ΓδT Cells Limits Tonic Signaling Associated with Chimeric Antigen Receptors. Sci. Signal 2019, 12, eaax1872. [Google Scholar] [CrossRef]
- Wallet, M.A.; Nishimura, T.; Del Casale, C.; Lebid, A.; Salantes, B.; Santostefano, K.; Bucher, S.; Mendonca, M.; Beqiri, M.; Thompson, L.J.; et al. Induced Pluripotent Stem Cell-Derived Gamma Delta CAR-T Cells for Cancer Immunotherapy. Blood 2021, 138, 2771–2771. [Google Scholar] [CrossRef]
- Watanabe, D.; Koyanagi-Aoi, M.; Taniguchi-Ikeda, M.; Yoshida, Y.; Azuma, T.; Aoi, T. The Generation of Human ΓδT Cell-Derived Induced Pluripotent Stem Cells from Whole Peripheral Blood Mononuclear Cell Culture. Stem Cells Transl. Med. 2018, 7, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Themeli, M.; Kloss, C.C.; Ciriello, G.; Fedorov, V.D.; Perna, F.; Gonen, M.; Sadelain, M. Generation of Tumor-Targeted Human T Lymphocytes from Induced Pluripotent Stem Cells for Cancer Therapy. Nat. Biotechnol. 2013, 31, 928–933. [Google Scholar] [CrossRef] [PubMed]
- Berglund, S.; Gaballa, A.; Sawaisorn, P.; Sundberg, B.; Uhlin, M. Expansion of Gammadelta T Cells from Cord Blood: A Therapeutical Possibility. Stem Cells Int. 2018, 2018, 8529104. [Google Scholar] [CrossRef]
- Xiao, L.; Chen, C.; Li, Z.; Zhu, S.; Tay, J.C.; Zhang, X.; Zha, S.; Zeng, J.; Tan, W.K.; Liu, X.; et al. Large-Scale Expansion of Vγ9Vδ2 T Cells with Engineered K562 Feeder Cells in G-Rex Vessels and Their Use as Chimeric Antigen Receptor–Modified Effector Cells. Cytotherapy 2018, 20, 420–435. [Google Scholar] [CrossRef] [PubMed]
- Rozenbaum, M.; Meir, A.; Aharony, Y.; Itzhaki, O.; Schachter, J.; Bank, I.; Jacoby, E.; Besser, M.J. Gamma-Delta CAR-T Cells Show CAR-Directed and Independent Activity Against Leukemia. Front. Immunol. 2020, 11, 1347. [Google Scholar] [CrossRef] [PubMed]
- Makkouk, A.; Yang, X.; Barca, T.; Lucas, A.; Turkoz, M.; Wong, J.T.S.; Nishimoto, K.P.; Brodey, M.M.; Tabrizizad, M.; Gundurao, S.R.Y.; et al. Off-the-Shelf Vδ1 Gamma Delta T Cells Engineered with Glypican-3 (GPC-3)-Specific Chimeric Antigen Receptor (CAR) and Soluble IL-15 Display Robust Antitumor Efficacy against Hepatocellular Carcinoma. J. Immunother. Cancer 2021, 9, e003441. [Google Scholar] [CrossRef] [PubMed]
- Ang, W.X.; Ng, Y.Y.; Xiao, L.; Chen, C.; Li, Z.; Chi, Z.; Tay, J.C.-K.; Tan, W.K.; Zeng, J.; Toh, H.C.; et al. Electroporation of NKG2D RNA CAR Improves Vγ9Vδ2 T Cell Responses against Human Solid Tumor Xenografts. Mol. Ther. Oncolytics 2020, 17, 421–430. [Google Scholar] [CrossRef]
- Izumi, T.; Kondo, M.; Takahashi, T.; Fujieda, N.; Kondo, A.; Tamura, N.; Murakawa, T.; Nakajima, J.; Matsushita, H.; Kakimi, K. Ex Vivo Characterization of Γδ T-Cell Repertoire in Patients after Adoptive Transfer of Vγ9Vδ2 T Cells Expressing the Interleukin-2 Receptor β-Chain and the Common γ-Chain. Cytotherapy 2013, 15, 481–491. [Google Scholar] [CrossRef]
- Capsomidis, A.; Benthall, G.; Van Acker, H.H.; Fisher, J.; Kramer, A.M.; Abeln, Z.; Majani, Y.; Gileadi, T.; Wallace, R.; Gustafsson, K.; et al. Chimeric Antigen Receptor-Engineered Human Gamma Delta T Cells: Enhanced Cytotoxicity with Retention of Cross Presentation. Mol. Ther. 2018, 26, 354–365. [Google Scholar] [CrossRef]
- Deniger, D.C.; Switzer, K.; Mi, T.; Maiti, S.; Hurton, L.; Singh, H.; Huls, H.; Olivares, S.; Lee, D.A.; Champlin, R.E.; et al. Bispecific T-Cells Expressing Polyclonal Repertoire of Endogenous Γδ T-Cell Receptors and Introduced CD19-Specific Chimeric Antigen Receptor. Mol. Ther. 2013, 21, 638–647. [Google Scholar] [CrossRef]
- Caron, J.; Ridgley, L.A.; Bodman-Smith, M. How to Train Your Dragon: Harnessing Gamma Delta T Cells Antiviral Functions and Trained Immunity in a Pandemic Era. Front. Immunol. 2021, 12, 666983. [Google Scholar] [CrossRef] [PubMed]
- Lertworapreecha, M.; Patumraj, S.; Niruthisard, S.; Hansasuta, P.; Bhattarakosol, P. Cytotoxic Function of Gamma Delta (Gamma/Delta) T Cells against Pamidronate-Treated Cervical Cancer Cells. Indian. J. Exp. Biol. 2013, 51, 597–605. [Google Scholar] [PubMed]
- Koizumi, H.; Liu, C.C.; Zheng, L.M.; Joag, S.V.; Bayne, N.K.; Holoshitz, J.; Young, J.D. Expression of Perforin and Serine Esterases by Human Gamma/Delta T Cells. J. Exp. Med. 1991, 173, 499–502. [Google Scholar] [CrossRef] [PubMed]
- Saura-Esteller, J.; de Jong, M.; King, L.A.; Ensing, E.; Winograd, B.; de Gruijl, T.D.; Parren, P.W.H.I.; van der Vliet, H.J. Gamma Delta T-Cell Based Cancer Immunotherapy: Past-Present-Future. Front. Immunol. 2022, 13, 915837. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Zhang, Y.; Xiao, H.; Zeng, X. Engineering Γδ T Cells: Recognizing and Activating on Their Own Way. Front. Immunol. 2022, 13, 889051. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, G.; Zhang, J.; Wu, X.; Chen, X. The Dual Roles of Human Γδ T Cells: Anti-Tumor or Tumor-Promoting. Front. Immunol. 2021, 11, 619954. [Google Scholar] [CrossRef]
- Braakman, E.; van de Winkel, J.G.J.; van Krimpen, B.A.; Jansze, M.; Bolhuis, R.L.H. CD16 on Human Γδ T Lymphocytes: Expression, Function, and Specificity for Mouse IgG Isotypes. Cell Immunol. 1992, 143, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Rincon-Orozco, B.; Kunzmann, V.; Wrobel, P.; Kabelitz, D.; Steinle, A.; Herrmann, T. Activation of Vγ9Vδ2 T Cells by NKG2D. J. Immunol. 2005, 175, 2144–2151. [Google Scholar] [CrossRef] [PubMed]
- Siemaszko, J.; Marzec-Przyszlak, A.; Bogunia-Kubik, K. NKG2D Natural Killer Cell Receptor—A Short Description and Potential Clinical Applications. Cells 2021, 10, 1420. [Google Scholar] [CrossRef]
- Pinheiro, P.F.; Justino, G.C.; Marques, M.M. NKp30—A Prospective Target for New Cancer Immunotherapy Strategies. Br. J. Pharmacol. 2020, 177, 4563–4580. [Google Scholar] [CrossRef]
- Zamai, L.; Del Zotto, G.; Buccella, F.; Gabrielli, S.; Canonico, B.; Artico, M.; Ortolani, C.; Papa, S. Understanding the Synergy of NKp46 and Co-Activating Signals in Various NK Cell Subpopulations: Paving the Way for More Successful NK-Cell-Based Immunotherapy. Cells 2020, 9, 753. [Google Scholar] [CrossRef] [PubMed]
- Cifaldi, L.; Doria, M.; Cotugno, N.; Zicari, S.; Cancrini, C.; Palma, P.; Rossi, P. DNAM-1 Activating Receptor and Its Ligands: How Do Viruses Affect the NK Cell-Mediated Immune Surveillance during the Various Phases of Infection? Int. J. Mol. Sci. 2019, 20, 3715. [Google Scholar] [CrossRef] [PubMed]
- Brandes, M.; Willimann, K.; Moser, B. Professional Antigen-Presentation Function by Human Γδ T Cells. Science 2005, 309, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Dong, G.; Guo, L.; Graves, D.T. The Function of Dendritic Cells in Modulating the Host Response. Mol. Oral. Microbiol. 2018, 33, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Randolph, G.J.; Angeli, V.; Swartz, M.A. Dendritic-Cell Trafficking to Lymph Nodes through Lymphatic Vessels. Nat. Rev. Immunol. 2005, 5, 617–628. [Google Scholar] [CrossRef]
- Wculek, S.K.; Cueto, F.J.; Mujal, A.M.; Melero, I.; Krummel, M.F.; Sancho, D. Dendritic Cells in Cancer Immunology and Immunotherapy. Nat. Rev. Immunol. 2020, 20, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, A.; Salvi, V.; Soriani, A.; Laffranchi, M.; Sozio, F.; Bosisio, D.; Sozzani, S. Dendritic Cell Subsets in Cancer Immunity and Tumor Antigen Sensing. Cell Mol. Immunol. 2023, 20, 432–447. [Google Scholar] [CrossRef]
- Liu, K. Dendritic Cells. In Encyclopedia of Cell Biology; Elsevier: Amsterdam, The Netherlands, 2016; pp. 741–749. [Google Scholar]
- Constantino, J.; Gomes, C.; Falcão, A.; Cruz, M.T.; Neves, B.M. Antitumor Dendritic Cell–Based Vaccines: Lessons from 20 Years of Clinical Trials and Future Perspectives. Transl. Res. 2016, 168, 74–95. [Google Scholar] [CrossRef]
- Gardner, A.; de Mingo Pulido, Á.; Ruffell, B. Dendritic Cells and Their Role in Immunotherapy. Front. Immunol. 2020, 11, 924. [Google Scholar] [CrossRef]
- Albert, M.L.; Sauter, B.; Bhardwaj, N. Dendritic Cells Acquire Antigen from Apoptotic Cells and Induce Class I-Restricted CTLs. Nature 1998, 392, 86–89. [Google Scholar] [CrossRef]
- Yamasaki, S.; Okino, T.; Chakraborty, N.G.; Adkisson, W.O.; Sampieri, A.; Padula, S.J.; Mauri, F.; Mukherji, B. Presentation of Synthetic Peptide Antigen Encoded by the MAGE-1 Gene by Granulocyte/Macrophage-Colony-Stimulating-Factor-Cultured Macrophages from HLA-A1 Melanoma Patients. Cancer Immunol. Immunother. 1995, 40, 268–271. [Google Scholar] [CrossRef]
- Mukherji, B.; Chakraborty, N.G.; Yamasaki, S.; Okino, T.; Yamase, H.; Sporn, J.R.; Kurtzman, S.K.; Ergin, M.T.; Ozols, J.; Meehan, J. Induction of Antigen-Specific Cytolytic T Cells in Situ in Human Melanoma by Immunization with Synthetic Peptide-Pulsed Autologous Antigen Presenting Cells. Proc. Natl. Acad. Sci. USA 1995, 92, 8078–8082. [Google Scholar] [CrossRef]
- Laureano, R.S.; Sprooten, J.; Vanmeerbeerk, I.; Borras, D.M.; Govaerts, J.; Naulaerts, S.; Berneman, Z.N.; Beuselinck, B.; Bol, K.F.; Borst, J.; et al. Trial Watch: Dendritic Cell (DC)-Based Immunotherapy for Cancer. Oncoimmunology 2022, 11, 2096363. [Google Scholar] [CrossRef]
- Suh, H.C.; Pohl, K.; Javier, A.P.L.; Slamon, D.J.; Chute, J.P. Effect of Dendritic Cells (DC) Transduced with Chimeric Antigen Receptor (CAR) on CAR T-Cell Cytotoxicity. J. Clin. Oncol. 2017, 35, 144–144. [Google Scholar] [CrossRef]
- Suh, H.C.; Pohl, K.A.; Termini, C.; Kan, J.; Timmerman, J.M.; Slamon, D.J.; Chute, J.P. Bioengineered Autologous Dendritic Cells Enhance CAR T Cell Cytotoxicity By Providing Cytokine Stimulation and Intratumoral Dendritic Cells. Blood 2018, 132, 3693–3693. [Google Scholar] [CrossRef]
- Kamath, A.T.; Henri, S.; Battye, F.; Tough, D.F.; Shortman, K. Developmental Kinetics and Lifespan of Dendritic Cells in Mouse Lymphoid Organs. Blood 2002, 100, 1734–1741. [Google Scholar] [CrossRef]
- Steinman, R.M.; Banchereau, J. Taking Dendritic Cells into Medicine. Nature 2007, 449, 419–426. [Google Scholar] [CrossRef]
- Banchereau, J.; Briere, F.; Caux, C.; Davoust, J.; Lebecque, S.; Liu, Y.-J.; Pulendran, B.; Palucka, K. Immunobiology of Dendritic Cells. Annu. Rev. Immunol. 2000, 18, 767–811. [Google Scholar] [CrossRef]
- Squadrito, M.L.; Cianciaruso, C.; Hansen, S.K.; De Palma, M. EVIR: Chimeric Receptors That Enhance Dendritic Cell Cross-Dressing with Tumor Antigens. Nat. Methods 2018, 15, 183–186. [Google Scholar] [CrossRef]
- Chang, W.-H.; Cerione, R.A.; Antonyak, M.A. Extracellular Vesicles and Their Roles in Cancer Progression. Methods Mol. Biol. 2021, 2174, 143–170. [Google Scholar]
- de Saint-Vis, B.; Fugier-Vivier, I.; Massacrier, C.; Gaillard, C.; Vanbervliet, B.; Aït-Yahia, S.; Banchereau, J.; Liu, Y.-J.; Lebecque, S.; Caux, C. The Cytokine Profile Expressed by Human Dendritic Cells Is Dependent on Cell Subtype and Mode of Activation. J. Immunol. 1998, 160, 1666–1676. [Google Scholar] [CrossRef]
- Lei, X.; Khatri, I.; de Wit, T.; de Rink, I.; Nieuwland, M.; Kerkhoven, R.; van Eenennaam, H.; Sun, C.; Garg, A.D.; Borst, J.; et al. CD4+ Helper T Cells Endow CDC1 with Cancer-Impeding Functions in the Human Tumor Micro-Environment. Nat. Commun. 2023, 14, 217. [Google Scholar] [CrossRef]
- Mierzejewska, J.; Węgierek-Ciura, K.; Rossowska, J.; Szczygieł, A.; Anger-Góra, N.; Szermer-Olearnik, B.; Geneja, M.; Pajtasz-Piasecka, E. The Beneficial Effect of IL-12 and IL-18 Transduced Dendritic Cells Stimulated with Tumor Antigens on Generation of an Antitumor Response in a Mouse Colon Carcinoma Model. J. Immunol. Res. 2022, 2022, 7508928. [Google Scholar] [CrossRef]
- Ghasemi, A.; Martinez-Usatorre, A.; Li, L.; Hicham, M.; Guichard, A.; Marcone, R.; Fournier, N.; Torchia, B.; Martinez Bedoya, D.; Davanture, S.; et al. Cytokine-Armed Dendritic Cell Progenitors for Antigen-Agnostic Cancer Immunotherapy. Nat. Cancer 2023, 5, 240–261. [Google Scholar] [CrossRef]
- Sozzani, S.; Allavena, P.; Vecchi, A.; Mantovan, A. The Role of Chemokines in the Regulation of Dendritic Cell Trafficking. J. Leukoc. Biol. 1999, 66, 1–9. [Google Scholar] [CrossRef]
- Cheng, H.; Chen, W.; Lin, Y.; Zhang, J.; Song, X.; Zhang, D. Signaling Pathways Involved in the Biological Functions of Dendritic Cells and Their Implications for Disease Treatment. Mol. Biomed. 2023, 4, 15. [Google Scholar] [CrossRef]
- Engering, A.; Geijtenbeek, T.B.H.; van Vliet, S.J.; Wijers, M.; van Liempt, E.; Demaurex, N.; Lanzavecchia, A.; Fransen, J.; Figdor, C.G.; Piguet, V.; et al. The Dendritic Cell-Specific Adhesion Receptor DC-SIGN Internalizes Antigen for Presentation to T Cells. J. Immunol. 2002, 168, 2118–2126. [Google Scholar] [CrossRef]
- Bonifacino, J.S.; Dell’Angelica, E.C. Molecular Bases for the Recognition of Tyrosine-Based Sorting Signals. J. Cell Biol. 1999, 145, 923–926. [Google Scholar] [CrossRef]
- Mahnke, K.; Guo, M.; Lee, S.; Sepulveda, H.; Swain, S.L.; Nussenzweig, M.; Steinman, R.M. The Dendritic Cell Receptor for Endocytosis, Dec-205, Can Recycle and Enhance Antigen Presentation via Major Histocompatibility Complex Class II–Positive Lysosomal Compartments. J. Cell Biol. 2000, 151, 673–684. [Google Scholar] [CrossRef]
- Amigorena, S. Fcγ Receptors and Cross-Presentation in Dendritic Cells. J. Exp. Med. 2002, 195, F1–F3. [Google Scholar] [CrossRef]
- Junker, F.; Gordon, J.; Qureshi, O. Fc Gamma Receptors and Their Role in Antigen Uptake, Presentation, and T Cell Activation. Front. Immunol. 2020, 11, 1393. [Google Scholar] [CrossRef]
- Geijtenbeek, T.B.H.; Gringhuis, S.I. Signalling through C-Type Lectin Receptors: Shaping Immune Responses. Nat. Rev. Immunol. 2009, 9, 465–479. [Google Scholar] [CrossRef]
- Hemmi, H.; Akira, S. TLR Signalling and the Function of Dendritic Cells. In Mechanisms of Epithelial Defense; KARGER: Basel, Switzerland, 2005; pp. 120–135. [Google Scholar]
- Duan, T.; Du, Y.; Xing, C.; Wang, H.Y.; Wang, R.-F. Toll-Like Receptor Signaling and Its Role in Cell-Mediated Immunity. Front. Immunol. 2022, 13, 812774. [Google Scholar] [CrossRef]
- Loo, Y.-M.; Gale, M. Immune Signaling by RIG-I-like Receptors. Immunity 2011, 34, 680–692. [Google Scholar] [CrossRef]
- Ma, D.Y.; Clark, E.A. The Role of CD40 and CD154/CD40L in Dendritic Cells. Semin. Immunol. 2009, 21, 265–272. [Google Scholar] [CrossRef]
- Strohm, L.; Ubbens, H.; Münzel, T.; Daiber, A.; Daub, S. Role of CD40(L)-TRAF Signaling in Inflammation and Resolution—A Double-Edged Sword. Front. Pharmacol. 2022, 13, 995061. [Google Scholar] [CrossRef]


| Cell Type | Effector Molecules | Signaling Domain/MOTIF | Antigen Recognition/ Autologous/Allogenic | Cell Source | ||
|---|---|---|---|---|---|---|
| Soluble | Membrane-Bound | |||||
| CAR-T | Perforin-1 | FasL | CD3ζ | ITAM | MHC-dependent | PB |
| GzmA/B | TRAIL | autologous | ||||
| Granulysin | TCR | HLA-matched | ||||
| IFN-γ | ||||||
| TNF-α | ||||||
| CAR-NK | Perforin-1 | FASL | CD3ζ | ITAM | MHC-independent | PB |
| GzmA/B/H/M | TRAIL | FcRγ | ITAM | autologous | UCB | |
| Granulysin | CD16 | DAP12 | ITAM | allogenic | iPSC | |
| TNF-α | NKG2D | DAP10 | YXXM | HLA-(mis)matched | Human embryonic SC | |
| IFN-γ | NCR | BM | ||||
| CXCL8/10 | YTS | |||||
| KHYG-1 | ||||||
| CCL2/3/5 | NK92 | |||||
| CAR-M | TNF-α | FcγR | CD3ζ | ITAM | MHC-independent | PB |
| IL-1α/1β/18 | CD36 CD14 | FcRγ | ITAM | autologous | UCB | |
| allogenic | iPSC | |||||
| IL-6/12/23 | TLR-3/4/7/8/11 | Megf10 | ITAM | HLA-(mis)matched | BM | |
| THP-1 | ||||||
| CCL2/3/4/8 | NLR | MerTK | U937 | |||
| CXCL8/10 | αvβ3-Integrin | Bai1 | J774A.1 | |||
| TIR | RAW264.7 | |||||
| MMP-3/9/10/11/12/13/14 | TAM | CD147 | ||||
| CR1/3 | P13K | |||||
| SR-A | ||||||
| MARCO | ||||||
| ROS/RNS/NO | ||||||
| Additional functions | Phagocytosis | |||||
| Antigen presentation (MHC II) | ||||||
| Apoptosis, pyroptosis, necroptosis | ||||||
| TME-remodeling | ||||||
| CAR-γδ T | Perforin-1 | Vδ1 TCR | CD3ζ | ITAM | MHC-independent | PB |
| GzmB | Vδ2 TCR | DAP10 | autologous | iPSC | ||
| Granulysin | NKG2D | Truncated/NS | allogenic | |||
| IFN-γ | NKp30 | HLA-(mis)matched | ||||
| TNF-α | NKp44 | |||||
| IL-4 | DNAM-1 | |||||
| IL-10 | TLR | |||||
| CCL5 | CD16 | |||||
| CXCL10 | FasL | |||||
| TRAIL | ||||||
| Additional functions | Antigen presentation MHC I/MHC II | |||||
| CAR-DC | TNF-α | DC-SIGN | CD3ζ | ITAM | MHC-dependent | PB |
| GM-CSF | CD205 | Truncated/NS | autologous HLA-matched | |||
| M-CSF | CD206 | |||||
| TGF-β | Dectin-1 | |||||
| IL-1α/1β/18 | CLEC9A | |||||
| IL-6/7/12/15 | RLR | |||||
| CCL-2/9/18/19/22/25 | CD40 | |||||
| TLR-1/2/3/4/5/6/8 | ||||||
| CXCL8 | ||||||
| Dex | ||||||
| Additional function | Macropinocytosis, receptor-mediated endocytosis, phagocytosis | |||||
| Antigen presentation (MHC I/MHC II) | ||||||
| Cross-dressing | ||||||
| Immune modulation | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, N.T.T.; Müller, R.; Briukhovetska, D.; Weber, J.; Feucht, J.; Künkele, A.; Hudecek, M.; Kobold, S. The Spectrum of CAR Cellular Effectors: Modes of Action in Anti-Tumor Immunity. Cancers 2024, 16, 2608. https://doi.org/10.3390/cancers16142608
Nguyen NTT, Müller R, Briukhovetska D, Weber J, Feucht J, Künkele A, Hudecek M, Kobold S. The Spectrum of CAR Cellular Effectors: Modes of Action in Anti-Tumor Immunity. Cancers. 2024; 16(14):2608. https://doi.org/10.3390/cancers16142608
Chicago/Turabian StyleNguyen, Ngoc Thien Thu, Rasmus Müller, Daria Briukhovetska, Justus Weber, Judith Feucht, Annette Künkele, Michael Hudecek, and Sebastian Kobold. 2024. "The Spectrum of CAR Cellular Effectors: Modes of Action in Anti-Tumor Immunity" Cancers 16, no. 14: 2608. https://doi.org/10.3390/cancers16142608
APA StyleNguyen, N. T. T., Müller, R., Briukhovetska, D., Weber, J., Feucht, J., Künkele, A., Hudecek, M., & Kobold, S. (2024). The Spectrum of CAR Cellular Effectors: Modes of Action in Anti-Tumor Immunity. Cancers, 16(14), 2608. https://doi.org/10.3390/cancers16142608







