Defensins: Exploring Their Opposing Roles in Colorectal Cancer Progression
Abstract
:Simple Summary
Abstract
1. Introduction
2. Colorectal Cancer
3. LncRNA in CRC
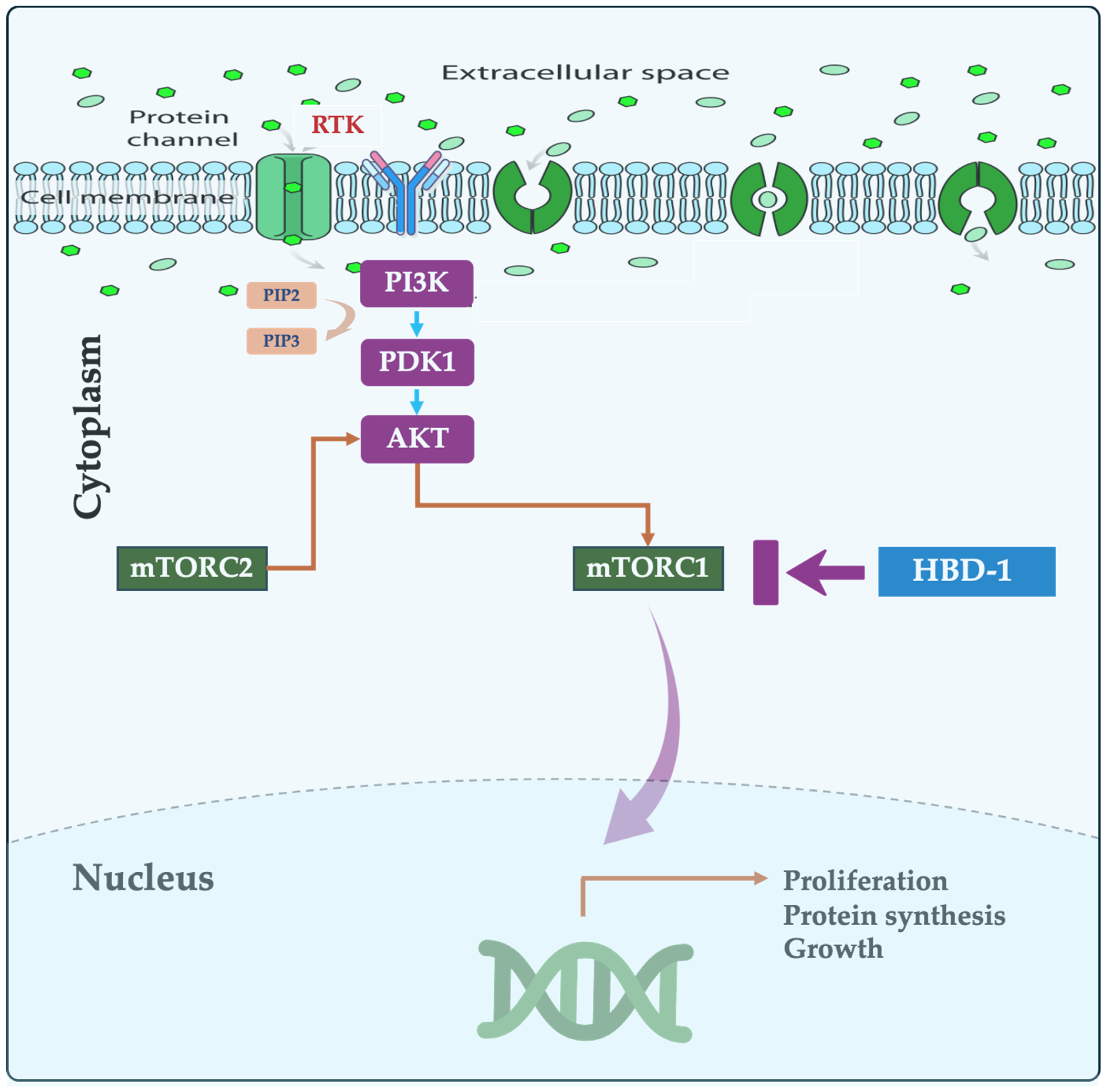
4. mTOR Pathway in CRC
5. Human β-Defensins
5.1. Structure and Function
5.2. Defensins in Cancer
5.3. β-Defensins as Proinflammatory Mediators
5.4. Defensins and the Gut Microbiota
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Eileen, M.; Melina, A.; Gini, A.; Lorenzoni, V.; Cabasag, C.J.; Mathieu, L.; Jerome, V.; Jacques, F.; Neil, M.; Freddie, B. Global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut 2023, 72, 338. [Google Scholar] [CrossRef]
- Garrett, W.S. The gut microbiota and colon cancer. Science 2019, 364, 1133–1135. [Google Scholar] [CrossRef]
- Chen, S.; Shen, X. Long noncoding RNAs: Functions and mechanisms in colon cancer. Mol. Cancer 2020, 19, 167. [Google Scholar] [CrossRef]
- Qian, J.; Cao, Y.; Zhang, J.; Li, L.; Wu, J.; Wei, G.; Yu, J.; Huo, J. Tanshinone IIA induces autophagy in colon cancer cells through MEK/ERK/mTOR pathway. Transl. Cancer Res. 2020, 9, 6919–6928. [Google Scholar] [CrossRef]
- Alzahrani, A.S. PI3K/Akt/mTOR inhibitors in cancer: At the bench and bedside. Semin. Cancer Biol. 2019, 59, 125–132. [Google Scholar] [CrossRef]
- Yu, L.; Wei, J.; Liu, P. Attacking the PI3K/Akt/mTOR signaling pathway for targeted therapeutic treatment in human cancer. Semin. Cancer Biol. 2022, 85, 69–94. [Google Scholar] [CrossRef]
- Cheng, H.; Walls, M.; Baxi, S.M.; Yin, M.J. Targeting the mTOR pathway in tumor malignancy. Curr. Cancer Drug Targets 2013, 13, 267–277. [Google Scholar] [CrossRef]
- Murugan, A.K. mTOR: Role in cancer, metastasis and drug resistance. Semin. Cancer Biol. 2019, 59, 92–111. [Google Scholar] [CrossRef]
- Droin, N.; Hendra, J.-B.; Ducoroy, P.; Solary, E. Human defensins as cancer biomarkers and antitumour molecules. J. Proteom. 2009, 72, 918–927. [Google Scholar] [CrossRef] [PubMed]
- Fruitwala, S.; El-Naccache, D.W.; Chang, T.L. Multifaceted immune functions of human defensins and underlying mechanisms. Semin. Cell Dev. Biol. 2019, 88, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Ding, J.; Liao, C.; Xu, J.; Liu, X.; Lu, W. Defensins: The natural peptide antibiotic. Adv. Drug Deliv. Rev. 2021, 179, 114008. [Google Scholar] [CrossRef]
- Zhao, X.; Lu, M.; Liu, Z.; Zhang, M.; Yuan, H.; Dan, Z.; Wang, D.; Ma, B.; Yang, Y.; Yang, F.; et al. Comprehensive analysis of alfa defensin expression and prognosis in human colorectal cancer. Front. Oncol. 2023, 12, 974654. [Google Scholar] [CrossRef]
- Wu, Z.; Ding, Z.; Cheng, B.; Cui, Z. The inhibitory effect of human DEFA5 in growth of gastric cancer by targeting BMI1. Cancer Sci. 2021, 112, 1075–1083. [Google Scholar] [CrossRef]
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 233–254. [Google Scholar] [CrossRef]
- Huang, G.; Long, K. Sensitization of colon cancer cells to cisplatin by Fbxw7 via negative regulation of the Nox1-mTOR pathway. Pathol. Res. Pract. 2023, 247, 154479. [Google Scholar] [CrossRef]
- Wei, P.L.; Lin, J.C.; Hung, C.S.; Makondi, P.T.; Batzorig, U.; Chang, T.C.; Huang, C.Y.; Chang, Y.J. Human α-defensin 6 (HD6) suppresses CRC proliferation and metastasis through abolished EGF/EGFR signaling pathway. Int. J. Med. Sci. 2022, 19, 34–46. [Google Scholar] [CrossRef]
- Panjeta, A.; Preet, S. Anticancer potential of human intestinal defensin 5 against 1, 2-dimethylhydrazine dihydrochloride induced colon cancer: A therapeutic approach. Peptides 2020, 126, 170263. [Google Scholar] [CrossRef]
- Breaux, W.A.; Bragg, M.A.; M’Koma, A.E. Ubiquitous Colonic Ileal Metaplasia Consistent with the Diagnosis of Crohn’s Colitis among Indeterminate Colitis Cohorts. Med. Res. Arch. 2023, 11, 4188. [Google Scholar] [CrossRef]
- Qiao, Q.; Bai, R.; Song, W.; Gao, H.; Zhang, M.; Lu, J.; Hong, M.; Zhang, X.; Sun, P.; Zhang, Q.; et al. Human α-defensin 5 suppressed colon cancer growth by targeting PI3K pathway. Exp. Cell Res. 2021, 407, 112809. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Song, W.; Shen, Y.; Wang, H.; Fan, Z. LncRNA KLK8 modulates stem cell characteristics in colon cancer. Pathol. Res. Pract. 2021, 224, 153437. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, L.; Wang, W.; Rao, X.; Lai, Y. Constructing a prognostic immune-related lncRNA model for colon cancer. Medicine 2022, 101, e30447. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Deng, L.; Wang, Y.D. Roles and Mechanisms of Long Non-Coding RNAs in Breast Cancer. Int. J. Mol. Sci. 2022, 24, 89. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Petinrin, O.O.; Toseef, M.; Chen, N.; Wong, K.C. Construction of Immune Infiltration-Related LncRNA Signatures Based on Machine Learning for the Prognosis in Colon Cancer. Biochem. Genet. 2023, 62, 1925–1952. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Shi, L.; Wang, B.; Wu, Z.; Zhao, H.; Zhao, T.; Shi, L. Role of oncogenic long noncoding RNA KCNQ1OT1 in colon cancer. Oncol. Res. 2024, 32, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Ren, Q.; Du, Y.; Ma, Y.; Gu, L.; Zhou, J.; Tian, W.; Deng, D. LncRNA miR663AHG represses the development of colon cancer in a miR663a-dependent manner. Cell Death Discov. 2023, 9, 220. [Google Scholar] [CrossRef] [PubMed]
- Tong, D.; Fan, L. LncRNA ZNF667-AS1 Targets miR-523-3p/KIF5C Axis to Hinder Colon Cancer Progression. Mol. Biotechnol. 2023, 66, 1464–1476. [Google Scholar] [CrossRef] [PubMed]
- Cui, K.; Wu, H.; Zhang, L.; Li, H.; Israr, G.; Li, Z. A novel lncRNA 495810 promotes the aerobic glycolysis in colorectal cancer by stabilizing pyruvate kinase isozyme M2. Int. J. Oncol. 2023, 62, 58. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.Y.; Sabatini, D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020, 21, 183–203. [Google Scholar] [CrossRef]
- Afzal, O.; Altamimi, A.S.A.; Mubeen, B.; Alzarea, S.I.; Almalki, W.H.; Al-Qahtani, S.D.; Atiya, E.M.; Al-Abbasi, F.A.; Ali, F.; Ullah, I.; et al. mTOR as a Potential Target for the Treatment of Microbial Infections, Inflammatory Bowel Diseases, and Colorectal Cancer. Int. J. Mol. Sci. 2022, 23, 12470. [Google Scholar] [CrossRef]
- Miricescu, D.; Totan, A.; Stanescu, S., II; Badoiu, S.C.; Stefani, C.; Greabu, M. PI3K/AKT/mTOR Signaling Pathway in Breast Cancer: From Molecular Landscape to Clinical Aspects. Int. J. Mol. Sci. 2020, 22, 173. [Google Scholar] [CrossRef]
- Glaviano, A.; Foo, A.S.C.; Lam, H.Y.; Yap, K.C.H.; Jacot, W.; Jones, R.H.; Eng, H.; Nair, M.G.; Makvandi, P.; Geoerger, B.; et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol. Cancer 2023, 22, 138. [Google Scholar] [CrossRef] [PubMed]
- Fattahi, S.; Amjadi-Moheb, F.; Tabaripour, R.; Ashrafi, G.H.; Akhavan-Niaki, H. PI3K/AKT/mTOR signaling in gastric cancer: Epigenetics and beyond. Life Sci. 2020, 262, 118513. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Azizian, N.G.; Sullivan, D.K.; Li, Y. mTOR inhibition attenuates chemosensitivity through the induction of chemotherapy resistant persisters. Nat. Commun. 2022, 13, 7047. [Google Scholar] [CrossRef] [PubMed]
- Huang, S. mTOR Signaling in Metabolism and Cancer. Cells 2020, 9, 2278. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.S.; Kim, J.; Ryu, Y.S.; Moon, J.H.; Shin, Y.J.; Kim, J.H.; Hong, S.W.; Jung, S.A.; Lee, S.; Kim, S.M.; et al. Mutant PIK3CA as a negative predictive biomarker for treatment with a highly selective PIM1 inhibitor in human colon cancer. Cancer Biol. Ther. 2023, 24, 2246208. [Google Scholar] [CrossRef] [PubMed]
- Rajabi, M.; Ericksen, B.; Wu, X.; de Leeuw, E.; Zhao, L.; Pazgier, M.; Lu, W. Functional determinants of human enteric α-defensin HD5: Crucial role for hydrophobicity at dimer interface. J. Biol. Chem. 2012, 287, 21615–21627. [Google Scholar] [CrossRef] [PubMed]
- Selsted, M.E.; Szklarek, D.; Lehrer, R.I. Purification and antibacterial activity of antimicrobial peptides of rabbit granulocytes. Infect Immun. 1984, 45, 150–154. [Google Scholar] [CrossRef]
- Pazgier, M.; Wei, G.; Ericksen, B.; Jung, G.; Wu, Z.; de Leeuw, E.; Yuan, W.; Szmacinski, H.; Lu, W.Y.; Lubkowski, J.; et al. Sometimes it takes two to tango: Contributions of dimerization to functions of human α-defensin HNP1 peptide. J. Biol. Chem. 2012, 287, 8944–8953. [Google Scholar] [CrossRef]
- Zhao, L.; Ericksen, B.; Wu, X.; Zhan, C.; Yuan, W.; Li, X.; Pazgier, M.; Lu, W. Invariant gly residue is important for α-defensin folding, dimerization, and function: A case study of the human neutrophil α-defensin HNP1. J. Biol. Chem. 2012, 287, 18900–18912. [Google Scholar] [CrossRef]
- Fischer, A.; Wannemacher, J.; Christ, S.; Koopmans, T.; Kadri, S.; Zhao, J.; Gouda, M.; Ye, H.; Mück-Häusl, M.; Krenn, P.W.; et al. Neutrophils direct preexisting matrix to initiate repair in damaged tissues. Nat. Immunol. 2022, 23, 518–531. [Google Scholar] [CrossRef]
- Chen, J.; Zhai, Z.; Long, H.; Yang, G.; Deng, B.; Deng, J. Inducible expression of defensins and cathelicidins by nutrients and associated regulatory mechanisms. Peptides 2020, 123, 170177. [Google Scholar] [CrossRef] [PubMed]
- Harwig, S.S.; Park, A.S.; Lehrer, R.I. Characterization of defensin precursors in mature human neutrophils. Blood 1992, 79, 1532–1537. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.E.; Bevins, C.L. Defensin-6 mRNA in human Paneth cells: Implications for antimicrobial peptides in host defense of the human bowel. FEBS Lett 1993, 315, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003, 3, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Lynn, D.J.; Higgs, R.; Gaines, S.; Tierney, J.; James, T.; Lloyd, A.T.; Fares, M.A.; Mulcahy, G.; O’Farrelly, C. Bioinformatic discovery and initial characterisation of nine novel antimicrobial peptide genes in the chicken. Immunogenetics 2004, 56, 170–177. [Google Scholar] [CrossRef]
- Choi, M.K.; Le, M.T.; Nguyen, D.T.; Choi, H.; Kim, W.; Kim, J.H.; Chun, J.; Hyeon, J.; Seo, K.; Park, C. Genome-level identification, gene expression, and comparative analysis of porcine ß-defensin genes. BMC Genet. 2012, 13, 98. [Google Scholar] [CrossRef] [PubMed]
- Meade, K.G.; Cormican, P.; Narciandi, F.; Lloyd, A.; O’Farrelly, C. Bovine β-defensin gene family: Opportunities to improve animal health? Physiol. Genom. 2014, 46, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Bonamy, C.; Sechet, E.; Amiot, A.; Alam, A.; Mourez, M.; Fraisse, L.; Sansonetti, P.J.; Sperandio, B. Expression of the human antimicrobial peptide β-defensin-1 is repressed by the EGFR-ERK-MYC axis in colonic epithelial cells. Sci. Rep. 2018, 8, 18043. [Google Scholar] [CrossRef] [PubMed]
- Adyns, L.; Proost, P.; Struyf, S. Role of Defensins in Tumor Biology. Int. J. Mol. Sci. 2023, 24, 5268. [Google Scholar] [CrossRef]
- Fijałkowska, M.; Bonczar, M.; Jastrzębski, I.; Ostrowski, P.; Antoszewski, B.; Koziej, M. The role of TGF-β and antimicrobial peptides in basal cell carcinoma: A systematic review. Postep. Dermatol. Alergol. 2023, 40, 384–389. [Google Scholar] [CrossRef]
- Koeninger, L.; Armbruster, N.S.; Brinch, K.S.; Kjaerulf, S.; Andersen, B.; Langnau, C.; Autenrieth, S.E.; Schneidawind, D.; Stange, E.F.; Malek, N.P.; et al. Human β-Defensin 2 Mediated Immune Modulation as Treatment for Experimental Colitis. Front. Immunol. 2020, 11, 93. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Chauhan, A.; Singh, K.; Kumar, K.; Kaur, R.; Masih, M.; Gautam, P.K. Immunomodulatory effects of β-defensin 2 on macrophages induced immuno-upregulation and their antitumor function in breast cancer. BMC Immunol. 2022, 23, 53. [Google Scholar] [CrossRef] [PubMed]
- Parvy, J.-P.; Yu, Y.; Dostalova, A.; Kondo, S.; Kurjan, A.; Bulet, P.; Lemaître, B.; Vidal, M.; Cordero, J.B. The antimicrobial peptide defensin cooperates with tumour necrosis factor to drive tumour cell death in Drosophila. eLife 2019, 8, e45061. [Google Scholar] [CrossRef]
- Hein, M.J.A.; Kvansakul, M.; Lay, F.T.; Phan, T.K.; Hulett, M.D. Defensin-lipid interactions in membrane targeting: Mechanisms of action and opportunities for the development of antimicrobial and anticancer therapeutics. Biochem. Soc. Trans. 2022, 50, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.Q.; Arnold, R.S.; Hsieh, C.L.; Dorin, J.R.; Lian, F.; Li, Z.; Petros, J.A. Discovery and mechanisms of host defense to oncogenesis: Targeting the β-defensin-1 peptide as a natural tumor inhibitor. Cancer Biol. Ther. 2019, 20, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Scola, N.; Gambichler, T.; Saklaoui, H.; Bechara, F.G.; Georgas, D.; Stücker, M.; Gläser, R.; Kreuter, A. The expression of antimicrobial peptides is significantly altered in cutaneous squamous cell carcinoma and precursor lesions. Br. J. Dermatol. 2012, 167, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Wang, R.; Sun, C.; Jin, X.; Liu, D.; Zhao, X.; Wang, L.; Ji, N.; Li, J.; Zhou, Y.; et al. Human Beta-Defensin-1 Suppresses Tumor Migration and Invasion and Is an Independent Predictor for Survival of Oral Squamous Cell Carcinoma Patients. PLoS ONE 2014, 9, e91867. [Google Scholar] [CrossRef]
- Li, X.; Song, W.; Zhang, M.; Zhao, P. Human β-defensin 1 Functions as a Tumor Suppressor via ER Stress-triggered JNK pathway in Hepatocellular Carcinoma. J. BUON 2021, 26, 1365–1372. [Google Scholar]
- Semple, F.; Dorin, J.R. β-Defensins: Multifunctional modulators of infection, inflammation and more? J. Innate Immun. 2012, 4, 337–348. [Google Scholar] [CrossRef]
- Ghosh, S.K.; McCormick, T.S.; Weinberg, A. Human Beta Defensins and Cancer: Contradictions and Common Ground. Front. Oncol. 2019, 9, 341. [Google Scholar] [CrossRef]
- Ling, Y.M.; Chen, J.Y.; Guo, L.; Wang, C.Y.; Tan, W.T.; Wen, Q.; Zhang, S.D.; Deng, G.H.; Lin, Y.; Kwok, H.F. β-defensin 1 expression in HCV infected liver/liver cancer: An important role in protecting HCV progression and liver cancer development. Sci. Rep. 2017, 7, 13404. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Zhang, B.; Liao, C.; Zhang, W.; Wang, W.; Chang, Y.; Shao, Y. Human beta-defensin 3 contributes to the carcinogenesis of cervical cancer via activation of NF-κB signaling. Oncotarget 2016, 7, 75902–75913. [Google Scholar] [CrossRef] [PubMed]
- DasGupta, T.; Nweze, E.I.; Yue, H.; Wang, L.; Jin, J.; Ghosh, S.K.; Kawsar, H.I.; Zender, C.; Androphy, E.J.; Weinberg, A.; et al. Human papillomavirus oncogenic E6 protein regulates human β-defensin 3 (hBD3) expression via the tumor suppressor protein p53. Oncotarget 2016, 7, 27430–27444. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.A.; Kim, K.H.; Lee, T.J.; Park, E.S.; Kim, M.K.; Myung, S.C. A role of human beta defensin-1 in predicting prostatic adenocarcinoma in cases of false-negative biopsy. Apmis 2017, 125, 1063–1069. [Google Scholar] [CrossRef] [PubMed]
- Bevins, C.L. Innate immune functions of α-defensins in the small intestine. Dig. Dis. 2013, 31, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Weinberg, A. Human antimicrobial peptides and cancer. Semin. Cell Dev. Biol. 2019, 88, 156–162. [Google Scholar] [CrossRef]
- Phan, T.K.; Lay, F.T.; Poon, I.K.; Hinds, M.G.; Kvansakul, M.; Hulett, M.D. Human β-defensin 3 contains an oncolytic motif that binds PI(4,5)P2 to mediate tumour cell permeabilisation. Oncotarget 2016, 7, 2054–2069. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Yang, Y.; Zhang, W.; Yang, C.; Xu, P. Human β-defensin-3 and nuclear factor-kappa B p65 synergistically promote the cell proliferation and invasion of oral squamous cell carcinoma. Transl. Oncol. 2023, 27, 101582. [Google Scholar] [CrossRef] [PubMed]
- Joly, S.; Compton, L.M.; Pujol, C.; Kurago, Z.B.; Guthmiller, J.M. Loss of human beta-defensin 1, 2, and 3 expression in oral squamous cell carcinoma. Oral Microbiol. Immunol. 2009, 24, 353–360. [Google Scholar] [CrossRef]
- Arimura, Y.; Ashitani, J.; Yanagi, S.; Tokojima, M.; Abe, K.; Mukae, H.; Nakazato, M. Elevated serum beta-defensins concentrations in patients with lung cancer. Anticancer Res. 2004, 24, 4051–4057. [Google Scholar]
- Gambichler, T.; Skrygan, M.; Huyn, J.; Bechara, F.G.; Sand, M.; Altmeyer, P.; Kreuter, A. Pattern of mRNA expression of beta-defensins in basal cell carcinoma. BMC Cancer 2006, 6, 163. [Google Scholar] [CrossRef]
- Shi, N.; Jin, F.; Zhang, X.; Clinton, S.K.; Pan, Z.; Chen, T. Overexpression of human β-defensin 2 promotes growth and invasion during esophageal carcinogenesis. Oncotarget 2014, 5, 11333–11344. [Google Scholar] [CrossRef]
- Donald, C.D.; Sun, C.Q.; Lim, S.D.; Macoska, J.; Cohen, C.; Amin, M.B.; Young, A.N.; Ganz, T.A.; Marshall, F.F.; Petros, J.A. Cancer-specific loss of beta-defensin 1 in renal and prostatic carcinomas. Lab. Investig. 2003, 83, 501–505. [Google Scholar] [CrossRef]
- Semlali, A.; Al Amri, A.; Azzi, A.; Al Shahrani, O.; Arafah, M.; Kohailan, M.; Aljebreen, A.M.; Alharbi, O.; Almadi, M.A.; Azzam, N.A.; et al. Expression and new exon mutations of the human Beta defensins and their association on colon cancer development. PLoS ONE 2015, 10, e0126868. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.E.; Harder, J.; Görögh, T.; Weise, J.B.; Schubert, S.; Janssen, D.; Maune, S. Human beta-defensin-2 in oral cancer with opportunistic Candida infection. Anticancer Res. 2004, 24, 1025–1030. [Google Scholar]
- Ye, Z.; Dong, H.; Li, Y.; Ma, T.; Huang, H.; Leong, H.S.; Eckel-Passow, J.; Kocher, J.A.; Liang, H.; Wang, L. Prevalent Homozygous Deletions of Type I Interferon and Defensin Genes in Human Cancers Associate with Immunotherapy Resistance. Clin. Cancer Res. 2018, 24, 3299–3308. [Google Scholar] [CrossRef]
- Berghmans, E.; Jacobs, J.; Deben, C.; Hermans, C.; Broeckx, G.; Smits, E.; Maes, E.; Raskin, J.; Pauwels, P.; Baggerman, G. Mass Spectrometry Imaging Reveals Neutrophil Defensins as Additional Biomarkers for Anti-PD-(L)1 Immunotherapy Response in NSCLC Patients. Cancers 2020, 12, 863. [Google Scholar] [CrossRef] [PubMed]
- Contreras, G.; Shirdel, I.; Braun, M.S.; Wink, M. Defensins: Transcriptional regulation and function beyond antimicrobial activity. Dev. Comp. Immunol. 2020, 104, 103556. [Google Scholar] [CrossRef]
- Zhu, S.; Gao, B. Evolutionary origin of β-defensins. Dev. Comp. Immunol. 2013, 39, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Zong, X.; Jin, M.; Min, J.; Wang, F.; Wang, Y. Mechanisms and regulation of defensins in host defense. Signal Transduct. Target Ther. 2023, 8, 300. [Google Scholar] [CrossRef] [PubMed]
- Oppenheim, J.J.; Biragyn, A.; Kwak, L.W.; Yang, D. Roles of antimicrobial peptides such as defensins in innate and adaptive immunity. Ann. Rheum. Dis. 2003, 62 (Suppl. S2), ii17–ii21. [Google Scholar] [CrossRef]
- Chen, Q.; Yang, Y.; Pan, Y.; Shen, L.; Zhang, Y.; Zheng, F.; Shu, Q.; Fang, X. Human Neutrophil Defensins Disrupt Liver Interendothelial Junctions and Aggravate Sepsis. Mediat. Inflamm. 2022, 2022, 7659282. [Google Scholar] [CrossRef]
- Liang, S.; Guo, X.K.; Ou, J.; Huang, R.; Xue, Q.; Zhang, B.; Chung, Y.; Wu, W.; Dong, C.; Yang, X.; et al. Nutrient Sensing by the Intestinal Epithelium Orchestrates Mucosal Antimicrobial Defense via Translational Control of Hes1. Cell Host Microbe 2019, 25, 706–718.e707. [Google Scholar] [CrossRef]
- Xu, D.; Lu, W. Defensins: A Double-Edged Sword in Host Immunity. Front. Immunol. 2020, 11, 764. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Lu, W. Defensins in innate immunity. Curr. Opin. Hematol. 2014, 21, 37–42. [Google Scholar] [CrossRef]
- Biragyn, A.; Ruffini, P.A.; Leifer, C.A.; Klyushnenkova, E.; Shakhov, A.; Chertov, O.; Shirakawa, A.K.; Farber, J.M.; Segal, D.M.; Oppenheim, J.J.; et al. Toll-like receptor 4-dependent activation of dendritic cells by beta-defensin 2. Science 2002, 298, 1025–1029. [Google Scholar] [CrossRef] [PubMed]
- Funderburg, N.T.; Jadlowsky, J.K.; Lederman, M.M.; Feng, Z.; Weinberg, A.; Sieg, S.F. The Toll-like receptor 1/2 agonists Pam(3) CSK(4) and human β-defensin-3 differentially induce interleukin-10 and nuclear factor-κB signalling patterns in human monocytes. Immunology 2011, 134, 151–160. [Google Scholar] [CrossRef]
- Németh, B.C.; Várkonyi, T.; Somogyvári, F.; Lengyel, C.; Fehértemplomi, K.; Nyiraty, S.; Kempler, P.; Mándi, Y. Relevance of α-defensins (HNP1-3) and defensin β-1 in diabetes. World J Gastroenterol. 2014, 20, 9128–9137. [Google Scholar] [CrossRef]
- Antikainen, A.A.V.; Sandholm, N.; Trégouët, D.A.; Charmet, R.; McKnight, A.J.; Ahluwalia, T.S.; Syreeni, A.; Valo, E.; Forsblom, C.; Gordin, D.; et al. Genome-wide association study on coronary artery disease in type 1 diabetes suggests beta-defensin 127 as a risk locus. Cardiovasc. Res. 2021, 117, 600–612. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef]
- Clapp, M.; Aurora, N.; Herrera, L.; Bhatia, M.; Wilen, E.; Wakefield, S. Gut microbiota’s effect on mental health: The gut-brain axis. Clin. Pract. 2017, 7, 987. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Miani, M.; Le Naour, J.; Waeckel-Enée, E.; Verma, S.C.; Straube, M.; Emond, P.; Ryffel, B.; van Endert, P.; Sokol, H.; Diana, J. Gut Microbiota-Stimulated Innate Lymphoid Cells Support β-Defensin 14 Expression in Pancreatic Endocrine Cells, Preventing Autoimmune Diabetes. Cell Metab. 2018, 28, 557–572.e556. [Google Scholar] [CrossRef] [PubMed]
- Saqib, Z.; De Palma, G.; Lu, J.; Surette, M.; Bercik, P.; Collins, S.M. Alterations in fecal β-defensin-3 secretion as a marker of instability of the gut microbiota. Gut Microbes 2023, 15, 2233679. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Nakamura, K.; Yokoi, Y.; Shimizu, Y.; Ohira, S.; Hagiwara, M.; Song, Z.; Gan, L.; Aizawa, T.; Hashimoto, D.; et al. Decreased Paneth cell α-defensins promote fibrosis in a choline-deficient L-amino acid-defined high-fat diet-induced mouse model of nonalcoholic steatohepatitis via disrupting intestinal microbiota. Sci. Rep. 2023, 13, 3953. [Google Scholar] [CrossRef] [PubMed]
- Shukla, P.K.; Rao, R.G.; Meena, A.S.; Giorgianni, F.; Lee, S.C.; Raju, P.; Shashikanth, N.; Shekhar, C.; Beranova, S.; Balazs, L.; et al. Paneth cell dysfunction in radiation injury and radio-mitigation by human α-defensin 5. Front. Immunol. 2023, 14, 1174140. [Google Scholar] [CrossRef]
- Sugi, Y.; Takahashi, K.; Kurihara, K.; Nakano, K.; Kobayakawa, T.; Nakata, K.; Tsuda, M.; Hanazawa, S.; Hosono, A.; Kaminogawa, S. α-Defensin 5 gene expression is regulated by gut microbial metabolites. Biosci. Biotechnol. Biochem. 2017, 81, 242–248. [Google Scholar] [CrossRef]
- Shimizu, Y.; Nakamura, K.; Kikuchi, M.; Ukawa, S.; Nakamura, K.; Okada, E.; Imae, A.; Nakagawa, T.; Yamamura, R.; Tamakoshi, A.; et al. Lower human defensin 5 in elderly people compared to middle-aged is associated with differences in the intestinal microbiota composition: The DOSANCO Health Study. Geroscience 2022, 44, 997–1009. [Google Scholar] [CrossRef]
- Puértolas-Balint, F.; Schroeder, B.O. Intestinal α-Defensins Play a Minor Role in Modulating the Small Intestinal Microbiota Composition as Compared to Diet. Microbiol. Spectr. 2023, 11, e0056723. [Google Scholar] [CrossRef]

| Cancer Type | HBD-1 | HBD-2 | HBD-3 | Source | Cases/Control | Ref. |
|---|---|---|---|---|---|---|
| Cervical | -- | Upregulated | Upregulated | Patients | 37/22 | [62] |
| Oral | Downregulated | Downregulated | Upregulated | Cell lines | 16/15 | [69] |
| Lung | Upregulated | Upregulated | -- | Patients | 56/46 | [70] |
| Skin | Downregulated | Upregulated | -- | Patients | 22/27 | [71] |
| Esophagus | -- | Upregulated | -- | Patients | 58 | [72] |
| Liver | Downregulated | -- | -- | Patients | 733/656 | [61] |
| Kidney | Downregulated | -- | -- | Patients | 48 | [73] |
| Colon | Downregulated | Downregulated | Downregulated | Patients | 40 | [74] |
| Prostate | -- | -- | -- | Patients | 100 | [73] |
| Tonsil | -- | Downregulated | -- | Patients | 8/8 | [75] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabit, H.; Pawlik, T.M.; Abdel-Ghany, S.; Arneth, B. Defensins: Exploring Their Opposing Roles in Colorectal Cancer Progression. Cancers 2024, 16, 2622. https://doi.org/10.3390/cancers16152622
Sabit H, Pawlik TM, Abdel-Ghany S, Arneth B. Defensins: Exploring Their Opposing Roles in Colorectal Cancer Progression. Cancers. 2024; 16(15):2622. https://doi.org/10.3390/cancers16152622
Chicago/Turabian StyleSabit, Hussein, Timothy M. Pawlik, Shaimaa Abdel-Ghany, and Borros Arneth. 2024. "Defensins: Exploring Their Opposing Roles in Colorectal Cancer Progression" Cancers 16, no. 15: 2622. https://doi.org/10.3390/cancers16152622






