Therapeutic Antisense Oligonucleotides in Oncology: From Bench to Bedside
Simple Summary
Abstract
1. Introduction
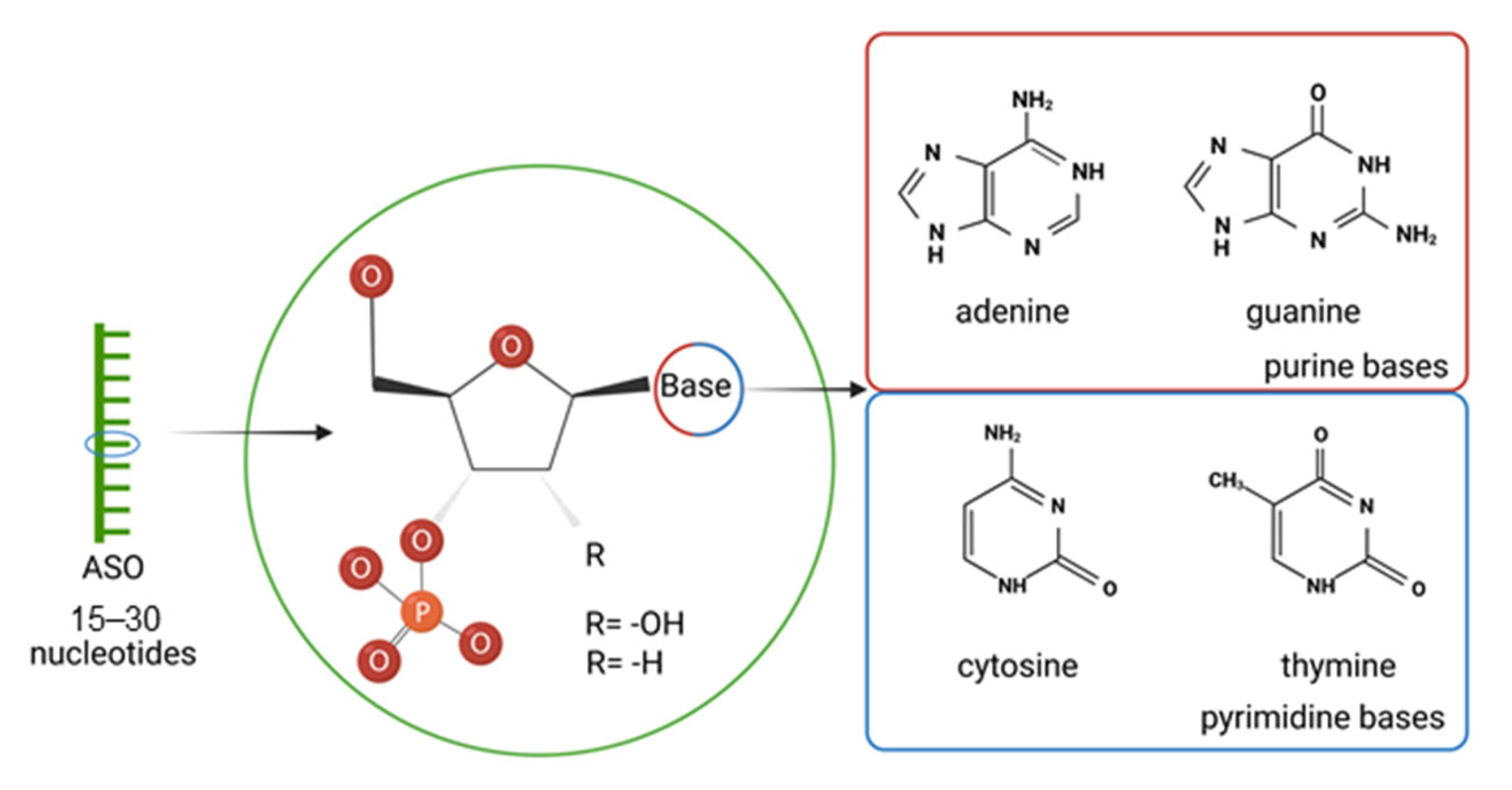
2. Antisense Oligonucleotides (ASOs)
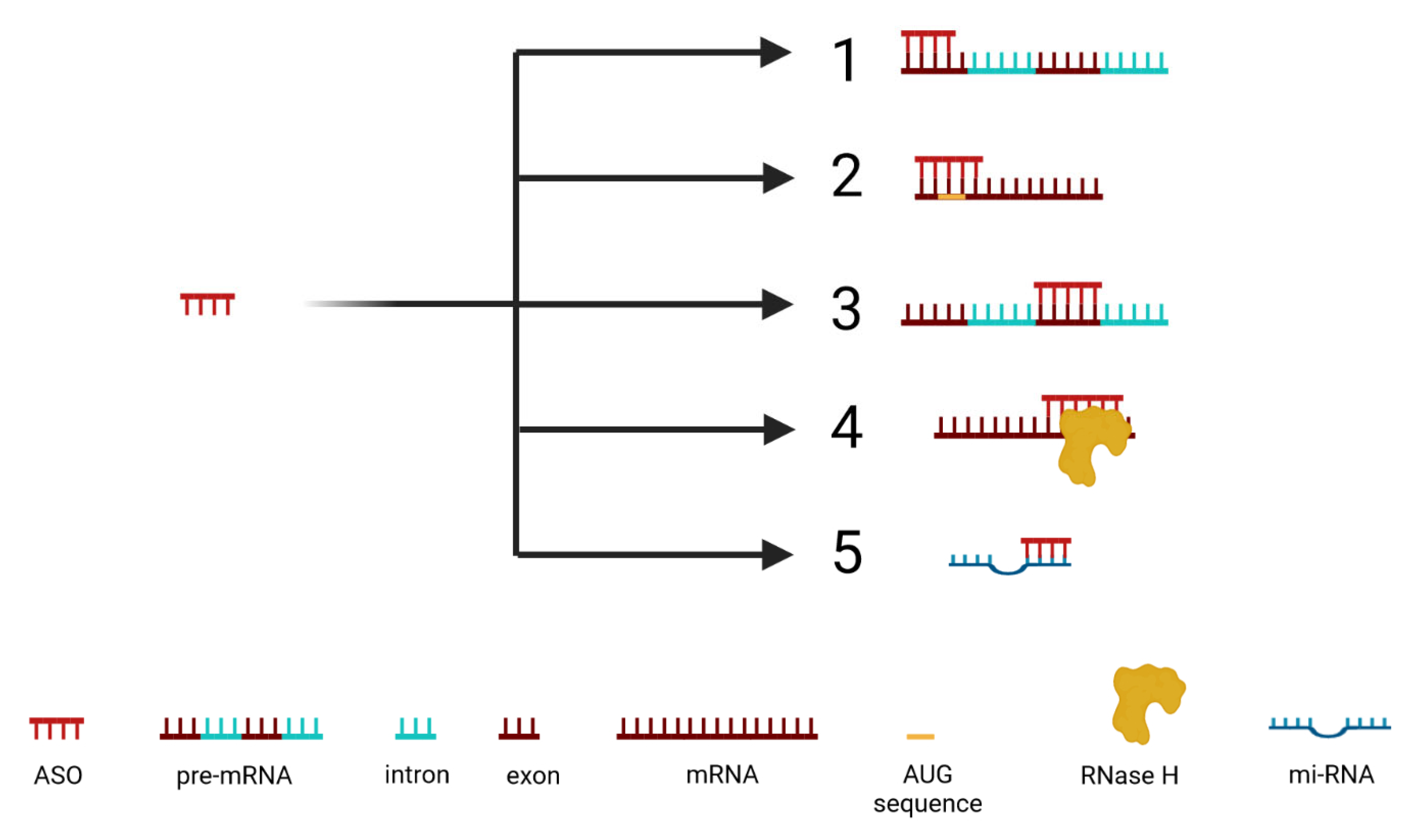
2.1. Mechanism of ASO Activity
2.1.1. Inhibition of 5′ Cap Formation
2.1.2. Steric Blocking of Translation
2.1.3. Alteration of Splicing
2.1.4. Activation of RNase H
2.1.5. Inhibition of miRNA Activity (antimiRs)
2.2. Methods of Delivery of ASOs
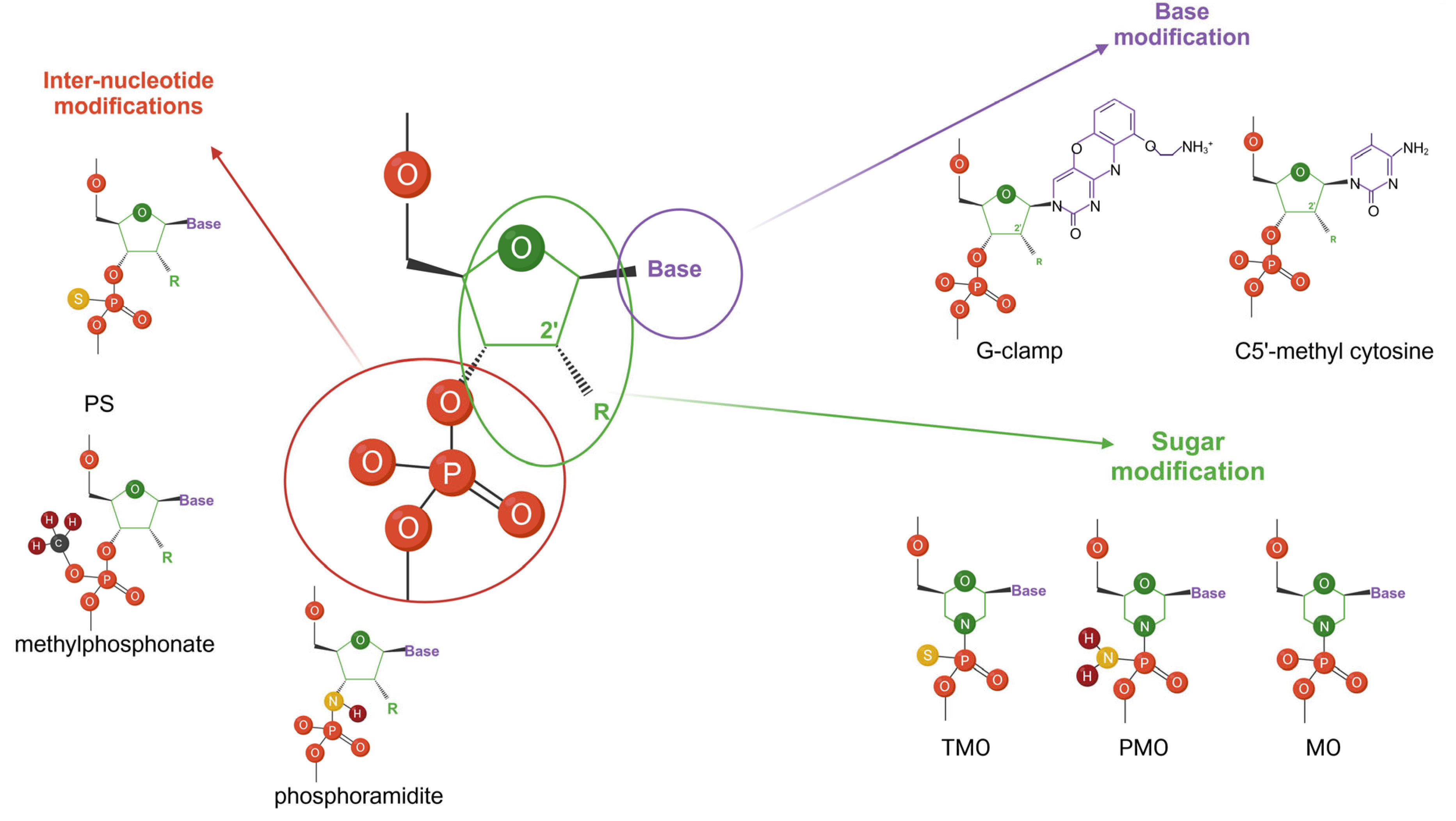
2.3. Designing ASOs
2.4. Clinical Future of ASOs
2.4.1. Breast Malignancies
2.4.2. Ovarian Malignancies
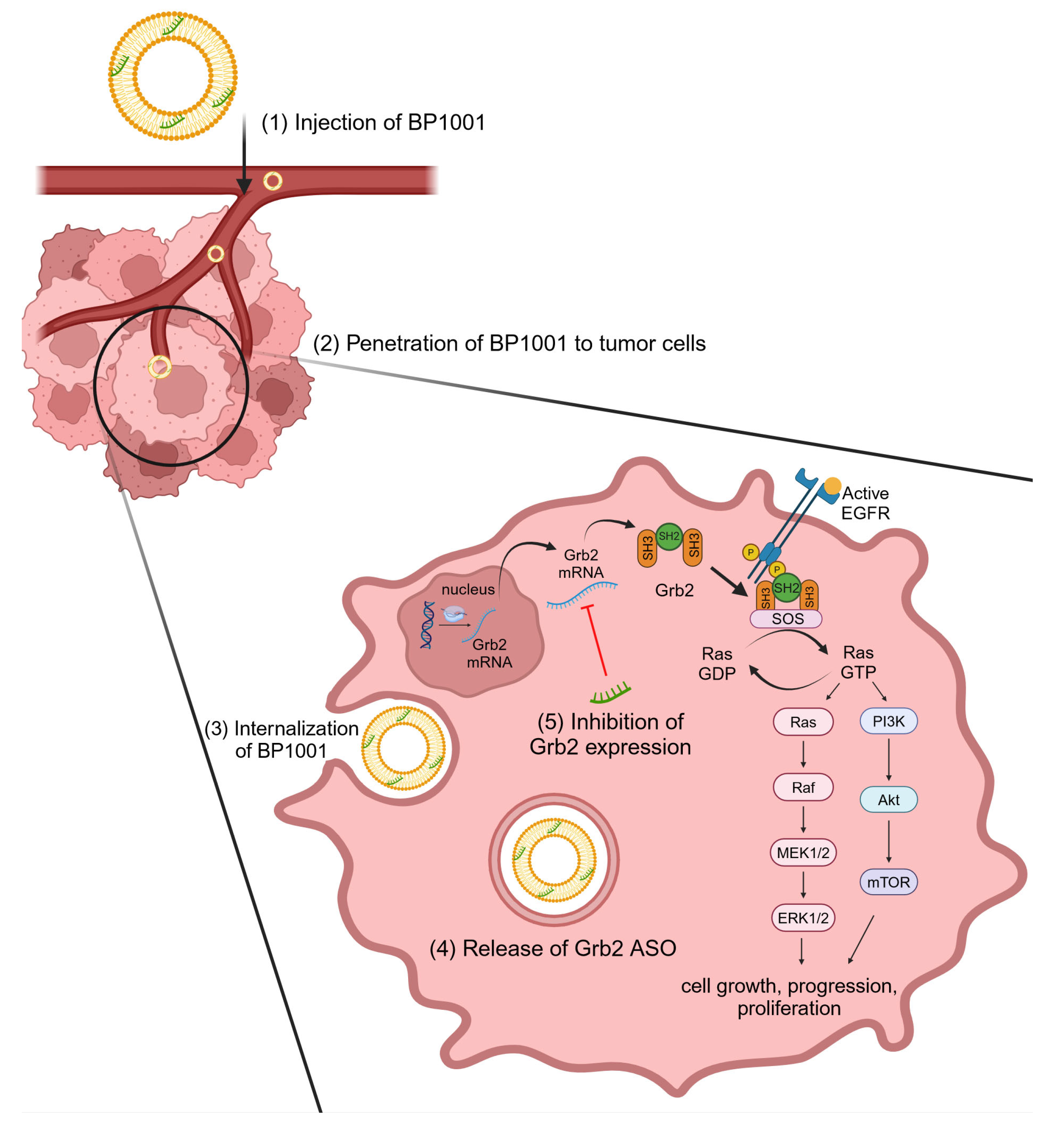
2.4.3. Hematologic Malignancies
2.4.4. Lung Cancer
2.4.5. Gastrointestinal Cancer
2.4.6. Potential Role of ASO in Immunotherapy and Anti-Angiogenic Therapies
3. Overcoming Challenges for Clinical Application of ASOs
4. Summary and Future Directions
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Correction Statement
Abbreviations
| ALSFRS-R | ALS functional rating scale revised |
| AML | Acute myeloid leukemia |
| ASO | Antisense oligonucleotide |
| BCL-2 | B-cell lymphoma 2 |
| BCL-XL | B-cell lymphoma-extra-large protein |
| cAMP | Adenosine 3′,5′-cyclic monophosphate |
| CLN7 | Neuronal ceroid lipofuscinosis 7 (CLN7) gene |
| c-Myb | Avian myeloblastosis viral oncogene homolog |
| CSF | Cerebrospinal fluid |
| DMD | Duchenne muscular dystrophy |
| DOPC | 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine |
| eIF-4α | Eukaryotic initiation factor 4 alpha |
| ERK | Extracellular signal-regulated kinase |
| FOXP3 | Transcription factor forkhead box protein 3 |
| Grb2 | Growth factor receptor-bound protein 2 |
| HER | Human epidermal growth factor receptor |
| Hsp27 | Heat shock protein 27 |
| IFN-γ | Interferon gamma |
| IGF-1 | Insulin-like growth factor 1 |
| IGF-1R | Insulin-like growth factor 1 receptor |
| IL-12 | Interleukin 12 |
| IL-6 | Interleukin 6 |
| Kb | Kilobase |
| KRAS | Kirsten rat sarcoma |
| LNA | Locked nucleic acid |
| MAPK | Mitogen-activated protein kinase |
| Mcl-1 | Myeloid cell leukemia-1 |
| miRNA | MicroRNA |
| MOE | 2′-O-methoxyethyl group |
| ncRNA | Non-coding RNA |
| NfL | Neurofilament Light Chain |
| Notch-1 | Neurogenic locus notch homolog protein 1 |
| NCI | The National Cancer Institute |
| NHGRI | The National Human Genome Research Institute |
| OMe | 2′-O-methyl group |
| P53 | Tumor protein |
| PCH3 | Phosphomethyl group |
| PDCD4 | Programmed cell death protein 4 |
| PI3K | Phospho-inositide-3-kinase |
| PKA | Protein kinase A |
| PKC | Protein kinase C |
| PNH2 | Phosphoamine group |
| PS | Phosphorothioate group |
| PTEN | Phosphatase and tensin homolog |
| Raf-1 | Rapidly accelerated fibrosarcoma |
| RAS | Rat sarcoma |
| RISC | RNA-induced silencing complex |
| SMA | Spinal muscular atrophy |
| STAT3 | Signal transducer and activator of transcription 3 |
| TCGA | Cancer Genome Atlas Research Network |
| TGF-β2 | Transforming growth factor beta 2 |
| TLR9 | Toll-like receptor 9 |
| TNF-α | Tumor necrosis factor alpha |
| TRPM-2 | Transient receptor potential melastatin 2 |
| VEGF | Vascular endothelial growth factor |
| XIAP | X-linked Inhibitor of Apoptosis Protein |
References
- American Cancer Society. Cancer Facts & Figures 2022; American Cancer Society: Atlanta, GA, USA, 2022. [Google Scholar]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Aunan, J.R.; Cho, W.C.; Søreide, K. The Biology of Aging and Cancer: A Brief Overview of Shared and Divergent Molecular Hallmarks. Aging Dis. 2017, 8, 628–642. [Google Scholar] [CrossRef]
- Chial, H. Proto-oncogenes to oncogenes to cancer. Nat. Educ. 2008, 1, 33. [Google Scholar]
- You, Y.; Lai, X.; Pan, Y.; Zheng, H.; Vera, J.; Liu, S.; Deng, S.; Zhang, L. Artificial intelligence in cancer target identification and drug discovery. Signal Transduct. Target. Ther. 2022, 7, 156. [Google Scholar] [CrossRef]
- Quemener, A.M.; Bachelot, L.; Forestier, A.; Donnou-Fournet, E.; Gilot, D.; Galibert, M.D. The powerful world of antisense oligonucleotides: From bench to bedside. Wiley Interdiscip. Rev. RNA 2020, 11, e1594. [Google Scholar] [CrossRef] [PubMed]
- Rayburn, E.R.; Zhang, R. Antisense, RNAi, and gene silencing strategies for therapy: Mission possible or impossible? Drug Discov. Today 2008, 13, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Bege, M.; Borbás, A. Rise and fall of fomivirsen, the first approved gene silencing medicine—A historical review. Acta Pharm. Hung. 2022, 92, 38–44. [Google Scholar] [CrossRef]
- Wong, E.; Goldberg, T. Mipomersen (kynamro): A novel antisense oligonucleotide inhibitor for the management of homozygous familial hypercholesterolemia. Pharm. Ther. 2014, 39, 119–122. [Google Scholar]
- FDA. KYNAMRO (Mipomersen Sodium) Label. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2019/203568s011lbl.pdf (accessed on 5 March 2024).
- EMA. Refusal of the Marketing Authorisation for Kynamro (Mipomersen). Available online: https://www.ema.europa.eu/en/documents/smop-initial/questions-and-answers-refusal-marketing-authorisation-kynamro-outcome-re-examination_en.pdf (accessed on 5 March 2024).
- Pacione, M.; Siskind, C.E.; Day, J.W.; Tabor, H.K. Perspectives on Spinraza (Nusinersen) Treatment Study: Views of Individuals and Parents of Children Diagnosed with Spinal Muscular Atrophy. J. Neuromuscul. Dis. 2019, 6, 119–131. [Google Scholar] [CrossRef]
- EMA. Spinraza (Nusinersen) Approval. Available online: https://www.ema.europa.eu/en/documents/overview/spinraza-epar-summary-public_en.pdf (accessed on 5 March 2024).
- Edinoff, A.N.; Nguyen, L.H.; Odisho, A.S.; Maxey, B.S.; Pruitt, J.W.; Girma, B.; Cornett, E.M.; Kaye, A.M.; Kaye, A.D. The Antisense Oligonucleotide Nusinersen for Treatment of Spinal Muscular Atrophy. Orthop. Rev. 2021, 13, 24934. [Google Scholar] [CrossRef]
- Aartsma-Rus, A.; Goemans, N. A Sequel to the Eteplirsen Saga: Eteplirsen Is Approved in the United States but Was Not Approved in Europe. Nucleic Acid. Ther. 2019, 29, 13–15. [Google Scholar] [CrossRef]
- EMA. Refusal of the Marketing Authorisation for Exondys (Eteplirsen). Available online: https://www.ema.europa.eu/en/documents/smop-initial/questions-and-answers-refusal-marketing-authorisation-exondys-eteplirsen-outcome-re-examination_en.pdf (accessed on 5 March 2024).
- Gales, L. Tegsedi (Inotersen): An Antisense Oligonucleotide Approved for the Treatment of Adult Patients with Hereditary Transthyretin Amyloidosis. Pharmaceuticals 2019, 12, 78. [Google Scholar] [CrossRef]
- EMA. Tegsedi (Inotersen) Approval. Available online: https://www.ema.europa.eu/en/documents/overview/tegsedi-epar-summary-public_en.pdf (accessed on 5 March 2024).
- Aartsma-Rus, A.; Corey, D.R. The 10th Oligonucleotide Therapy Approved: Golodirsen for Duchenne Muscular Dystrophy. Nucleic Acid. Ther. 2020, 30, 67–70. [Google Scholar] [CrossRef]
- Kim, J.; Hu, C.; Moufawad El Achkar, C.; Black, L.E.; Douville, J.; Larson, A.; Pendergast, M.K.; Goldkind, S.F.; Lee, E.A.; Kuniholm, A.; et al. Patient-Customized Oligonucleotide Therapy for a Rare Genetic Disease. N. Engl. J. Med. 2019, 381, 1644–1652. [Google Scholar] [CrossRef] [PubMed]
- Scharner, J.; Aznarez, I. Clinical Applications of Single-Stranded Oligonucleotides: Current Landscape of Approved and In-Development Therapeutics. Mol. Ther. 2021, 29, 540–554. [Google Scholar] [CrossRef] [PubMed]
- Aoki, Y.; Wood, M.J.A. Emerging Oligonucleotide Therapeutics for Rare Neuromuscular Diseases. J. Neuromuscul. Dis. 2021, 8, 869–884. [Google Scholar] [CrossRef] [PubMed]
- Esan, O.; Wierzbicki, A.S. Volanesorsen in the Treatment of Familial Chylomicronemia Syndrome or Hypertriglyceridaemia: Design, Development and Place in Therapy. Drug Des. Dev. Ther. 2020, 14, 2623–2636. [Google Scholar] [CrossRef]
- Paik, J.; Duggan, S. Volanesorsen: First Global Approval. Drugs 2019, 79, 1349–1354. [Google Scholar] [CrossRef]
- Ionis Pharmaceuticals, Inc. Akcea and Ionis Announce Approval of WAYLIVRA® (Volanesorsen) in the European Union. Available online: https://ir.ionispharma.com/news-releases/news-release-details/akcea-and-ionis-announce-approval-waylivrar-volanesorsen (accessed on 5 March 2024).
- Dhillon, S. Viltolarsen: First Approval. Drugs 2020, 80, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- FDA. Viltepso (Viltolarsen) Label. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/212154s000lbl.pdf (accessed on 5 March 2024).
- EMA. Viltolarsen Approval of Investigation Plan. Available online: https://www.ema.europa.eu/en/documents/pip-decision/p00832022-ema-decision-11-march-2022-agreement-paediatric-investigation-plan-and-granting-deferral-and-granting-waiver-viltolarsen-emea-002853-pip01-20_en.pdf (accessed on 5 March 2024).
- Shirley, M. Casimersen: First Approval. Drugs 2021, 81, 875–879. [Google Scholar] [CrossRef]
- Nie, T. Eplontersen: First Approval. Drugs 2024, 84, 473–478. [Google Scholar] [CrossRef] [PubMed]
- FDA. Wainua (Eplontersen) Label. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2023/217388s000lbl.pdf (accessed on 5 March 2024).
- EMA. Approval of Qalsody (Tofersen). Available online: https://www.ema.europa.eu/en/documents/smop-initial/chmp-summary-positive-opinion-qalsody_en.pdf (accessed on 5 March 2024).
- Saini, A.; Chawla, P.A. Breaking barriers with tofersen: Enhancing therapeutic opportunities in amyotrophic lateral sclerosis. Eur. J. Neurol. 2024, 31, e16140. [Google Scholar] [CrossRef] [PubMed]
- Batista-Duharte, A.; Sendra, L.; Herrero, M.J.; Téllez-Martínez, D.; Carlos, I.Z.; Aliño, S.F. Progress in the Use of Antisense Oligonucleotides for Vaccine Improvement. Biomolecules 2020, 10, 316. [Google Scholar] [CrossRef] [PubMed]
- Collotta, D.; Bertocchi, I.; Chiapello, E.; Collino, M. Antisense oligonucleotides: A novel Frontier in pharmacological strategy. Front. Pharmacol. 2023, 14, 1304342. [Google Scholar] [CrossRef] [PubMed]
- EMA. Tegsedi Summary of Product Characteristic. Available online: https://www.ema.europa.eu/en/documents/product-information/tegsedi-epar-product-information_en.pdf (accessed on 5 March 2024).
- Dias, N.; Stein, C.A. Antisense oligonucleotides: Basic concepts and mechanisms. Mol. Cancer Ther. 2002, 1, 347–355. [Google Scholar] [PubMed]
- DeVos, S.L.; Miller, T.M. Antisense oligonucleotides: Treating neurodegeneration at the level of RNA. Neurotherapeutics 2013, 10, 486–497. [Google Scholar] [CrossRef]
- Deleavey, G.F.; Damha, M.J. Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol. 2012, 19, 937–954. [Google Scholar] [CrossRef]
- Khan, P.; Siddiqui, J.; Lakshmanan, I.; Ganti, A.; Salgia, R.; Jain, M.; Batra, S.; Nasser, M. RNA-based therapies: A cog in the wheel of lung cancer defense. Mol. Cancer 2021, 20, 54. [Google Scholar] [CrossRef]
- Majlessi, M.; Nelson, N.C.; Becker, M.M. Advantages of 2′-O-methyl oligoribonucleotide probes for detecting RNA targets. Nucleic Acids Res. 1998, 26, 2224–2229. [Google Scholar] [CrossRef]
- Raguraman, P.; Balachandran, A.A.; Chen, S.; Diermeier, S.D.; Veedu, R.N. Antisense Oligonucleotide-Mediated Splice Switching: Potential Therapeutic Approach for Cancer Mitigation. Cancers 2021, 13, 5555. [Google Scholar] [CrossRef]
- Soler-Bistué, A.; Zorreguieta, A.; Tolmasky, M.E. Bridged Nucleic Acids Reloaded. Molecules 2019, 24, 2297. [Google Scholar] [CrossRef] [PubMed]
- Bege, M.; Borbás, A. The Medicinal Chemistry of Artificial Nucleic Acids and Therapeutic Oligonucleotides. Pharmaceuticals 2022, 15, 909. [Google Scholar] [CrossRef] [PubMed]
- Vandermeeren, M.; Préveral, S.; Janssens, S.; Geysen, J.; Saison-Behmoaras, E.; Van Aerschot, A.; Herdewijn, P. Biological activity of hexitol nucleic acids targeted at Ha-ras and intracellular adhesion molecule-1 mRNA. Biochem. Pharmacol. 2000, 59, 655–663. [Google Scholar] [CrossRef]
- Du, L.; Gatti, R.A. Potential therapeutic applications of antisense morpholino oligonucleotides in modulation of splicing in primary immunodeficiency diseases. J. Immunol. Methods 2011, 365, 1–7. [Google Scholar] [CrossRef]
- Gan, L.; Wu, L.C.L.; Wood, J.A.; Yao, M.; Treleaven, C.M.; Estrella, N.L.; Wentworth, B.M.; Hanson, G.J.; Passini, M.A. A cell-penetrating peptide enhances delivery and efficacy of phosphorodiamidate morpholino oligomers in mdx mice. Mol. Ther. Nucleic Acids 2022, 30, 17–27. [Google Scholar] [CrossRef]
- Hyrup, B.; Nielsen, P.E. Peptide Nucleic Acids (PNA): Synthesis, properties and potential applications. Bioorg. Med. Chem. 1996, 4, 5–23. [Google Scholar] [CrossRef]
- Pradeep, S.P.; Malik, S.; Slack, F.J.; Bahal, R. Unlocking the potential of chemically modified peptide nucleic acids for RNA-based therapeutics. RNA 2023, 29, 434–445. [Google Scholar] [CrossRef]
- Le, B.T.; Filichev, V.V.; Veedu, R.N. Investigation of twisted intercalating nucleic acid (TINA)-modified antisense oligonucleotides for splice modulation by induced exon-skipping in vitro. RSC Adv. 2016, 6, 95169–95172. [Google Scholar] [CrossRef]
- Liu, L.S.; Leung, H.M.; Tam, D.Y.; Lo, T.W.; Wong, S.W.; Lo, P.K. α-l-Threose Nucleic Acids as Biocompatible Antisense Oligonucleotides for Suppressing Gene Expression in Living Cells. ACS Appl. Mater. Interfaces 2018, 10, 9736–9743. [Google Scholar] [CrossRef]
- Matsuda, S.; Bala, S.; Liao, J.-Y.; Datta, D.; Mikami, A.; Woods, L.; Harp, J.M.; Gilbert, J.A.; Bisbe, A.; Manoharan, R.M.; et al. Shorter Is Better: The α-(l)-Threofuranosyl Nucleic Acid Modification Improves Stability, Potency, Safety, and Ago2 Binding and Mitigates Off-Target Effects of Small Interfering RNAs. J. Am. Chem. Soc. 2023, 145, 19691–19706. [Google Scholar] [CrossRef]
- Wang, F.; Liu, L.S.; Lau, C.H.; Han Chang, T.J.; Tam, D.Y.; Leung, H.M.; Tin, C.; Lo, P.K. Synthetic α-l-Threose Nucleic Acids Targeting BcL-2 Show Gene Silencing and in Vivo Antitumor Activity for Cancer Therapy. ACS Appl. Mater. Interfaces 2019, 11, 38510–38518. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.; Wang, Y.; England, W.E.; Chaput, J.C.; Spitale, R.C. Allele-Specific RNA Knockdown with a Biologically Stable and Catalytically Efficient XNAzyme. J. Am. Chem. Soc. 2021, 143, 4519–4523. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Nguyen, K.; Spitale, R.C.; Chaput, J.C. A biologically stable DNAzyme that efficiently silences gene expression in cells. Nat. Chem. 2021, 13, 319–326. [Google Scholar] [CrossRef]
- Crooke, S.T. Potential roles of antisense technology in cancer chemotherapy. Oncogene 2000, 19, 6651–6659. [Google Scholar] [CrossRef][Green Version]
- Agrawal, S. Importance of nucleotide sequence and chemical modifications of antisense oligonucleotides. BBA-Gene Struct. Expr. 1999, 1489, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.H.; Lim, S.; Wong, W.S. Antisense oligonucleotides: From design to therapeutic application. Clin. Exp. Pharmacol. Physiol. 2006, 33, 533–540. [Google Scholar] [CrossRef]
- Kilanowska, A.; Studzińska, S. In vivo and in vitro studies of antisense oligonucleotides—A review. RSC Adv. 2020, 10, 34501–34516. [Google Scholar] [CrossRef]
- Kole, R.; Krainer, A.R.; Altman, S. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012, 11, 125–140. [Google Scholar] [CrossRef]
- Mansoor, M.; Melendez, A.J. Advances in antisense oligonucleotide development for target identification, validation, and as novel therapeutics. Gene Regul. Syst. Biol. 2008, 2, 275–295. [Google Scholar] [CrossRef]
- Yokota, T.; Maruyama, R. Gapmers: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Inoue, H.; Hayase, Y.; Iwai, S.; Ohtsuka, E. Sequence-dependent hydrolysis of RNA using modified oligonucleotide splints and RNase H. FEBS Lett. 1987, 215, 327–330. [Google Scholar] [CrossRef]
- Marrosu, E.; Ala, P.; Muntoni, F.; Zhou, H. Gapmer Antisense Oligonucleotides Suppress the Mutant Allele of COL6A3 and Restore Functional Protein in Ullrich Muscular Dystrophy. Mol. Ther. Nucleic Acids 2017, 8, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Maranon, D.G.; Wilusz, J. Mind the Gapmer: Implications of Co-transcriptional Cleavage by Antisense Oligonucleotides. Mol. Cell 2020, 77, 932–933. [Google Scholar] [CrossRef] [PubMed]
- Burel, S.A.; Hart, C.E.; Cauntay, P.; Hsiao, J.; Machemer, T.; Katz, M.; Watt, A.; Bui, H.H.; Younis, H.; Sabripour, M.; et al. Hepatotoxicity of high affinity gapmer antisense oligonucleotides is mediated by RNase H1 dependent promiscuous reduction of very long pre-mRNA transcripts. Nucleic Acids Res. 2016, 44, 2093–2109. [Google Scholar] [CrossRef]
- Arun, G.; Diermeier, S.; Akerman, M.; Chang, K.C.; Wilkinson, J.E.; Hearn, S.; Kim, Y.; MacLeod, A.R.; Krainer, A.R.; Norton, L.; et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes. Dev. 2016, 30, 34–51. [Google Scholar] [CrossRef] [PubMed]
- Chery, J. RNA therapeutics: RNAi and antisense mechanisms and clinical applications. Postdoc J. 2016, 4, 35–50. [Google Scholar]
- Crooke, S.T. Molecular mechanisms of action of antisense drugs. Biochim. Biophys. Acta (BBA)—Gene Struct. Expr. 1999, 1489, 31–43. [Google Scholar] [CrossRef]
- Crooke, S.T. Progress in antisense technology: The end of the beginning. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2000; Volume 313, pp. 3–45. [Google Scholar]
- Graff, J.R.; Konicek, B.W.; Vincent, T.M.; Lynch, R.L.; Monteith, D.; Weir, S.N.; Schwier, P.; Capen, A.; Goode, R.L.; Dowless, M.S.; et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J. Clin. Investig. 2007, 117, 2638–2648. [Google Scholar] [CrossRef] [PubMed]
- MD Anderson Cancer Center. LY2275796 in Advanced Cancer; MD Anderson Cancer Center: Houston, TX, USA, 2006. [Google Scholar]
- Ionis Pharmaceuticals Inc. Safety and Tolerability Study of ISIS EIF4E Rx in Combination with Docetaxel and Prednisone (CRPC); Ionis Pharmaceuticals Inc.: Carlsbad, CA, USA, 2010. [Google Scholar]
- National Cancer Institute; National Institutes of Health Clinical Center. ISIS 183750 with Irinotecan for Advanced Solid. Tumors or Colorectal Cancer; National Cancer Institute: Bethesda, MD, USA, 2012.
- Holgersen, E.M.; Gandhi, S.; Zhou, Y.; Kim, J.; Vaz, B.; Bogojeski, J.; Bugno, M.; Shalev, Z.; Cheung-Ong, K.; Gonçalves, J.; et al. Transcriptome-Wide Off-Target Effects of Steric-Blocking Oligonucleotides. Nucleic Acid. Ther. 2021, 31, 392–403. [Google Scholar] [CrossRef]
- El Boujnouni, N.; van der Bent, M.L.; Willemse, M.; Ac’t Hoen, P.; Brock, R.; Wansink, D.G. Block or degrade? Balancing on- and off-target effects of antisense strategies against transcripts with expanded triplet repeats in DM1. Mol. Ther.—Nucleic Acids 2023, 32, 622–636. [Google Scholar] [CrossRef]
- Li, Y.; Chen, S.; Rahimizadeh, K.; Zhang, Z.; Veedu, R.N. Inhibition of survivin by 2′-O-methyl phosphorothioate-modified steric-blocking antisense oligonucleotides. RSC Adv. 2024, 14, 13336–13341. [Google Scholar] [CrossRef]
- Dhuri, K.; Bechtold, C.; Quijano, E.; Pham, H.; Gupta, A.; Vikram, A.; Bahal, R. Antisense Oligonucleotides: An Emerging Area in Drug Discovery and Development. J. Clin. Med. 2020, 9, 2004. [Google Scholar] [CrossRef]
- Singh, N.N.; Luo, D.; Singh, R.N. Pre-mRNA Splicing Modulation by Antisense Oligonucleotides. Methods Mol. Biol. 2018, 1828, 415–437. [Google Scholar]
- Wahl, M.C.; Will, C.L.; Lührmann, R. The Spliceosome: Design Principles of a Dynamic RNP Machine. Cell 2009, 136, 701–718. [Google Scholar] [CrossRef]
- Svasti, S.; Suwanmanee, T.; Fucharoen, S.; Moulton, H.M.; Nelson, M.H.; Maeda, N.; Smithies, O.; Kole, R. RNA repair restores hemoglobin expression in IVS2-654 thalassemic mice. Proc. Natl. Acad. Sci. USA 2009, 106, 1205–1210. [Google Scholar] [CrossRef]
- Xie, S.Y.; Li, W.; Ren, Z.R.; Huang, S.Z.; Zeng, F.; Zeng, Y.T. Correction of β654-thalassaemia mice using direct intravenous injection of siRNA and antisense RNA vectors. Int. J. Hematol. 2011, 93, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Sahashi, K.; Rigo, F.; Hung, G.; Horev, G.; Bennett, C.F.; Krainer, A.R. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature 2011, 478, 123–126. [Google Scholar] [CrossRef]
- Hua, Y.; Sahashi, K.; Hung, G.; Rigo, F.; Passini, M.A.; Bennett, C.F.; Krainer, A.R. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes. Dev. 2010, 24, 1634–1644. [Google Scholar] [CrossRef] [PubMed]
- Passini, M.A.; Bu, J.; Richards, A.M.; Kinnecom, C.; Sardi, S.P.; Stanek, L.M.; Hua, Y.; Rigo, F.; Matson, J.; Hung, G.; et al. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci. Transl. Med. 2011, 3, 72ra18. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.L.; Rabinowitz, A.; Chen, Y.C.; Yokota, T.; Yin, H.; Alter, J.; Jadoon, A.; Bou-Gharios, G.; Partridge, T. Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles. Proc. Natl. Acad. Sci. USA 2005, 102, 198–203. [Google Scholar] [CrossRef]
- Goemans, N.M.; Tulinius, M.; van den Akker, J.T.; Burm, B.E.; Ekhart, P.F.; Heuvelmans, N.; Holling, T.; Janson, A.A.; Platenburg, G.J.; Sipkens, J.A.; et al. Systemic administration of PRO051 in Duchenne’s muscular dystrophy. N. Engl. J. Med. 2011, 364, 1513–1522. [Google Scholar] [CrossRef]
- Cirak, S.; Arechavala-Gomeza, V.; Guglieri, M.; Feng, L.; Torelli, S.; Anthony, K.; Abbs, S.; Garralda, M.E.; Bourke, J.; Wells, D.J.; et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: An open-label, phase 2, dose-escalation study. Lancet 2011, 378, 595–605. [Google Scholar] [CrossRef]
- Zanetta, C.; Nizzardo, M.; Simone, C.; Monguzzi, E.; Bresolin, N.; Comi, G.P.; Corti, S. Molecular therapeutic strategies for spinal muscular atrophies: Current and future clinical trials. Clin. Ther. 2014, 36, 128–140. [Google Scholar] [CrossRef]
- Singh, N.N.; Howell, M.D.; Androphy, E.J.; Singh, R.N. How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy. Gene Ther. 2017, 24, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, Q.; Han, L.; Tian, N.; Liang, Q.; Li, Y.; Zhao, X.; Du, C.; Tian, Y. Pro-apoptotic effects of splice-switching oligonucleotides targeting Bcl-x pre-mRNA in human glioma cell lines. Oncol. Rep. 2016, 35, 1013–1019. [Google Scholar] [CrossRef] [PubMed]
- Evers, M.M.; Toonen, L.J.A.; van Roon-Mom, W.M.C. Antisense oligonucleotides in therapy for neurodegenerative disorders. Adv. Drug Deliv. Rev. 2015, 87, 90–103. [Google Scholar] [CrossRef]
- Vickers, T.A.; Crooke, S.T. The rates of the major steps in the molecular mechanism of RNase H1-dependent antisense oligonucleotide induced degradation of RNA. Nucleic Acids Res. 2015, 43, 8955–8963. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, Y.; Wang, M.; Li, Q.; Qu, Z.; Shi, V.; Kraft, P.; Kim, S.; Gao, Y.; Pak, J.; et al. Downregulation of HER3 by a Novel Antisense Oligonucleotide, EZN-3920, Improves the Antitumor Activity of EGFR and HER2 Tyrosine Kinase Inhibitors in Animal Models. Mol. Cancer Ther. 2013, 12, 427–437. [Google Scholar] [CrossRef]
- Stenvang, J.; Kauppinen, S. MicroRNAs as targets for antisense-based therapeutics. Expert. Opin. Biol. Ther. 2008, 8, 59–81. [Google Scholar] [CrossRef]
- Ding, T.; Cui, P.; Zhou, Y.; Chen, C.; Zhao, J.; Wang, H.; Guo, M.; He, Z.; Xu, L. Antisense Oligonucleotides against miR-21 Inhibit the Growth and Metastasis of Colorectal Carcinoma via the DUSP8 Pathway. Mol. Ther. Nucleic Acids 2018, 13, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.H.; Tsao, C.J. Emerging role of microRNA-21 in cancer. Biomed. Rep. 2016, 5, 395–402. [Google Scholar] [CrossRef]
- Shang, M.; Wu, Y.; Wang, Y.; Cai, Y.; Jin, J.; Yang, Z. Dual antisense oligonucleotide targeting miR-21/miR-155 synergize photodynamic therapy to treat triple-negative breast cancer and inhibit metastasis. Biomed. Pharmacother. 2022, 146, 112564. [Google Scholar] [CrossRef]
- Huang, S.; Hao, X.-Y.; Li, Y.-J.; Wu, J.Y.; Xiang, D.-X.; Luo, S. Nonviral delivery systems for antisense oligonucleotide therapeutics. Biomater. Res. 2022, 26, 49. [Google Scholar] [CrossRef]
- Chen, S.; Heendeniya, S.N.; Le, B.T.; Rahimizadeh, K.; Rabiee, N.; Zahra, Q.U.A.; Veedu, R.N. Splice-Modulating Antisense Oligonucleotides as Therapeutics for Inherited Metabolic Diseases. BioDrugs 2024, 38, 177–203. [Google Scholar] [CrossRef]
- Crooke, S.T.; Baker, B.F.; Xia, S.; Yu, R.Z.; Viney, N.J.; Wang, Y.; Tsimikas, S.; Geary, R.S. Integrated Assessment of the Clinical Performance of GalNAc(3)-Conjugated 2′-O-Methoxyethyl Chimeric Antisense Oligonucleotides: I. Human Volunteer Experience. Nucleic Acid. Ther. 2019, 29, 16–32. [Google Scholar] [CrossRef]
- Miller, T.M.; Pestronk, A.; David, W.; Rothstein, J.; Simpson, E.; Appel, S.H.; Andres, P.L.; Mahoney, K.; Allred, P.; Alexander, K.; et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: A phase 1, randomised, first-in-man study. Lancet Neurol. 2013, 12, 435–442. [Google Scholar] [CrossRef]
- A Dose Escalation Phase I Study of LNA-i-miR-221 for the Treatment of Refractory Multiple Myeloma and Advanced Solid Tumors. 2021. Available online: https://adisinsight.springer.com/trials/700335224 (accessed on 11 August 2024).
- Paterson, B.M.; Roberts, B.E.; Kuff, E.L. Structural gene identification and mapping by DNA-mRNA hybrid-arrested cell-free translation. Proc. Natl. Acad. Sci. USA 1977, 74, 4370–4374. [Google Scholar] [CrossRef]
- Zamecnik, P.C.; Stephenson, M.L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl. Acad. Sci. USA 1978, 75, 280–284. [Google Scholar] [CrossRef]
- Mohs, R.C.; Greig, N.H. Drug discovery and development: Role of basic biological research. Alzheimers Dement. 2017, 3, 651–657. [Google Scholar] [CrossRef]
- Szymanowska, A.; Rodriguez-Aguayo, C.; Lopez-Berestein, G.; Amero, P. Non-Coding RNAs: Foes or Friends for Targeting Tumor Microenvironment. Noncoding RNA 2023, 9, 52. [Google Scholar] [CrossRef]
- Jin, X.; Mei, Y.; Yang, P.; Huang, R.; Zhang, H.; Wu, Y.; Wang, M.; He, X.; Jiang, Z.; Zhu, W.; et al. Prioritization of therapeutic targets for cancers using integrative multi-omics analysis. Hum. Genom. 2024, 18, 42. [Google Scholar] [CrossRef]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 19, A68–A77. [Google Scholar] [CrossRef]
- NIH. The Cancer Genome Atlas Program (TCGA). Available online: https://www.cancer.gov/ccg/research/genome-sequencing/tcga (accessed on 5 March 2024).
- Uttarkar, A. Protocol for In-silico Design, Docking and Molecular Dynamic Simulation of Antisense Oligonucleotides v1. 2022. Available online: https://www.protocols.io/view/protocol-for-in-silico-design-docking-and-molecula-ewov1nr4ogr2/v1 (accessed on 5 March 2024).
- Yasuhara, H.; Yoshida, T.; Sasaki, K.; Obika, S.; Inoue, T. Reduction of Off-Target Effects of Gapmer Antisense Oligonucleotides by Oligonucleotide Extension. Mol. Diagn. Ther. 2022, 26, 117–127. [Google Scholar] [CrossRef]
- Krotz, A.H.; Carty, R.L.; Scozzari, A.N.; Cole, D.L.; Ravikumar, V.T. Large-Scale Synthesis of Antisense Oligonucleotides without Chlorinated Solvents. Org. Process Res. Dev. 2000, 4, 190–193. [Google Scholar] [CrossRef]
- Andrews, B.I.; Antia, F.D.; Brueggemeier, S.B.; Diorazio, L.J.; Koenig, S.G.; Kopach, M.E.; Lee, H.; Olbrich, M.; Watson, A.L. Sustainability Challenges and Opportunities in Oligonucleotide Manufacturing. J. Org. Chem. 2021, 86, 49–61. [Google Scholar] [CrossRef]
- Crooke, S.T. Antisense Drug Technology: Principles, Strategies and Applications, 2nd ed.; Crooke, S.T., Ed.; CRC Press: Boca Raton, FL, USA, 2008. [Google Scholar]
- Doxtader Lacy, K.A.; Liang, X.H.; Zhang, L.; Crooke, S.T. RNA modifications can affect RNase H1-mediated PS-ASO activity. Mol. Ther. Nucleic Acids 2022, 28, 814–828. [Google Scholar] [CrossRef]
- Stulz, R.; Lerche, M.; Luige, O.; Taylor, A.; Geschwindner, S.; Ghidini, A. An enhanced biophysical screening strategy to investigate the affinity of ASOs for their target RNA. RSC Chem. Biol. 2023, 4, 1123–1130. [Google Scholar] [CrossRef]
- Mathews, D.H.; Moss, W.N.; Turner, D.H. Folding and finding RNA secondary structure. Cold Spring Harb. Perspect. Biol. 2010, 2, a003665. [Google Scholar] [CrossRef]
- Reeder, J.; Höchsmann, M.; Rehmsmeier, M.; Voss, B.; Giegerich, R. Beyond Mfold: Recent advances in RNA bioinformatics. J. Biotechnol. 2006, 124, 41–55. [Google Scholar] [CrossRef]
- Ding, Y.; Lawrence, C.E. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 2003, 31, 7280–7301. [Google Scholar] [CrossRef]
- Matveeva, O.V.; Tsodikov, A.D.; Giddings, M.; Freier, S.M.; Wyatt, J.R.; Spiridonov, A.N.; Shabalina, S.A.; Gesteland, R.F.; Atkins, J.F. Identification of sequence motifs in oligonucleotides whose presence is correlated with antisense activity. Nucleic Acids Res. 2000, 28, 2862–2865. [Google Scholar] [CrossRef][Green Version]
- Aartsma-Rus, A.; van Vliet, L.; Hirschi, M.; Janson, A.A.; Heemskerk, H.; de Winter, C.L.; de Kimpe, S.; van Deutekom, J.C.; Ac’t Hoen, P.; van Ommen, G.J. Guidelines for antisense oligonucleotide design and insight into splice-modulating mechanisms. Mol. Ther. 2009, 17, 548–553. [Google Scholar] [CrossRef]
- Siwak, D.R.; Tari, A.M.; Lopez-Berestein, G. Liposomal antisense oligonucleotides for cancer therapy. Methods Enzymol. 2004, 387, 241–253. [Google Scholar] [PubMed]
- Stewart, A. Antisense against protein kinase C-alpha mRNA makes sense for cancer therapy? Mol. Med. Today 1997, 3, 324. [Google Scholar] [CrossRef] [PubMed]
- Tari, A.M.; Lopez-Berestein, G. GRB2: A pivotal protein in signal transduction. Semin. Oncol. 2001, 28, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Roberts, P.J.; Der, C.J. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 2007, 26, 3291–3310. [Google Scholar] [CrossRef]
- Tari, A.M.; Hung, M.C.; Li, K.Y.; Lopez-Berestein, G. Growth inhibition of breast cancer cells by Grb2 downregulation is correlated with inactivation of mitogen-activated protein kinase in EGFR, but not in ErbB2, cells. Oncogene 1999, 18, 1325–1332. [Google Scholar] [CrossRef][Green Version]
- Bio-Path Holdings, Inc. Clinical Trial of BP1001 (L-Grb-2 Antisense Oligonucleotide) in CML, AML, ALL & MDS; Bio-Path Holdings, Inc.: Houston, TX, USA, 2010. [Google Scholar]
- Bio-Path Holdings, Inc. Clinical Trial of BP1001 in Combination with Venetoclax Plus Decitabine in AML; Bio-Path Holdings, Inc.: Houston, TX, USA, 2016. [Google Scholar]
- Bio-Path Holdings, Inc. BP1001-A in Patients with Advanced or Recurrent Solid Tumors; Bio-Path Holdings, Inc.: Houston, TX, USA, 2022. [Google Scholar]
- Dean, N.M.; Bennett, C.F. Antisense oligonucleotide-based therapeutics for cancer. Oncogene 2003, 22, 9087–9096. [Google Scholar] [CrossRef]
- Roshmi, R.R.; Yokota, T. Viltolarsen: From Preclinical Studies to FDA Approval. Methods Mol. Biol. 2023, 2587, 31–41. [Google Scholar]
- Kaur, S.J.; McKeown, S.R.; Rashid, S. Mutant SOD1 mediated pathogenesis of Amyotrophic Lateral Sclerosis. Gene 2016, 577, 109–118. [Google Scholar] [CrossRef]
- Biogen. A Study of BIIB067 (Tofersen) Initiated in Clinically Presymptomatic Adults with a Confirmed Superoxide Dismutase 1 Mutation; Biogen: Cambridge, MA, USA, 2021. [Google Scholar]
- Ambulanzpartner Soziotechnologie APST GmbH. Registry Study of Assistive Devices, Medicines and Healthcare Measures in ALS, SMA and Other Neurological Diseases; Ambulanzpartner Soziotechnologie APST GmbH: Berlin, Germany, 2015. [Google Scholar]
- Ross, S.J.; Revenko, A.S.; Hanson, L.L.; Ellston, R.; Staniszewska, A.; Whalley, N.; Pandey, S.K.; Revill, M.; Rooney, C.; Buckett, L.K.; et al. Targeting KRAS-dependent tumors with AZD4785, a high-affinity therapeutic antisense oligonucleotide inhibitor of KRAS. Sci. Transl. Med. 2017, 9, eaal5253. [Google Scholar] [CrossRef]
- Roh, H.; Pippin, J.A.; Green, D.W.; Boswell, C.B.; Hirose, C.T.; Mokadam, N.; Drebin, J.A. HER2/neu antisense targeting of human breast carcinoma. Oncogene 2000, 19, 6138–6143. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sapio, L.; Di Maiolo, F.; Illiano, M.; Esposito, A.; Chiosi, E.; Spina, A.; Naviglio, S. Targeting protein kinase A in cancer therapy: An update. EXCLI J. 2014, 13, 843–855. [Google Scholar] [PubMed]
- Yang, D.C.; Elliott, R.L.; Head, J.F. Gene targets of antisense therapies in breast cancer. Expert. Opin. Ther. Targets 2002, 6, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Adewunmi, O.; Shen, Y.; Zhang, X.H.F.; Rosen, J.M. Targeted Inhibition of lncRNA Malat1 Alters the Tumor Immune Microenvironment in Preclinical Syngeneic Mouse Models of Triple-Negative Breast Cancer. Cancer Immunol. Res. 2023, 11, 1462–1479. [Google Scholar] [CrossRef]
- Neuenschwander, S.; Roberts, C.T., Jr.; LeRoith, D. Growth inhibition of MCF-7 breast cancer cells by stable expression of an insulin-like growth factor I receptor antisense ribonucleic acid. Endocrinology 1995, 136, 4298–4303. [Google Scholar] [CrossRef]
- Roychowdhury, D.; Lahn, M. Antisense therapy directed to protein kinase C-alpha (affinitak, LY900003/ISIS 3521): Potential role in breast cancer. Semin. Oncol. 2003, 30, 30–33. [Google Scholar] [CrossRef]
- Lara, O.D.; Bayraktar, E.; Amero, P.; Ma, S.; Ivan, C.; Hu, W.; Wang, Y.; Mangala, L.S.; Dutta, P.; Bhattacharya, P.; et al. Therapeutic efficacy of liposomal Grb2 antisense oligodeoxynucleotide (L-Grb2) in preclinical models of ovarian and uterine cancer. Oncotarget 2020, 11, 2819–2833. [Google Scholar] [CrossRef]
- Cunningham, C.C.; Holmlund, J.T.; Schiller, J.H.; Geary, R.S.; Kwoh, T.J.; Dorr, A.; Nemunaitis, J.J. A phase I trial of c-Raf kinase antisense oligonucleotide ISIS 5132 administered as a continuous intravenous infusion in patients with advanced cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2000, 6, 1626–1631. [Google Scholar]
- Oza, A.M.; Elit, L.; Swenerton, K.; Faught, W.; Ghatage, P.; Carey, M.; McIntosh, L.; Dorr, A.; Holmlund, J.T.; Eisenhauer, E.; et al. Phase II study of CGP 69846A (ISIS 5132) in recurrent epithelial ovarian cancer: An NCIC clinical trials group study (NCIC IND.116). Gynecol. Oncol. 2003, 89, 129–133. [Google Scholar] [CrossRef]
- NCIC Clinical Trials Group; Canadian Cancer Trials Group. ISIS 5132 in Treating Patients with Metastatic or Recurrent Ovarian Cancer; Canadian Cancer Trials Group: Kingston, ON, Canada, 1999. [Google Scholar]
- Lu, J.; Zhang, Y.; Wang, S.; Bi, Y.; Huang, T.; Luo, X.; Cai, Y.D. Analysis of Four Types of Leukemia Using Gene Ontology Term and Kyoto Encyclopedia of Genes and Genomes Pathway Enrichment Scores. Comb. Chem. High. Throughput Screen. 2020, 23, 295–303. [Google Scholar] [CrossRef]
- Puil, L.; Liu, J.; Gish, G.; Mbamalu, G.; Bowtell, D.; Pelicci, P.G.; Arlinghaus, R.; Pawson, T. Bcr-Abl oncoproteins bind directly to activators of the Ras signalling pathway. EMBO J. 1994, 13, 764–773. [Google Scholar] [CrossRef] [PubMed]
- Timsah, Z.; Ahmed, Z.; Ivan, C.; Berrout, J.; Gagea, M.; Zhou, Y.; Pena, G.N.; Hu, X.; Vallien, C.; Kingsley, C.V.; et al. Grb2 depletion under non-stimulated conditions inhibits PTEN, promotes Akt-induced tumor formation and contributes to poor prognosis in ovarian cancer. Oncogene 2016, 35, 2186–2196. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Shinohara, N.; Moriya, K.; Sazawa, A.; Kobayashi, Y.; Ogiso, Y.; Takiguchi, M.; Yasuda, J.; Koyanagi, T.; Kuzumaki, N.; et al. Significance of the Grb2 and Son of sevenless (Sos) proteins in human bladder cancer cell lines. IUBMB Life 2000, 49, 317–320. [Google Scholar] [CrossRef] [PubMed]
- Giubellino, A.; Burke, T.R., Jr.; Bottaro, D.P. Grb2 signaling in cell motility and cancer. Expert. Opin. Ther. Targets 2008, 12, 1021–1033. [Google Scholar] [CrossRef]
- Lopez-Berestein, G.; Tari, A.M.; Arlinghaus, R.B. Inhibition of Chronic Myelogenous Leukemic Cell Growth by Liposomal-Antisense Oligodeoxy-Nucleotides Targeting to Grb2 or Crk1. U.S. US7309692B1, 18 December 2007. [Google Scholar]
- Gagliardi, M.; Ashizawa, A.T. The Challenges and Strategies of Antisense Oligonucleotide Drug Delivery. Biomedicines 2021, 9, 433. [Google Scholar] [CrossRef]
- Tari, A.M.; Gutiérrez-Puente, Y.; Monaco, G.; Stephens, C.; Sun, T.; Rosenblum, M.; Belmont, J.; Arlinghaus, R.; Lopez-Berestein, G. Liposome-incorporated Grb2 antisense oligodeoxynucleotide increases the survival of mice bearing bcr-abl-positive leukemia xenografts. Int. J. Oncol. 2007, 31, 1243–1250. [Google Scholar][Green Version]
- Carter, B.Z.; Mak, P.Y.; Mu, H.; Zhou, H.; Mak, D.H.; Schober, W.; Leverson, J.D.; Zhang, B.; Bhatia, R.; Huang, X.; et al. Combined targeting of BCL-2 and BCR-ABL tyrosine kinase eradicates chronic myeloid leukemia stem cells. Sci. Transl. Med. 2016, 8, 355ra117. [Google Scholar] [CrossRef]
- Kang, M.H.; Reynolds, C.P. Bcl-2 inhibitors: Targeting mitochondrial apoptotic pathways in cancer therapy. Clin. Cancer Res. 2009, 15, 1126–1132. [Google Scholar] [CrossRef]
- Morris, M.J.; Tong, W.P.; Cordon-Cardo, C.; Drobnjak, M.; Kelly, W.K.; Slovin, S.F.; Terry, K.L.; Siedlecki, K.; Swanson, P.; Rafi, M.; et al. Phase I trial of BCL-2 antisense oligonucleotide (G3139) administered by continuous intravenous infusion in patients with advanced cancer. Clin. Cancer Res. 2002, 8, 679–683. [Google Scholar]
- Thomas, S.; Quinn, B.A.; Das, S.K.; Dash, R.; Emdad, L.; Dasgupta, S.; Wang, X.Y.; Dent, P.; Reed, J.C.; Pellecchia, M.; et al. Targeting the Bcl-2 family for cancer therapy. Expert. Opin. Ther. Targets 2013, 17, 61–75. [Google Scholar] [CrossRef]
- Carter, B.Z.; Wang, R.Y.; Schober, W.D.; Milella, M.; Chism, D.; Andreeff, M. Targeting Survivin expression induces cell proliferation defect and subsequent cell death involving mitochondrial pathway in myeloid leukemic cells. Cell Cycle 2003, 2, 488–493. [Google Scholar] [CrossRef][Green Version]
- Erba, H.P.; Sayar, H.; Juckett, M.; Lahn, M.; Andre, V.; Callies, S.; Schmidt, S.; Kadam, S.; Brandt, J.T.; Van Bockstaele, D.; et al. Safety and pharmacokinetics of the antisense oligonucleotide (ASO) LY2181308 as a single-agent or in combination with idarubicin and cytarabine in patients with refractory or relapsed acute myeloid leukemia (AML). Investig. New Drugs 2013, 31, 1023–1034. [Google Scholar] [CrossRef]
- Chen, K.; Zhang, Y.; Qian, L.; Wang, P. Emerging strategies to target RAS signaling in human cancer therapy. J. Hematol. Oncol. 2021, 14, 116. [Google Scholar] [CrossRef] [PubMed]
- Sacco, A.; Federico, C.; Todoerti, K.; Ziccheddu, B.; Giacomini, A.; Ravelli, C.; Maccarinelli, F.; Bianchi, G.; Belotti, A.; Ribolla, R.; et al. Specific Targeting of KRAS Using a Novel High-Affinity KRAS Antisense Oligonucleotide in Multiple Myeloma. Blood 2019, 134, 3104. [Google Scholar] [CrossRef]
- Shimojo, M.; Kasahara, Y.; Inoue, M.; Tsunoda, S.-i.; Shudo, Y.; Kurata, T.; Obika, S. A gapmer antisense oligonucleotide targeting SRRM4 is a novel therapeutic medicine for lung cancer. Sci. Rep. 2019, 9, 7618. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Barsoumian, H.B.; Puebla-Osorio, N.; Hu, Y.; Sezen, D.; Wasley, M.D.; Bertolet, G.; Zhang, J.; Leuschner, C.; Yang, L.; et al. Inhibition of STAT6 with Antisense Oligonucleotides Enhances the Systemic Antitumor Effects of Radiotherapy and Anti–PD-1 in Metastatic Non–Small Cell Lung Cancer. Cancer Immunol. Res. 2023, 11, 486–500. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, H.; Zhao, Z.; Jin, Y.; Zhang, Z.; Tan, J.; Hu, F. Gene-microRNA Network Analysis Identified Seven Hub Genes in Association with Progression and Prognosis in Non-Small Cell Lung Cancer. Genes 2022, 13, 1480. [Google Scholar] [CrossRef]
- Hiromi, I.; Yuuya, K.; Harumi, Y.; Susumu, K.; Akihiro, K.; Satoshi, O.; Susumu, N. Administration of Gapmer-type Antisense Oligonucleotides Targeting γ-Glutamylcyclotransferase Suppresses the Growth of A549 Lung Cancer Xenografts. Anticancer Res. 2022, 42, 1221. [Google Scholar]
- Wan, J.-L.; Wang, B.; Wu, M.-L.; Li, J.; Gong, R.-M.; Song, L.-N.; Zhang, H.-S.; Zhu, G.-Q.; Chen, S.-P.; Cai, J.-L.; et al. MTDH antisense oligonucleotides reshape the immunosuppressive tumor microenvironment to sensitize Hepatocellular Carcinoma to immune checkpoint blockade therapy. Cancer Lett. 2022, 541, 215750. [Google Scholar] [CrossRef]
- Jiang, T.; Zhou, C.; Ren, S. Role of IL-2 in cancer immunotherapy. Oncoimmunology 2016, 5, e1163462. [Google Scholar] [CrossRef]
- Lee, H.K.; Nam, M.-W.; Go, R.-E.; Koo, J.; Kim, T.H.; Park, J.-E.; Choi, K.-C. TGF-β2 antisense oligonucleotide enhances T-cell mediated anti-tumor activities by IL-2 via attenuation of fibrotic reaction in a humanized mouse model of pancreatic ductal adenocarcinoma. Biomed. Pharmacother. 2023, 159, 114212. [Google Scholar] [CrossRef] [PubMed]
- Bayraktar, E.; Bayraktar, R.; Oztatlici, H.; Lopez-Berestein, G.; Amero, P.; Rodriguez-Aguayo, C. Targeting miRNAs and Other Non-Coding RNAs as a Therapeutic Approach: An Update. Noncoding RNA 2023, 9, 27. [Google Scholar] [CrossRef]
- Xu, L.; Dai, W.Q.; Xu, X.F.; Wang, F.; He, L.; Guo, C.Y. Effects of multiple-target anti-microRNA antisense oligodeoxyribonucleotides on proliferation and migration of gastric cancer cells. Asian Pac. J. Cancer Prev. 2012, 13, 3203–3207. [Google Scholar] [CrossRef]
- Codiak BioSciences. A Study of exoASO-STAT6 (CDK-004) in Patients with Advanced Hepatocellular Carcinoma (HCC) and Patients with Liver Metastases from EIther Primary Gastric Cancer or Colorectal Cancer (CRC); Codiak BioSciences: Cambridge, MA, USA, 2022. [Google Scholar]
- Zhou, B.; Gao, Y.; Zhang, P.; Chu, Q. Acquired Resistance to Immune Checkpoint Blockades: The Underlying Mechanisms and Potential Strategies. Front. Immunol. 2021, 12, 693609. [Google Scholar] [CrossRef]
- Robert, D.L.; Leisha, A.E. Targeting adenosine for cancer immunotherapy. J. ImmunoTherapy Cancer 2018, 6, 57. [Google Scholar]
- Kashyap, A.S.; Thelemann, T.; Klar, R.; Kallert, S.M.; Festag, J.; Buchi, M.; Hinterwimmer, L.; Schell, M.; Michel, S.; Jaschinski, F.; et al. Antisense oligonucleotide targeting CD39 improves anti-tumor T cell immunity. J. Immunother. Cancer 2019, 7, 67. [Google Scholar] [CrossRef] [PubMed]
- Revenko, A.; Carnevalli, L.S.; Sinclair, C.; Johnson, B.; Peter, A.; Taylor, M.; Hettrick, L.; Chapman, M.; Klein, S.; Solanki, A.; et al. Direct targeting of FOXP3 in Tregs with AZD8701, a novel antisense oligonucleotide to relieve immunosuppression in cancer. J. Immunother. Cancer 2022, 10, e003892. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Rodriguez, L.; Cianciaruso, C.; Bill, R.; Trefny, M.P.; Klar, R.; Kirchhammer, N.; Buchi, M.; Festag, J.; Michel, S.; Kohler, R.H.; et al. Dual TLR9 and PD-L1 targeting unleashes dendritic cells to induce durable antitumor immunity. J. Immunother. Cancer 2023, 11, e006714. [Google Scholar] [CrossRef]
- Kamerkar, S.; Leng, C.; Burenkova, O.; Jang, S.C.; McCoy, C.; Zhang, K.; Dooley, K.; Kasera, S.; Zi, T.; Sisó, S.; et al. Exosome-mediated genetic reprogramming of tumor-associated macrophages by exoASO-STAT6 leads to potent monotherapy antitumor activity. Sci. Adv. 2022, 8, eabj7002. [Google Scholar] [CrossRef]
- Du, H.; Zhao, J.; Hai, L.; Wu, J.; Yi, H.; Shi, Y. The roles of vasohibin and its family members: Beyond angiogenesis modulators. Cancer Biol. Ther. 2017, 18, 827–832. [Google Scholar] [CrossRef]
- Horie, S.; Suzuki, Y.; Yamamoto, T.; Obika, S.; Mohri, K.; Kiyota, C.; Ren, Q.; Warashina, S.; Wada, Y.; Watanabe, Y.; et al. Novel strategy of liver cancer treatment with modified antisense oligonucleotides targeting human vasohibin-2. Cancer Sci. 2023, 114, 3740–3749. [Google Scholar] [CrossRef] [PubMed]
- Setoguchi, K.; Cui, L.; Hachisuka, N.; Obchoei, S.; Shinkai, K.; Hyodo, F.; Kato, K.; Wada, F.; Yamamoto, T.; Harada-Shiba, M.; et al. Antisense Oligonucleotides Targeting Y-Box Binding Protein-1 Inhibit Tumor Angiogenesis by Downregulating Bcl-xL-VEGFR2/-Tie Axes. Mol. Ther. Nucleic Acids 2017, 9, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, K.; Shintani, T.; Hayashido, Y.; Hamada, A.; Higaki, M.; Yoshioka, Y.; Sakamoto, A.; Yanamoto, S.; Okamoto, T. VEGF-A promotes the motility of human melanoma cells through the VEGFR1-PI3K/Akt signaling pathway. In Vitro Cell. Dev. Biol. Anim. 2022, 58, 758–770. [Google Scholar] [CrossRef] [PubMed]
- University of Southern California. A Study of VEGF-Antisense Oligonucleotide in Combination with Pemetrexed and Cisplatin for the Treatment of Advanced Malignant Mesothelioma; University of Southern California: Los Angeles, CA, USA, 2011. [Google Scholar]
- Zhejiang Haichang Biotech Co., Ltd. A Phase I First in Human Study to Evaluate the Safety, Tolerability, and Pharmacokinetics of WGI-0301 in Patients with Advanced Solid Tumors; Zhejiang Haichang Biotech Co., Ltd.: Hangzhou, China, 2022. [Google Scholar]
- Bio-Path Holdings, Inc. A Clinical Trial of BP1002 in Patients with Advanced Lymphoid Malignancies; Bio-Path Holdings, Inc.: Houston, TX, USA, 2020. [Google Scholar]
- Bio-Path Holdings, Inc. A Clinical Trial of BP1002 in Patients with Refractory/Relapsed Acute Myeloid Leukemia (AML); Bio-Path Holdings, Inc.: Houston, TX, USA, 2022. [Google Scholar]
- Genta Incorporated. Phase I/II Study of Genasense in Patients with Chronic Lymphocytic Leukemia; Genta Incorporated: Berkeley Heights, NJ, USA, 2001. [Google Scholar]
- National Cancer Institute. Oblimersen and Interferon Alfa in Treating Patients with Metastatic Renal Cell Cancer; National Cancer Institute: Bethesda, MD, USA, 2003.
- University of Chicago; National Cancer Institute. Bcl-2 Antisense Oligodeoxynucleotide G3139 and Paclitaxel in Treating Patients with Recurrent Small Cell Lung Cancer; University of Chicago: Chicago, IL, USA, 2000. [Google Scholar]
- Genta Incorporated; National Cancer Institute. Dacarbazine with or without Oblimersen (G3139) in Treating Patients with Advanced Malignant Melanoma; National Cancer Institute: Bethesda, MD, USA, 2000.
- National Cancer Institute. Combination Chemotherapy Plus Oblimersen in Treating Patients with Advanced Solid Tumors; National Cancer Institute: Bethesda, MD, USA, 2002.
- National Cancer Institute. Combination Chemotherapy Plus Oblimersen in Treating Patients with Previously Untreated Extensive-Stage Small Cell Lung Cancer; National Cancer Institute: Bethesda, MD, USA, 2001.
- National Cancer Institute. Oblimersen Plus Doxorubicin and Docetaxel in Treating Patients with Metastatic or Locally Advanced Breast Cancer; National Cancer Institute: Bethesda, MD, USA, 2003.
- British Columbia Cancer Agency; National Cancer Institute. Oblimersen, Rituximab, Cyclophosphamide, Doxorubicin, Vincristine, and Prednisone in Treating Patients with Stage II, Stage III, or Stage IV Diffuse Large B-Cell Lymphoma; National Cancer Institute: Bethesda, MD, USA, 2003.
- Genta Incorporated; National Cancer Institute. Dexamethasone with or without Oblimersen in Treating Patients with Relapsed or Refractory Multiple Myeloma; National Cancer Institute: Bethesda, MD, USA, 2000.
- Jonsson Comprehensive Cancer Center; National Cancer Institute. Oblimersen and Dacarbazine in Treating Patients with Advanced Malignant Melanoma That Has Responded to Treatment on Clinical Trial GENTA-GM301; National Cancer Institute: Bethesda, MD, USA, 2003.
- Genta Incorporated. A Phase I Study of G3139 Subcutaneous in Solid Tumors; Genta Incorporated: Berkeley Heights, NJ, USA, 2005. [Google Scholar]
- Genta Incorporated; National Cancer Institute. Docetaxel with or without Oblimersen in Treating Patients with Non-Small Cell Lung Cancer; Genta Incorporated: Berkeley Heights, NJ, USA, 2001. [Google Scholar]
- European Organization for Research Treatment of Cancer. Docetaxel with or without Oblimersen in Treating Patients with Hormone-Refractory Adenocarcinoma (Cancer) of the Prostate. 2004. Available online: https://ctv.veeva.com/study/docetaxel-with-or-without-oblimersen-in-treating-patients-with-hormone-refractory-adenocarcinoma-ca (accessed on 16 August 2024).
- Genta Incorporated; National Cancer Institute. Fludarabine and Cyclophosphamide with or without Oblimersen in Treating Patients with Relapsed or Refractory Chronic Lymphocytic Leukemia; National Cancer Institute: Bethesda, MD, USA, 2001.
- NCIC Clinical Trials Group; Canadian Cancer Trials Group. Hormone Therapy and OGX-011 Before Radical Prostatectomy in Treating Patients with Prostate Cancer; Canadian Cancer Trials Group: Kingston, ON, Canada, 2002. [Google Scholar]
- The University of Texas Health Science Center at San Antonio; National Cancer Institute. Oblimersen and Irinotecan in Treating Patients with Metastatic or Recurrent Colorectal Cancer; National Cancer Institute: Bethesda, MD, USA, 2000.
- National Cancer Institute. S0349 Rituximab, Cyclophosphamide, Doxorubicin, Vincristine, and Prednisone with or without Oblimersen in Treating Patients with Advanced Diffuse Large B-Cell Non-Hodgkin’s Lymphoma; National Cancer Institute: Bethesda, MD, USA, 2004.
- NCIC Clinical Trials Group; Canadian Cancer Trials Group. OGX-011 and Docetaxel in Treating Patients with Metastatic or Locally Recurrent Solid Tumors; Canadian Cancer Trials Group: Kingston, ON, Canada, 2003. [Google Scholar]
- NCIC Clinical Trials Group; Canadian Cancer Trials Group. OGX-011 and Docetaxel in Treating Women with Locally Advanced or Metastatic Breast Cancer; Canadian Cancer Trials Group: Kingston, ON, Canada, 2005. [Google Scholar]
- Achieve Life Sciences; Teva Pharmaceuticals USA. A Study Evaluating the Pain Palliation Benefit of Adding Custirsen to Docetaxel Retreatment or Cabazitaxel as Second Line Therapy in Men with Metastatic Castrate Resistant Prostate Cancer (mCRPC); Achieve Life Sciences: Bothell, WA, USA, 2010. [Google Scholar]
- Abramson Cancer Center of the University of Pennsylvania. Infusional C-myb ASODN in Advanced Hematologic Malignancies. 2002. Available online: https://clin.larvol.com/trial-detail/NCT00780052 (accessed on 16 August 2024).
- AstraZeneca. First Time in Human Study of AZD8701 with or without Durvalumab in Participants with Advanced Solid Tumours; AstraZeneca: Cambridge, UK, 2020. [Google Scholar]
- Bio-Path Holdings, Inc. Clinical Trial of BP1001 (Liposomal Grb2 Antisense Oligonucleotide) in Combination With Dasatinib in Patients With Ph + CML Who Have Failed TKI, Ph+ AML, Ph+ MDS. 01-15-2023. 2017. Available online: https://ckb.jax.org/clinicalTrial/show?nctId=NCT02923986 (accessed on 16 August 2024).
- Enzon Pharmaceuticals, Inc. Phase 1 Study of EZN-2968 Weekly in Adult Patients with Advanced Solid Tumors or Lymphoma; Enzon Pharmaceuticals, Inc.: Cranford, NJ, USA, 2007. [Google Scholar]
- National Cancer Institute; National Institutes of Health Clinical Center. A Pilot Study of EZN-2968, an Antisense Oligonucleotide Inhibitor of HIF-1alpha, in Adults with Advanced Solid Tumors with Liver Metastases; National Cancer Institute: Bethesda, MD, USA, 2010.
- Chi, K.N.; Yu, E.Y.; Jacobs, C.; Bazov, J.; Kollmannsberger, C.; Higano, C.S.; Mukherjee, S.D.; Gleave, M.E.; Stewart, P.S.; Hotte, S.J. A phase I dose-escalation study of apatorsen (OGX-427), an antisense inhibitor targeting heat shock protein 27 (Hsp27), in patients with castration-resistant prostate cancer and other advanced cancers. Ann. Oncol. 2016, 27, 1116–1122. [Google Scholar] [CrossRef] [PubMed]
- Achieve Life Sciences. Safety Study of an Antisense Product in Prostate, Ovarian, NSCL, Breast or Bladder Cancer; Achieve Life Sciences: Bothell, WA, USA, 2007. [Google Scholar]
- British Columbia Cancer Agency; Achieve Life Sciences. OGX-427 in Castration Resistant Prostate Cancer Patients; Achieve Life Sciences: Bothell, WA, USA, 2010. [Google Scholar]
- Noah Hahn; Achieve Life Sciences; Hoosier Cancer Research Network. Phase 2 Study of Docetaxel +/− OGX-427 in Patients with Relapsed or Refractory Metastatic Bladder Cancer; Achieve Life Sciences: Bothell, WA, USA, 2013. [Google Scholar]
- Imvax, A. Phase 2b Clinical Study with a Combination Immunotherapy in Newly Diagnosed Patients with Glioblastoma—The ImmuneSense Study; Imvax A: Philadelphia, PA, USA, 2022. [Google Scholar]
- AstraZeneca. Phase I Dose-Escalation Study of AZD4785 in Patients with Advanced Solid Tumours; AstraZeneca: Cambridge, UK, 2017. [Google Scholar]
- Eleos, Inc. Aezea® (Cenersen) and Chemotherapy for AML Subjects ≥ 55 Years of Age with No Response to Frontline Induction Course; Eleos, Inc.: San Ramon, CA, USA, 2012. [Google Scholar]
- Eleos, Inc. A Study of Aezea® (Cenersen) in Transfusion Dependent Anemia Associated with Myelodysplastic Syndrome (MDS); Eleos, Inc.: San Ramon, CA, USA, 2014. [Google Scholar]
- Eleos, Inc. Dosing Study of Ara-C/EL625/Idarubicin in Refractory and Relapsed AML; Eleos, Inc.: San Ramon, CA, USA, 2004. [Google Scholar]
- Eleos, Inc. EL625 in Persistent Chronic Lymphocytic Leukemia or Small Lymphocytic Lymphoma; Eleos, Inc.: San Ramon, CA, USA, 2008. [Google Scholar]
- Aptose Biosciences Inc.; Wake Forest University; University of Chicago. Combination of Capecitabine and GTI-2040 in the Treatment of Renal Cell Carcinoma; Aptose Biosciences Inc.: Toronto, ON, Canada, 2002. [Google Scholar]
- Lee, Y.; Vassilakos, A.; Feng, N.; Lam, V.; Xie, H.; Wang, M.; Jin, H.; Xiong, K.; Liu, C.; Wright, J.; et al. GTI-2040, an antisense agent targeting the small subunit component (R2) of human ribonucleotide reductase, shows potent antitumor activity against a variety of tumors. Cancer Res. 2003, 63, 2802–2811. [Google Scholar]
- Aptose Biosciences Inc.; The Ohio State University. Combination of GTI-2040 and Cytarabine in the Treatment of Refractory and Relapsed Acute Myeloid Leukemia (AML); The Ohio State University: Columbus, OH, USA, 2007. [Google Scholar]
- National Cancer Institute. GTI-2040 and High-Dose Cytarabine in Treating Patients with Refractory or Relapsed Acute Myeloid Leukemia; National Cancer Institute: Bethesda, MD, USA, 2003.
- National Cancer Institute. GTI-2040 and Docetaxel in Treating Patients with Recurrent, Metastatic, or Unresectable Locally Advanced Non-Small Cell Lung Cancer, Prostate Cancer, or Other Solid Tumors; National Cancer Institute: Bethesda, MD, USA, 2003.
- National Cancer Institute. GTI-2040, Oxaliplatin, and Capecitabine in Treating Patients with Locally Advanced or Metastatic Colorectal Cancer or Other Solid Tumors; National Cancer Institute: Bethesda, MD, USA, 2004.
- National Cancer Institute. GTI-2040 and Gemcitabine in Treating Patients with Metastatic or Unresectable Solid Tumors; National Cancer Institute: Bethesda, MD, USA, 2004.
- National Cancer Institute. GTI-2040 in Treating Patients with Relapsed, Refractory, or High-Risk Acute Leukemia, High-Grade Myelodysplastic Syndromes, or Refractory or Blastic Phase Chronic Myelogenous Leukemia; National Cancer Institute: Bethesda, MD, USA, 2007.
- University Health Network; National Cancer Institute. GTI-2040, Docetaxel, and Prednisone in Treating Patients with Prostate Cancer; National Cancer Institute: Bethesda, MD, USA, 2005.
- National Cancer Institute. GTI-2040 and Capecitabine in Treating Patients with Metastatic Breast Cancer; National Cancer Institute: Bethesda, MD, USA, 2003.
- Eastern Cooperative Oncology Group; National Cancer Institute. Chemotherapy in Treating Women with Previously Treated Metastatic Breast Cancer; National Cancer Institute: Bethesda, MD, USA, 1998.
- INSYS Therapeutics Inc. Study to Determine the Maximum Tolerated Dose of LErafAON in Patients with Advanced Cancer; INSYS: Phoenix, AZ, USA, 2004. [Google Scholar]
- INSYS Therapeutics Inc.; Georgetown University. Study to Determine the Maximum Tolerated Dose of LErafAON in Patients with Advanced Solid Tumors; INSYS: Phoenix, AZ, USA, 2001. [Google Scholar]
- INSYS Therapeutics Inc.; Georgetown University. Study to Determine Maximum Tolerated Dose of LErafAON Combined with Radiotherapy in Patients with Advanced Malignancies; INSYS: Phoenix, AZ, USA, 2001. [Google Scholar]
- Ionis Pharmaceuticals, Inc.; AstraZeneca. Phase 1/2, Open-label, Dose-Escalation Study of IONIS-STAT3Rx, Administered to Patients with Advanced Cancers; AstraZeneca: Cambridge, UK, 2012. [Google Scholar]
- AstraZeneca; Ionis Pharmaceuticals, Inc. A Phase I/Ib Study of AZD9150 (ISIS-STAT3Rx) in Patients with Advanced/Metastatic Hepatocellular Carcinoma; AstraZeneca: Cambridge, UK, 2013. [Google Scholar]
- AstraZeneca. AZD9150 Plus Durvalumab Alone or in Combination with Chemotherapy in Patients with Advanced, Solid Tumours and in Patients with Non-Small-Cell Lung Cancer; AstraZeneca: Cambridge, UK, 2018. [Google Scholar]
- AstraZeneca. Study of AZD9150 and MEDI4736 (Durvalumab) in Japanese Adult Patients with Advanced Solid Malignancies; AstraZeneca: Cambridge, UK, 2018. [Google Scholar]
- MedImmune, LLC. MEDI4736 Alone and in Combination with Tremelimumab or AZD9150 in Adult Subjects with Relapsed/Refractory DLBCL (D4190C00023); MedImmune, LLC: Gaithersburg, MD, USA, 2016. [Google Scholar]
- National Cancer Institute; National Institutes of Health Clinical Center. AZD9150, a STAT3 Antisense Oligonucleotide, in People with Malignant Ascites; National Cancer Institute: Bethesda, MD, USA, 2015.
- AstraZeneca. Study to Assess MEDI4736 with Either AZD9150 or AZD5069 in Advanced Solid Tumors & Relapsed Metastatic Squamous Cell Carcinoma of Head & Neck; AstraZeneca: Cambridge, UK, 2015. [Google Scholar]
- Montefiore Medical Center; Malaysia Digital Arrival Card (MDAC); Flamingo Therapeutics. Danvatirsen Monotherapy Followed by Combination with Venetoclax in Relapsed/Refractory MDS & AML; Malaysia Digital Arrival Card (MDAC): Putrajaya, Malaysia, 2024. [Google Scholar]
- AP B.V.; AstraZeneca. Platform Study for the Treatment of Relapsed or Refractory Aggressive Non-Hodgkin’s Lymphoma (PRISM Study); AstraZeneca: Cambridge, UK, 2018. [Google Scholar]
- AstraZeneca. Open-Label, Randomised, Multi-Drug, Biomarker-Directed, Phase 1b Study in Pts w/ Muscle Invasive Bladder Cancer; AstraZeneca: Cambridge, UK, 2016. [Google Scholar]
- AstraZeneca. Phase II Umbrella Study of Novel Anti-cancer Agents in Patients with NSCLC Who Progressed on an Anti-PD-1/PD-L1 Containing Therapy; AstraZeneca: Cambridge, UK, 2017. [Google Scholar]
- Autotelicbio. TASO-001 in Combination with Recombinant Interleukin-2(Aldesleukin) in Advanced or Metastatic Solid Tumor; Autotelicbio: Cheongju-si, Republic of Korea, 2021. [Google Scholar]
- Levy Restaurants; Bristol-Myers Squibb; Stanford University. SD-101 and BMS-986178 in Treating Patients with Advanced or Metastatic Solid Malignancies; Levy Restaurants: Chicago, IL, USA, 2019. [Google Scholar]
- TriSalus Life Sciences, Inc. Pressure Enabled Intrapancreatic Delivery of SD-101 with Checkpoint Blockade for Locally Advanced Pancreatic Adenocarcinoma; TriSalus Life Sciences, Inc.: Westminster, CO, USA, 2023. [Google Scholar]
- University of California, Davis; National Cancer Institute; Bristol Myers Squibb; DT Corporation. SD-101, Nivolumab, and Radiation Therapy in Treating Patients with Chemotherapy-Refractory Metastatic Pancreatic Cancer; National Cancer Institute: Bethesda, MD, USA, 2019.
- Merck Sharp & Dohme LLC. Dose Evaluation of MK-1966 in Combination with SD-101 in Participants with Advanced Malignancies (MK-1966-001); Merck Sharp & Dohme LLC: Rahway, NJ, USA, 2016. [Google Scholar]
- TriSalus Life Sciences, Inc. Pressure Enabled Delivery of SD-101 with Checkpoint Blockade for Primary Liver Tumors; TriSalus Life Sciences, Inc.: Westminster, CO, USA, 2022. [Google Scholar]
- Levy Restaurants; National Cancer Institute; Stanford University. TLR9 Agonist SD-101, Anti-OX40 Antibody BMS 986178, and Radiation Therapy in Treating Patients with Low-Grade B-Cell Non-Hodgkin Lymphomas; National Cancer Institute: Bethesda, MD, USA, 2018.
- Lowsky, R.; Janssen, L.; National Cancer Institute; Leukemia, T.; Society, L.; Foundation, R.T.; Stanford University. TLR9 Agonist SD-101, Ibrutinib, and Radiation Therapy in Treating Patients with Relapsed or Refractory Grade 1-3A Follicular Lymphoma; Stanford University: Stanford, CA, USA, 2016. [Google Scholar]
- TriSalus Life Sciences, Inc. Intrahepatic Delivery of SD-101 by Pressure-Enabled Regional Immuno-Oncology (PERIO), with Checkpoint Blockade in Adults with Metastatic Uveal Melanoma; TriSalus Life Sciences, Inc.: Westminster, CO, USA, 2021. [Google Scholar]
- Idera Pharmaceuticals, Inc. Study of IMO-2055 in Metastatic or Locally Recurrent Clear Cell Renal Carcinoma; Idera Pharmaceuticals, Inc.: Cambridge, MA, USA, 2004. [Google Scholar]
- EMD Serono. Study of FOLFIRI Plus Cetuximab Plus IMO-2055 in Patients with Colorectal Cancer; EMD Serono: Rockland, MA, USA, 2009. [Google Scholar]
- EMD Serono. Safety of Adding IMO-2055 to Erlotinib + Bevacizumab in 2nd Line Treatment for Patients with NSCLC; EMD Serono: Rockland, MA, USA, 2007. [Google Scholar]
- EMD Serono. EMD 1201081 in Combination with Cetuximab in Second-Line Cetuximab-Naïve Subjects with Recurrent or Metastatic Squamous Cell Carcinoma of the Head and Neck; EMD Serono: Rockland, MA, USA, 2009. [Google Scholar]
- Merck Sharp & Dohme LLC. EMD 1201081 + 5-FU + Cisplatin + Cetuximab in Subjects with Recurrent Metastatic Squamous Cell Carcinoma of the Head and Neck; Merck Sharp & Dohme LLC: Rahway, NJ, USA, 2010. [Google Scholar]
- Aegera Therapeutics. A Phase 1–2, XIAP Antisense AEG35156 with Gemcitabine in Patients with Advanced Pancreatic Cancer; Aegera Therapeutics: Montreal, QC, Canada, 2007. [Google Scholar]
- Aegera Therapeutics. A Phase 1–2, XIAP Antisense AEG35156 with Weekly Paclitaxel in Patients with Advanced Breast Cancer; Aegera Therapeutics: Montreal, QC, Canada, 2007. [Google Scholar]
- NCIC Clinical Trials Group; Canadian Cancer Trials Group. AEG35156 and Docetaxel in Treating Patients with Solid Tumors; Canadian Cancer Trials Group: Kingston, ON, Canada, 2005. [Google Scholar]
- NCIC Clinical Trials Group; Canadian Cancer Trials Group. AEG35156 and Docetaxel in Treating Patients with Locally Advanced, Metastatic, or Recurrent Solid Tumors; Canadian Cancer Trials Group: Kingston, ON, Canada, 2006. [Google Scholar]
- Aegera Therapeutics. Study of XIAP Antisense for Advanced Cancers; Aegera Therapeutics: Montreal, QC, Canada, 2006. [Google Scholar]
- Aegera Therapeutics. AEG35156 in Combination with High-Dose Cytarabine and Idarubicin in AML Following Failure of a Single Standard Dose Cytarabine Based Frontline Induction Regimen; Aegera Therapeutics: Montreal, QC, Canada, 2009. [Google Scholar]
- Aegera Therapeutics; Leukemia, T.; Society, L. A Phase 1–2, Multicenter, Open-Label Study of AEG35156 in Patients with Relapsed or Refractory Chronic Lymphocytic Leukemia and Indolent B-Cell Lymphomas; Aegera Therapeutics: Montreal, QC, Canada, 2008. [Google Scholar]
- Aegera Therapeutics. Study of XIAP Antisense Given with Chemotherapy for Refractory/Relapsed AML; Aegera Therapeutics: Montreal, QC, Canada, 2005. [Google Scholar]
- Aegera Therapeutics. XIAP Antisense AEG35156 in Combination with Sorafenib in Patients with Advanced Hepatocellular Carcinoma (HCC); Aegera Therapeutics: Montreal, QC, Canada, 2009. [Google Scholar]
- Aegera Therapeutics. A Phase 1–2 XIAP Antisense AEG35156 with Carboplatin and Paclitaxel in Patients with Advanced Non-Small Cell Lung Cancer; Aegera Therapeutics: Montreal, QC, Canada, 2007. [Google Scholar]
- Oncotelic Inc.; Mateon Therapeutics. OT-101 in Combination with Pembrolizumab in Subjects with Malignant Pleural Mesothelioma Failing to Respond to Checkpoint Inhibition; Oncotelic Inc.: Agoura Hills, CA, USA, 2023. [Google Scholar]
- Oncotelic Inc.; Mateon Therapeutics. A Study of OT-101 with FOLFIRINOX in Patients with Advanced and Unresectable or Metastatic Pancreatic Cancer; Oncotelic Inc.: Agoura Hills, CA, USA, 2023. [Google Scholar]
- University of Washington; Genentech, Inc.; Oncotelic Therapeutics, Inc. OT-101 in Combination with Atezolizumab for the Treatment of Metastatic or Recurrent Non-Small Cell Lung Cancer; University of Washington: Seattle, WA, USA, 2023. [Google Scholar]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef]
- Kuijper, E.C.; Bergsma, A.J.; Pijnappel, W.W.M.P.; Aartsma-Rus, A. Opportunities and challenges for antisense oligonucleotide therapies. J. Inherit. Metab. Dis. 2021, 44, 72–87. [Google Scholar] [CrossRef]
- Yoshida, T.; Naito, Y.; Yasuhara, H.; Sasaki, K.; Kawaji, H.; Kawai, J.; Naito, M.; Okuda, H.; Obika, S.; Inoue, T. Evaluation of off-target effects of gapmer antisense oligonucleotides using human cells. Genes. Cells 2019, 24, 827–835. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Guo, Y.; Wang, P.; Su, Y.; Jin, X.; Zhu, X.; Zhang, C. Drug-grafted DNA as a novel chemogene for targeted combinatorial cancer therapy. Exploration 2022, 2, 20210172. [Google Scholar] [CrossRef]
- Nishina, T.; Numata, J.; Nishina, K.; Yoshida-Tanaka, K.; Nitta, K.; Piao, W.; Iwata, R.; Ito, S.; Kuwahara, H.; Wada, T.; et al. Chimeric Antisense Oligonucleotide Conjugated to α-Tocopherol. Mol. Ther.—Nucleic Acids 2015, 4, e220. [Google Scholar] [CrossRef]
- Shadidi, M.; Sioud, M. Identification of novel carrier peptides for the specific delivery of therapeutics into cancer cells. FASEB J. 2003, 17, 256–258. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Okuda, T.; Kasahara, Y.; Obika, S. Base-modified aptamers obtained by cell-internalization SELEX facilitate cellular uptake of an antisense oligonucleotide. Mol. Ther. Nucleic Acids 2021, 23, 440–449. [Google Scholar] [CrossRef] [PubMed]
- Gong, N.; Teng, X.; Li, J.; Liang, X.J. Antisense Oligonucleotide-Conjugated Nanostructure-Targeting lncRNA MALAT1 Inhibits Cancer Metastasis. ACS Appl. Mater. Interfaces 2019, 11, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Future Market Insights, Inc. Antisense Oligonucleotides Market Outlook. Available online: https://www.futuremarketinsights.com/reports/antisense-oligonucleotides-market (accessed on 5 March 2024).
- Alcyone Therapeutics, Inc. Study of an Intrathecal Port and Catheter System for Subjects with Spinal Muscular Atrophy; Alcyone Therapeutics, Inc.: Lowell, MA, USA, 2023. [Google Scholar]
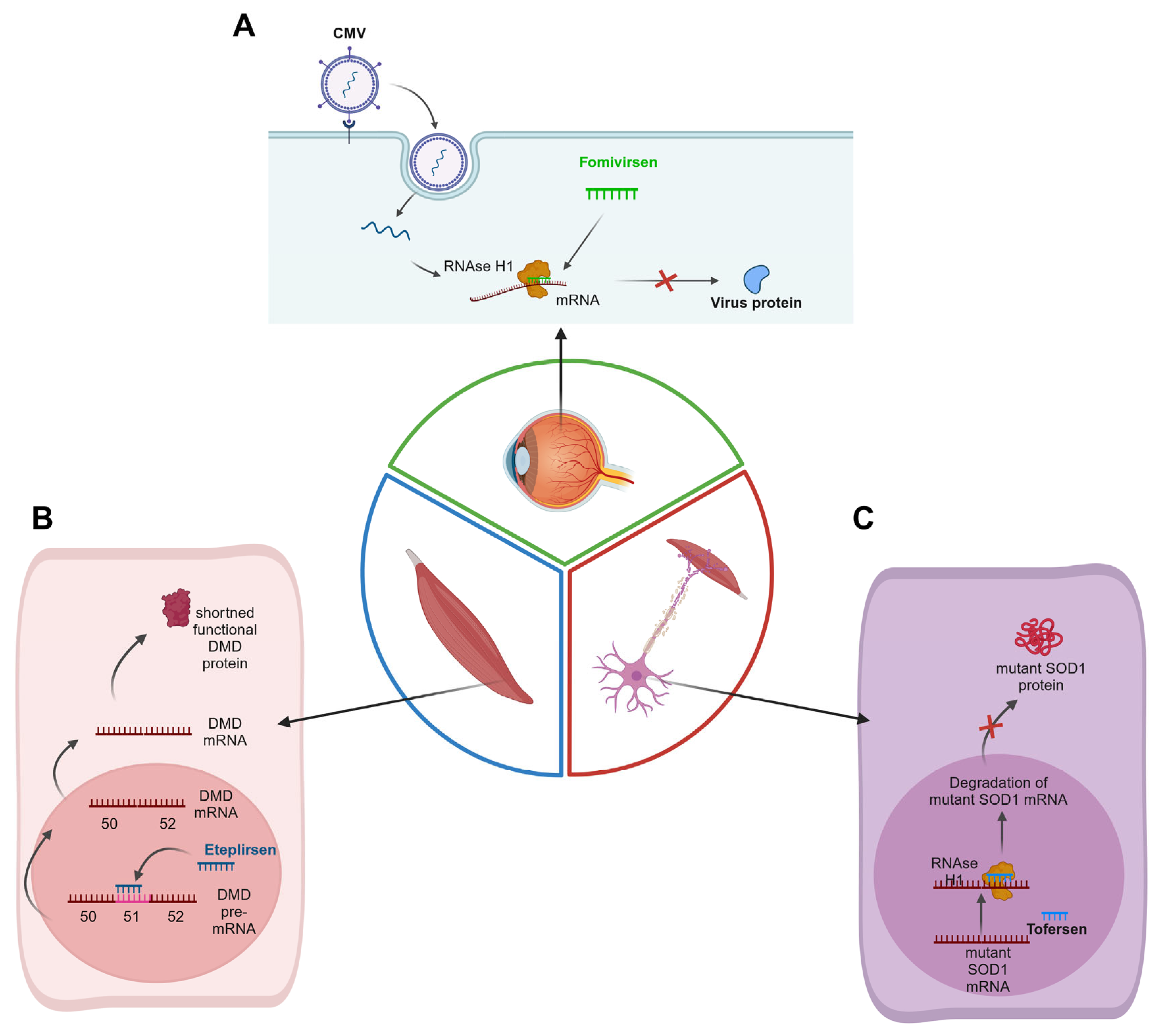
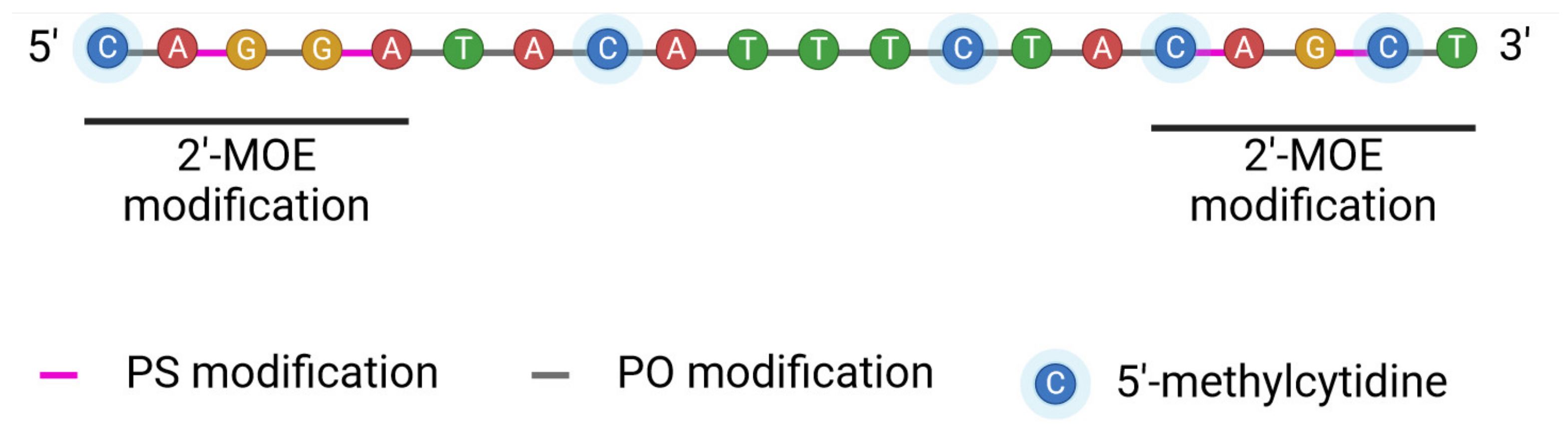
| ASO | Drug Name (Company) | Structure | Target | FDA Approval Year | EMA Approval Year | FDA/EMA Approved Use | Reference |
|---|---|---|---|---|---|---|---|
| Fomivirsen | Vitravene (Isis Pharmaceuticals, Novartis, Carlsbad, CA, USA) | 21-mer | UL123 mRNA | 1998 (withdrawn in 2001 due to reduced incidence of AIDS-related CMVR) | 1999 (withdrawn in 2002 due to reduced incidence of AIDS-related CMVR) | Cytomegalovirus (CMV) retinitis in AIDS patients | [8] |
| Mipomirsen | Kynamro (Genzyme, Isis Pharmaceuticals, Carlsbad, CA, USA) | 20-mer | apoB-100 mRNA | 2013 | Not approved (possible liver damage, risk of cardiovascular events) | Familial hypercholesterolemia | [9,10,11] |
| Nusinersen | Spiranza (Biogen Inc. Cambridge, MA, USA) | 18-mer | 7th exon SMN2 | 2016 | 2017 | Spinal Muscular Atrophy (SMA) in pediatric and adult patients | [12,13,14] |
| Eteplirsen | Exondys 51 (Sarepta Therapeutics, Cambridge, MA, USA) | 30-mer | 51st exon of the DMD gene | 2016 | Not approved (not satisfactionary effect of the drug compared to placebo) | Duchenne Muscular Dystrophy (DMD) | [15,16] |
| Inotersen | Tegsedi (Ionis Pharmaceuticals, Akcea Therapeutics, Carlsbad, CA, USA) | 20-mer | TTR mRNA | 2018 | 2014 (approved as orphan drug) | Adult Patients with Hereditary Transthyretin Amyloidosis | [17,18] |
| Goldodirsen | Vyondys 53 (Sarepta Therapeutics, Cambridge, MA, USA) | 25-mer | 53rd exon of dystrophin pre-mRNA | 2019 | - | Duchenne Muscular Dystrophy (DMD) | [19] |
| Milasen | Milasen (TriLink, Brammer Bio, Cambridge, MA, USA) | 22-mer | 6th exon of MFSD8 | 2019 (personalized medicine for single patient) | - | Spinal Muscular Atrophy (SMA) | [20,21,22] |
| Volanesorsen | Waylivra (Ionis Pharmaceuticals, Akcea Therapeutics, Carlsbad, CA, USA) | 20-mer | apoC-III mRNA | Not approved (safety issues—thrombocytopenia) | 2019 | Familial Chylomicronemia Syndrome or Hypertriglyceridaemia | [23,24,25] |
| Viltolarsen | Viltepso (NS Pharma, Inc., Nippon Shinyaku, Kyoto City, Kyoto Prefecture, Japan) | 21-mer | 53rd exon of DMD gene | 2020 | 2022 (decision of approval of investigation plan) | Duchenne Muscular Dystrophy (DMD) | [26,27,28] |
| Casimersen | Amondys 45 (Sarepta Therapeutics, Cambridge, MA, USA) | 22-mer | 45th exon of DMD gene | 2021 | - | Duchenne Muscular Dystrophy (DMD) | [29] |
| Eplontersen | Wainua (Ionis Pharmaceuticals and AstraZeneca, Carlsbad, CA, USA) | 20-mer linked with N-acetyl galactosamine | TTR pre-mRNA | 2023 | 2023 (approved as orphan drug) | Polyneuropathy of Hereditary Transthyretin Amyloidosis (ATTR) | [30,31] |
| Tofersen | Qalsody (Biogen, Cambridge, MA, USA) | 20-mer | SOD1 | 2023 | 2024 (positive opinion) | Amyotrophic Lateral Sclerosis (ALS) | [32,33] |
| ASO Generation | Modification | Resistance to Nucleases | Half-Life | Charge | References |
|---|---|---|---|---|---|
| Not modified | Not applicable | + | ~1 h | Negative | [59,60] |
| I | An oxygen atom in the -P=O position is replaced by sulfur, methyl, or amine group | ++ | ~4–6 h | Negative | [59,60] |
| II | Modification of sugar in 2′-position including insertion of methyl or methoxylethyl group | +++ | 22 days | Multiple negative charges | [61] |
| III | Sugar modification and phosphate linkages | ++++ | ~20 h | Neutral | [59,60] |
| Type of Cancer | Gene or Protein Associated with Cancer | ASO | Recruitment Status and Phase of Clinical Trial | Reference |
|---|---|---|---|---|
| Solid tumors | Akt-1 | WGI-0301 | Recruiting, phase 1 | [184] |
| Lymphomas | Bcl-2 | BP1002 | Recruiting, phase 1 | [185] |
| Acute Myeloid Leukemia | Bcl-2 | BP1002 | Recruiting, phase 1 | [186] |
| Leukemia | Bcl-2 | G3139 (oblimersen) | Completed, phase 1/2 | [187] |
| Renal cancer | Bcl-2 | G3139 (oblimersen) | Completed, phase 2 | [188] |
| Lung cancer | Bcl-2 | G3139 (oblimersen) | Completed, phase 1/2 | [189] |
| Melanoma | Bcl-2 | G3139 (oblimersen) | Completed, phase 3 | [190] |
| Solid tumors | Bcl-2 | G3139 (oblimersen) | Completed, phase 1 | [191] |
| Small cell lung cancer | Bcl-2 | G3139 (oblimersen) | Completed, phase 1 | [192] |
| Breast cancer | Bcl-2 | G3139 (oblimersen) | Terminated, phase 1/2 | [193] |
| Lymphoma | Bcl-2 | G3139 (oblimersen) | Completed, phase 1 | [194] |
| Multiple myeloma | Bcl-2 | G3139 (oblimersen) | Completed, phase 3 | [195] |
| Melanoma | Bcl-2 | G3139 (oblimersen) | Unknown, not applicable | [196] |
| Solid tumors | Bcl-2 | G3139 (oblimersen) | Completed, phase 1 | [197] |
| Lung cancer | Bcl-2 | G3139 (oblimersen) | Unknown, phase 2/3 | [198] |
| Prostate cancer | Bcl-2 | G3139 (oblimersen) | Completed, phase 2 | [199] |
| Leukemia | Bcl-2 | G3139 (oblimersen) | Completed, phase 3 | [200] |
| Prostate cancer | Bcl-2 | OGX-011 | Completed, phase 1 | [201] |
| Colorectal cancer | Bcl-2 | G3139 (oblimersen) | Completed, phase 1/2 | [202] |
| Lymphoma | Bcl-2 | G3139 (oblimersen) | Terminated, phase 2 | [203] |
| Solid tumors | Clusterin | OGX-011 (custirsen sodium) | Completed, phase1 | [204] |
| Breast cancer | Clusterin | OGX-011 (custirsen sodium) | Completed, phase 2 | [205] |
| Prostate cancer | Clusterin | OGX-011 (custirsen sodium) | Terminated, phase 3 | [206] |
| Hematologic neoplasms | c-Myb | c-Myb AS ODN | Completed, phase 1 | [207] |
| Solid tumors | FOXP3 | AZD8701 | Active, not recruiting, phase 1 | [208] |
| Acute myeloid leukemia | Grb2 | BP1001 (Prexigebersen) | Recruiting, phase 2 | [129] |
| Lymphomas | Grb2 | BP1001 (Prexigebersen) | Completed, phase 1 | [128] |
| Lymphomas | Grb2 | BP1001 (Prexigebersen) | Withdrawn, phase 1/2 | [209] |
| Solid tumors | Grb2 | BP1001 (Prexigebersen) | Recruiting, phase 1 | [130] |
| Lymphomas | HIF-1α | EZN-2968 | Completed, phase 1 | [210] |
| Solid tumors | HIF-1α | EZN-2968 | Completed, phase 1 | [211] |
| Bladder cancer | Hsp27 | OGX-427 | Unknown, phase 1 | [212] |
| Cancers | Hsp27 | OGX-427 | Completed, phase 1 | [213] |
| Prostate cancer | Hsp27 | OGX-427 | Completed, phase 2 | [214] |
| Bladder carcinoma | Hsp27 | OGX-427 | Completed, phase 2 | [215] |
| Glioblastoma | IGF-1R | IMV-001 | Not yet recruiting, phase 2 | [216] |
| Solid tumors | KRAS | AZD4785 | Completed, phase 1 | [217] |
| Solid tumors | miRNA-221 | LNA-i-Mir-221 | Completed, phase 1 | [103] |
| Acute myeloid leukemia | p53 | Cenersen | Withdrawn, phase 2 | [218] |
| Myelodysplastic Syndromes | p53 | Cenersen | Terminated, phase 1 | [219] |
| Acute Myelogenous Leukemia | p53 | Cenersen | Completed, phase 2 | [220] |
| Chronic or Small Lymphocytic Leukemia | p53 | Cenersen | Terminated, phase 2 | [221] |
| Renal carcinoma | R2 subunit of ribonucleotide reductase | GTI-2040 | Completed, phase 1/2 | [222,223] |
| Acute Myeloid Leukemia | R2 subunit of ribonucleotide reductase | GTI-2040 | Completed, phase 2 | [224] |
| Acute Myeloid Leukemia | R2 subunit of ribonucleotide reductase | GTI-2040 | Completed, phase 1 | [225] |
| Solid tumors | R2 subunit of ribonucleotide reductase | GTI-2040 | Completed, phase 1/2 | [226] |
| Colorectal cancer and solid tumors | R2 subunit of ribonucleotide reductase | GTI-2040 | Completed, phase 1 | [227] |
| Solid tumors | R2 subunit of ribonucleotide reductase | GTI-2040 | Completed, phase 1 | [228] |
| Acute Leukemia | R2 subunit of ribonucleotide reductase | GTI-2040 | Completed, phase 1 | [229] |
| Prostate cancer | R2 subunit of ribonucleotide reductase | GTI-2040 | Completed phase 2 | [230] |
| Breast cancer | R2 subunit of ribonucleotide reductase | GTI-2040 | Completed, Phase 2 | [231] |
| Breast cancer | Raf-1 | ISIS 5132 | Completed, phase 2 | [232] |
| Ovarian Cancer | Raf-1 | ISIS 5132 | Completed, phase 2 | [146] |
| Cancers | Raf-1 | LErafAON-ETU | Completed, phase 1 | [233] |
| Cancers | Raf-1 | LErafAON | Completed, phase 1 | [234,235] |
| Lymphomas | STAT3 | ISIS 481464 | Completed, phase 1/2 | [236] |
| Gastrointestinal cancers | STAT3 | AZD9150 (Danvatirsen) | Terminated (not enough patients), phase 2 | [237] |
| Advanced Solid Tumors | STAT3 | AZD9150 (Danvatirsen) | Active nor recruiting, phase 1 | [238] |
| Advanced Solid Malignancies | STAT3 | AZD9150 (Danvatirsen) | Completed, phase 1 | [239] |
| Diffuse Large B-Cell Lymphoma | STAT3 | AZD9150 (Danvatirsen) | Completed, phase 1 | [240] |
| Ovarian Cancer | STAT3 | AZD9150 (Danvatirsen) | Terminated, phase 2 | [241] |
| Solid Tumors | STAT3 | AZD9150 (Danvatirsen) | Active not recruiting, phase 1/2 | [242] |
| AML/MDS | STAT3 | AZD9150 (Danvatirsen) | Active recruiting, phase 1 | [243] |
| NHL DLBCL Non-hodgkin’s Lymphoma | STAT3 | AZD9150 (Danvatirsen) | Completed, phase 1 | [244] |
| Muscle Invasive Bladder Cancer | STAT3 | AZD9150 (Danvatirsen) | Active not recruiting, phase 1 | [245] |
| Non-Small Cell Lung Cancer | STAT3 | AZD9150 (Danvatirsen) | Active not recruiting, phase 2 | [246] |
| Solid tumors | TGF-B2 | TGF-B2 (TASO-001) | Recruiting, phase 1 | [247] |
| Solid tumors | TLR9 | SD-101 | Active not recruiting, phase 1 | [248] |
| Pancreatic adenocarcinoma | TLR9 | SD-101 | Recruiting, phase 1 | [249] |
| Pancreatic adenocarcinoma | TLR9 | SD-101 | Completed, phase 1 | [250] |
| Advanced malignancies | TLR9 | SD-101 | Terminated, phase 1 | [251] |
| Liver tumors | TLR9 | SD-101 | Recruiting, phase 1/2 | [252] |
| Low Grade B cell Non Hodgkin Lymphomas | TLR9 | SD-101 | Active not recruiting, phase 1 | [253] |
| Follicular lymphoma | TLR9 | SD-101 | Active recruiting, phase 1/2 | [254] |
| Uveal Melanoma with metastasis in liver | TLR9 | SD-101 | Recruiting, phase 1 | [255] |
| Renal carcinoma | TLR9 | IMO-2055 (EMD 1201081) | Completed, phase 2 | [256] |
| Colorectal cancer | TLR9 | IMO-2055 (EMD 1201081) | Terminated, phase 1 | [257] |
| Non-Small Cell Lung Cancer | TLR9 | IMO-2055 (EMD 1201081) | Completed, phase 1 | [258] |
| Squamous Cell Carcinoma of the Head and Neck Cancer | TLR9 | IMO-2055 (EMD 1201081) | Completed, phase 2 | [259] |
| Squamous Cell Carcinoma of the Head and Neck | TLR9 | IMO-2055 (EMD 1201081) | Terminated, phase 1 | [260] |
| Mesothelioma | VEGF | VEGF-AS | Withdrawn, phase 1/2 | [183] |
| Pancreatic carcinoma | XIAP | AEG35156 | Terminated, phase 1/2 | [261] |
| Breast cancer | XIAP | AEG35156 | Terminated, phase 1/2 | [262] |
| Solid tumors | XIAP | AEG35156 | Completed, phase 1 | [263] |
| Solid tumors | XIAP | AEG35156 | Completed, phase 1 | [264] |
| Advanced cancers | XIAP | AEG35156 | Terminated, phase 1 | [265] |
| Leukemia | XIAP | AEG35156 | Terminated, phase 2 | [266] |
| Leukemia | XIAP | AEG35156 | Terminated, phase 1/2 | [267] |
| Leukemia | XIAP | AEG35156 | Completed, phase 1/2 | [268] |
| Hepatocellular Carcinoma | XIAP | AEG35156 | Completed, phase 1/2 | [269] |
| Non-Small-Cell Lung | XIAP | AEG35156 | Terminate, phase 1/2 | [270] |
| Malignant Pleural Mesothelioma | TGF-β | OT-101 (Trabedersen) | Not yet recruiting, phase 2 | [271] |
| Pancreatic cancer | TGF-β | OT-101 (Trabedersen) | Not yet recruiting, phase 2/3 | [272] |
| Lung Non-Small Cell Carcinoma | TGF-β | OT-101 (Trabedersen) | Withdrawn, phase 2 | [273] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Çakan, E.; Lara, O.D.; Szymanowska, A.; Bayraktar, E.; Chavez-Reyes, A.; Lopez-Berestein, G.; Amero, P.; Rodriguez-Aguayo, C. Therapeutic Antisense Oligonucleotides in Oncology: From Bench to Bedside. Cancers 2024, 16, 2940. https://doi.org/10.3390/cancers16172940
Çakan E, Lara OD, Szymanowska A, Bayraktar E, Chavez-Reyes A, Lopez-Berestein G, Amero P, Rodriguez-Aguayo C. Therapeutic Antisense Oligonucleotides in Oncology: From Bench to Bedside. Cancers. 2024; 16(17):2940. https://doi.org/10.3390/cancers16172940
Chicago/Turabian StyleÇakan, Elif, Olivia D. Lara, Anna Szymanowska, Emine Bayraktar, Arturo Chavez-Reyes, Gabriel Lopez-Berestein, Paola Amero, and Cristian Rodriguez-Aguayo. 2024. "Therapeutic Antisense Oligonucleotides in Oncology: From Bench to Bedside" Cancers 16, no. 17: 2940. https://doi.org/10.3390/cancers16172940
APA StyleÇakan, E., Lara, O. D., Szymanowska, A., Bayraktar, E., Chavez-Reyes, A., Lopez-Berestein, G., Amero, P., & Rodriguez-Aguayo, C. (2024). Therapeutic Antisense Oligonucleotides in Oncology: From Bench to Bedside. Cancers, 16(17), 2940. https://doi.org/10.3390/cancers16172940









