Enhancing Preoperative Outcome Prediction: A Comparative Retrospective Case–Control Study on Machine Learning versus the International Esodata Study Group Risk Model for Predicting 90-Day Mortality in Oncologic Esophagectomy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Setting and Study Population
2.2. IESG Risk Model Validation
2.3. ML Training and Validation
2.4. Feature Importance Analysis
2.5. Comparison between IESG Model and ML Classifiers
3. Results
3.1. Patient Baseline Characteristics
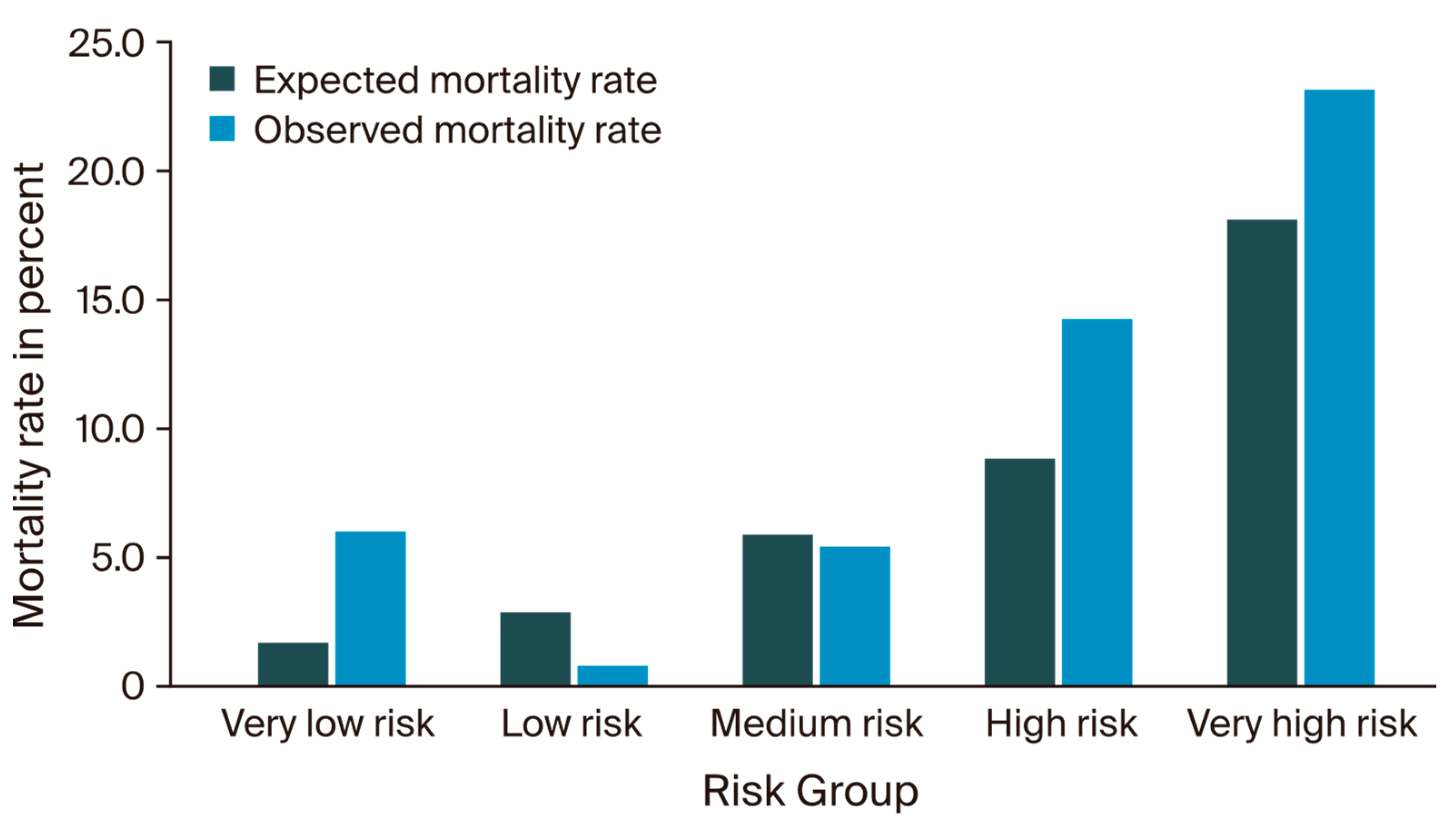
3.2. IESG Risk Model Evaluation
3.3. Machine Learning Model Evaluation
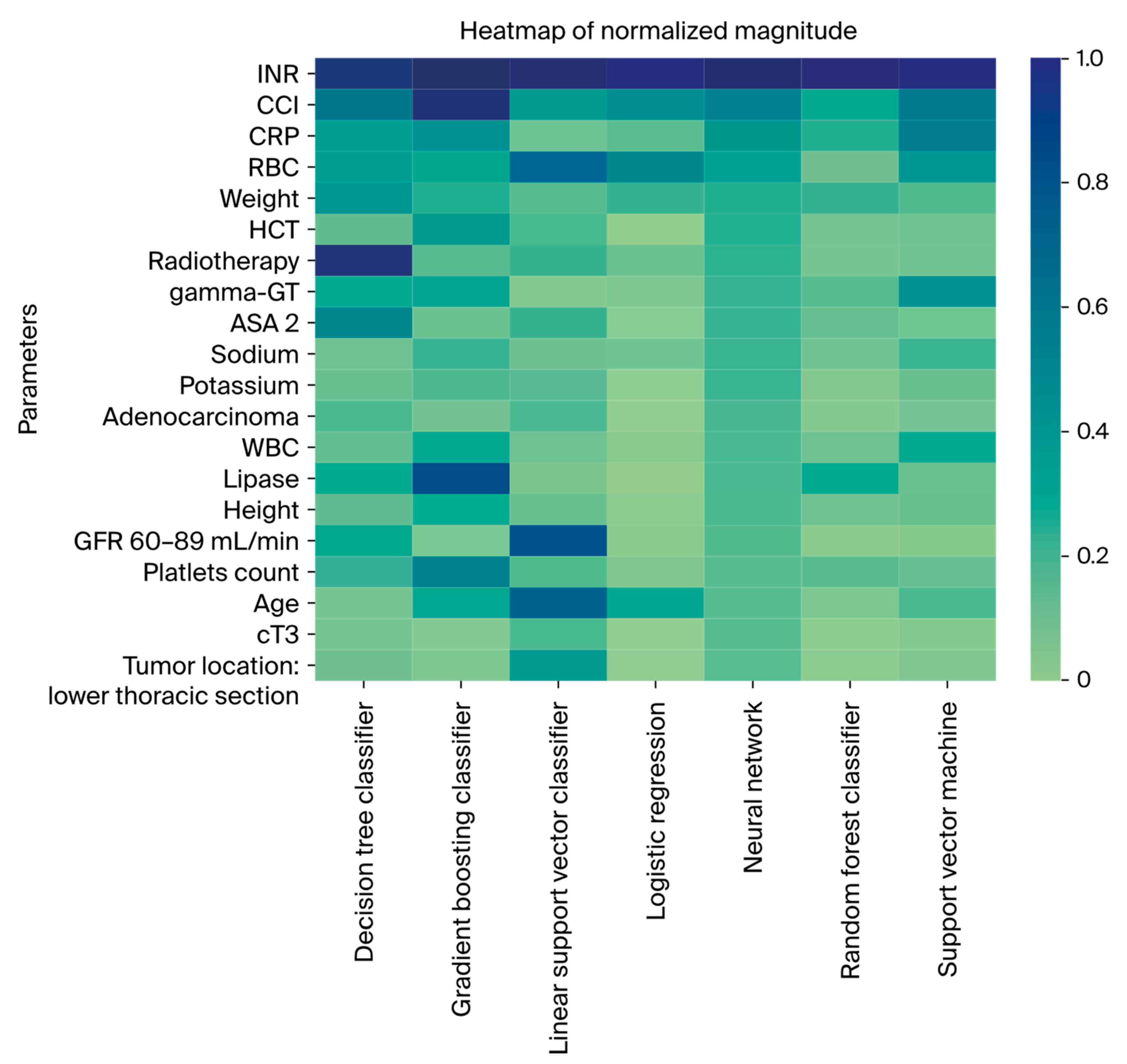
3.4. Feature Importance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Obermannová, R.; Alsina, M.; Cervantes, A.; Leong, T.; Lordick, F.; Nilsson, M.; van Grieken, N.; Vogel, A.; Smyth, E. Oesophageal cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2022, 33, 992–1004. [Google Scholar] [CrossRef]
- Leitlinienprogramm Onkologie (Deutsche Krebsgesellschaft, Deutsche Krebshilfe, AWMF): Diagnostik und Therapie der Plattenepithelkarzinome und Adenokarzinome des Ösophagus; Langversion 3. 1, 2022, AWMF—Registernummer: 021/023OL. 2022. Available online: https://www.leitlinienprogrammonkologie.de/leitlinien/oesophaguskarzinom/ (accessed on 8 August 2024).
- GBD 2017 Oesophageal Cancer Collaborators. The global, regional, and national burden of oesophageal cancer and its attributable risk factors in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 582–597. [Google Scholar] [CrossRef] [PubMed]
- D’Journo, X.B.; Boulate, D.; Fourdrain, A.; Loundou, A.; van Berge Henegouwen, M.I.; Gisbertz, S.S.; O’neill, J.R.; Hoelscher, A.; Piessen, G.; van Lanschot, J.; et al. Risk Prediction Model of 90-Day Mortality After Esophagectomy for Cancer. JAMA Surg. 2021, 156, 836–845. [Google Scholar] [CrossRef]
- Busweiler, L.A.; Henneman, D.; Dikken, J.L.; Fiocco, M.; van Berge Henegouwen, M.I.; Wijnhoven, B.P.; van Hillegersberg, R.; Rosman, C.; Wouters, M.; van Sandick, J.; et al. Failure-to-rescue in patients undergoing surgery for esophageal or gastric cancer. Eur. J. Surg. Oncol. 2017, 43, 1962–1969. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.M.; Gisbertz, S.S.; Moons, J.; Rouvelas, I.; Kauppi, J.; Brown, A.; Asti, E.; Luyer, M.; Lagarde, S.M.; Berlth, F.; et al. Defining benchmarks for transthoracic esophagectomy: A multicenter analysis of total minimally invasive esophagectomy in low risk patients. Ann. Surg. 2017, 266, 814–821. [Google Scholar] [CrossRef]
- Dhungel, B.; Diggs, B.S.; Hunter, J.G.; Sheppard, B.C.; Vetto, J.T.; Dolan, J.P. Patient and peri-operative predictors of morbidity and mortality after esophagectomy: American College of Surgeons National Surgical Quality Improvement Program (ACS-NSQIP), 2005–2008. J. Gastrointest. Surg. 2010, 14, 1492–1501. [Google Scholar] [CrossRef] [PubMed]
- Baum, P.; Diers, J.; Lichthardt, S.; Kastner, C.; Schlegel, N.; Germer, C.-T.; Wiegering, A. Mortality and complications following visceral surgery: A nationwide analysis based on the diagnostic categories used in german hospital invoicing data. Dtsch. Arztebl. Int. 2019, 116, 739–746. [Google Scholar] [PubMed]
- Rees, J.; Hurt, C.N.; Gollins, S.; Mukherjee, S.; Maughan, T.; Falk, S.J.; Staffurth, J.; Ray, R.; Bashir, N.; Geh, J.I.; et al. Patient-reported outcomes during and after definitive chemoradiotherapy for oesophageal cancer. Br. J. Cancer 2015, 113, 603–610. [Google Scholar] [CrossRef]
- Wittmann, E.; Beaton, C.; Lewis, W.G.; Hopper, A.N.; Zamawi, F.; Jackson, C.; Dave, B.; Bowen, R.; Willacombe, A.; Blackshaw, G.; et al. Comparison of patients’ needs and doctors’ perceptions of information requirements related to a diagnosis of oesophageal or gastric cancer. Eur. J. Cancer Care 2011, 20, 187–195. [Google Scholar] [CrossRef]
- Jacobs, M.; Macefield, R.C.; Elbers, R.G.; Sitnikova, K.; Korfage, I.J.; Smets, E.M.A.; Henselmans, I.; Henegouwen, M.I.v.B.; de Haes, J.C.J.M.; Blazeby, J.M.; et al. Meta-analysis shows clinically relevant and long-lasting deterioration in health-related quality of life after esophageal cancer surgery. Qual. Life Res. 2014, 23, 1155–1176. [Google Scholar] [CrossRef]
- Noordman, B.J.; de Bekker-Grob, E.W.; Coene, P.P.L.O.; van der Harst, E.; Lagarde, S.M.; Shapiro, J.; Wijnhoven, B.P.L.; van Lanschot, J.J.B. Patients’ preferences for treatment after neoadjuvant chemoradiotherapy for oesophageal cancer. Br. J. Surg. 2018, 105, 1630–1638. [Google Scholar] [CrossRef]
- Noordman, B.J.; Wijnhoven, B.P.L.; Lagarde, S.M.; Boonstra, J.J.; Coene, P.P.L.O.; Dekker, J.W.T.; Doukas, M.; van der Gaast, A.; Heisterkamp, J.; Kouwenhoven, E.A.; et al. Neoadjuvant chemoradiotherapy plus surgery versus active surveillance for oesophageal cancer: A stepped-wedge cluster randomised trial. BMC Cancer 2018, 18, 142. [Google Scholar] [CrossRef] [PubMed]
- Booka, E.; Takeuchi, H.; Suda, K.; Fukuda, K.; Nakamura, R.; Wada, N.; Kawakubo, H.; Kitagawa, Y. Meta-analysis of the impact of postoperative complications on survival after oesophagectomy for cancer. BJS Open 2018, 2, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Sesti, J.; Almaz, B.; Bell, J.; Nguyen, A.; Bamboat, Z.; Lackey, A.; Langan, R.C.; Turner, A.L.; Hilden, P.; Paul, S. Impact of postoperative complications on long-term survival after esophagectomy in older adults: A SEER-Medicare analysis. J. Surg. Oncol. 2021, 124, 751–766. [Google Scholar] [CrossRef] [PubMed]
- Karran, A.; Blake, P.; Chan, D.; Reid, T.D.; Davies, I.L.; Kelly, M.; Roberts, S.A.; Crosby, T.; Lewis, W.G. Propensity score analysis of oesophageal cancer treatment with surgery or definitive chemoradiotherapy. Br. J. Surg. 2014, 101, 502–510. [Google Scholar] [CrossRef]
- Crosby, T.; Hurt, C.N.; Falk, S.; Gollins, S.; Staffurth, J.; Ray, R.; Bridgewater, J.A.; Geh, J.I.; Cunningham, D.; Blazeby, J.; et al. Long-term results and recurrence patterns from SCOPE-1: A phase II/III randomised trial of definitive chemoradiotherapy +/− cetuximab in oesophageal cancer. Br. J. Cancer 2017, 116, 709–716. [Google Scholar] [CrossRef]
- Mantziari, S.; Hübner, M.; Demartines, N.; Schäfer, M. Impact of preoperative risk factors on morbidity after esophagectomy: Is there room for improvement? World J. Surg. 2014, 38, 2882–2890. [Google Scholar] [CrossRef]
- Griffin, S.M.; Shaw, I.H.; Dresner, S.M. Early complications after Ivor Lewis subtotal esophagectomy with two-field lymphadenectomy: Risk factors and management. J. Am. Coll. Surg. 2002, 194, 285–297. [Google Scholar] [CrossRef]
- Yoshida, N.; Watanabe, M.; Baba, Y.; Iwagami, S.; Ishimoto, T.; Iwatsuki, M.; Sakamoto, Y.; Miyamoto, Y.; Ozaki, N.; Baba, H. Risk factors for pulmonary complications after esophagectomy for esophageal cancer. Surg. Today 2014, 44, 526–532. [Google Scholar] [CrossRef]
- Kamarajah, S.K.; Lin, A.; Tharmaraja, T.; Bharwada, Y.; Bundred, J.R.; Nepogodiev, D.; Evans, R.P.T.; Singh, P.; Griffiths, E.A. Risk factors and outcomes associated with anastomotic leaks following esophagectomy: A systematic review and meta-analysis. Dis. Esophagus 2020, 33, doz089. [Google Scholar] [CrossRef]
- Takeuchi, H.; Miyata, H.; Gotoh, M.; Kitagawa, Y.; Baba, H.; Kimura, W.; Tomita, N.; Nakagoe, T.; Shimada, M.; Sugihara, K.; et al. A risk model for esophagectomy using data of 5354 patients included in a Japanese nationwide web-based database. Ann. Surg. 2014, 260, 259–266. [Google Scholar] [CrossRef]
- Tekkis, P.P.; McCulloch, P.; Poloniecki, J.D.; Prytherch, D.R.; Kessaris, N.; Steger, A.C. Risk-adjusted prediction of operative mortality in oesophagogastric surgery with O-POSSUM. Br. J. Surg. 2004, 91, 288–295. [Google Scholar] [CrossRef]
- Internullo, E.; Moons, J.; Nafteux, P.; Coosemans, W.; Decker, G.; De Leyn, P.; Van Raemdonck, D.; Lerut, T. Outcome after esophagectomy for cancer of the esophagus and GEJ in patients aged over 75 years. Eur. J. Cardiothorac. Surg. 2008, 33, 1096–1104. [Google Scholar] [CrossRef] [PubMed]
- Bilimoria, K.Y.; Liu, Y.; Paruch, J.L.; Zhou, L.; Kmiecik, T.E.; Ko, C.Y.; Cohen, M.E. Development and evaluation of the universal ACS NSQIP surgical risk calculator: A decision aid and informed consent tool for patients and surgeons. J. Am. Coll. Surg. 2013, 217, 833–842.e1. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, N.; Watanabe, M.; Baba, Y.; Iwagami, S.; Ishimoto, T.; Iwatsuki, M.; Sakamoto, Y.; Miyamoto, Y.; Ozaki, N.; Baba, H. Estimation of physiologic ability and surgical stress (E-PASS) can assess short-term outcome after esophagectomy for esophageal cancer. Esophagus 2013, 10, 86–94. [Google Scholar] [CrossRef]
- Bertsimas, D.; Dunn, J. Optimal classification trees. Mach. Learn. 2017, 106, 1039–1082. [Google Scholar] [CrossRef]
- Bertsimas, D.; Dunn, J.; Velmahos, G.C.; Kaafarani, H.M.A. Surgical Risk Is Not Linear: Derivation and Validation of a Novel, User-friendly, and Machine-learning-based Predictive OpTimal Trees in Emergency Surgery Risk (POTTER) Calculator. Ann. Surg. 2018, 268, 574–583. [Google Scholar] [CrossRef]
- Seely, A.J.; Christou, N.V. Multiple organ dysfunction syndrome: Exploring the paradigm of complex nonlinear systems. Crit. Care Med. 2000, 28, 2193–2200. [Google Scholar] [CrossRef]
- Akcay, M.; Etiz, D.; Celik, O. Prediction of survival and recurrence patterns by machine learning in gastric cancer cases undergoing radiation therapy and chemotherapy. Adv. Radiat. Oncol. 2020, 5, 1179–1187. [Google Scholar] [CrossRef]
- Rahman, S.A.; Walker, R.C.; Lloyd, M.A.; Grace, B.L.; van Boxel, G.I.; Kingma, B.F.; Ruurda, J.P.; van Hillegersberg, R.; Harris, S.; Mercer, S.; et al. Machine learning to predict early recurrence after oesophageal cancer surgery. Br. J. Surg. 2020, 107, 1042–1052. [Google Scholar] [CrossRef]
- Mathew, G.; Agha, R.; Albrecht, J.; Goel, P.; Mukherjee, I.; Pai, P.; D’Cruz, A.K.; Nixon, I.J.; Roberto, K.; Enam, S.A.; et al. STROCSS 2021: Strengthening the reporting of cohort, cross-sectional and case-control studies in surgery. Int. J. Surg. 2021, 96, 106165. [Google Scholar] [CrossRef] [PubMed]
- Collins, G.S.; Moons, K.G.M.; Dhiman, P.; Riley, R.D.; Beam, A.L.; Van Calster, B.; Ghassemi, M.; Liu, X.; Reitsma, J.B.; van Smeden, M.; et al. TRIPOD+AI statement: Updated guidance for reporting clinical prediction models that use regression or machine learning methods. BMJ 2024, 385, e078378. [Google Scholar] [CrossRef] [PubMed]
- Newcombe, R.G. Two-sided confidence intervals for the single proportion: Comparison of seven methods. Stat. Med. 1998, 17, 857–872. [Google Scholar] [CrossRef]
- Brierley, J.D.; Gospodarowicz, M.K.; Wittekind, C. (Eds.) TNM Classification of Malignant Tumours, 8th ed.; Wiley: Hoboken, NJ, USA, 2016; Available online: https://www.wiley.com/en-gb/TNM+Classification+of+Malignant+Tumours%2C+8th+Edition-p-9781119263579 (accessed on 26 April 2024).
- Quan, H.; Sundararajan, V.; Halfon, P.; Fong, A.; Burnand, B.; Luthi, J.-C.; Saunders, L.D.; Beck, C.A.; Feasby, T.E.; Ghali, W.A. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med. Care 2005, 43, 1130–1139. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Lundberg, S.M.; Lee, S.-I. Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2017. [Google Scholar]
- Joung, R.H.-S.; Merkow, R.P. Is it Time to Abandon 30-Day Mortality as a Quality Measure? Ann. Surg. Oncol. 2021, 28, 1263–1264. [Google Scholar] [CrossRef]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef]
- Jung, J.-O.; Pisula, J.I.; Bozek, K.; Popp, F.; Fuchs, H.F.; Schröder, W.; Bruns, C.J.; Schmidt, T. Prediction of postoperative complications after oesophagectomy using machine-learning methods. Br. J. Surg. 2023, 110, 1361–1366. [Google Scholar] [CrossRef]
- Zhao, Z.; Cheng, X.; Sun, X.; Ma, S.; Feng, H.; Zhao, L. Prediction model of anastomotic leakage among esophageal cancer patients after receiving an esophagectomy: Machine learning approach. JMIR Med. Inform. 2021, 9, e27110. [Google Scholar] [CrossRef]
- Pecqueux, M.; Riediger, C.; Distler, M.; Oehme, F.; Bork, U.; Kolbinger, F.R.; Schöffski, O.; van Wijngaarden, P.; Weitz, J.; Schweipert, J.; et al. The use and future perspective of Artificial Intelligence-A survey among German surgeons. Front. Public Health 2022, 10, 982335. [Google Scholar] [CrossRef]
- Grass, J.K.; Küsters, N.; Kemper, M.; Tintrup, J.; Piecha, F.; Izbicki, J.R.; Perez, D.; Melling, N.; Bockhorn, M.; Reeh, M. Risk stratification of cirrhotic patients undergoing esophagectomy for esophageal cancer: A single-centre experience. PLoS ONE 2022, 17, e0265093. [Google Scholar] [CrossRef] [PubMed]
- van den Bosch, T.; Warps, A.-L.K.; de Nerée Tot Babberich, M.P.M.; Stamm, C.; Geerts, B.F.; Vermeulen, L.; Wouters, M.W.J.M.; Dekker, J.W.T.; Tollenaar, R.A.E.M.; Tanis, P.J.; et al. Predictors of 30-Day Mortality Among Dutch Patients Undergoing Colorectal Cancer Surgery, 2011–2016. JAMA Netw. Open 2021, 4, e217737. [Google Scholar] [CrossRef] [PubMed]
- Pera, M.; Gibert, J.; Gimeno, M.; Garsot, E.; Eizaguirre, E.; Miró, M.; Castro, S.; Miranda, C.; Reka, L.; Leturio, S.; et al. Machine Learning Risk Prediction Model of 90-day Mortality After Gastrectomy for Cancer. Ann. Surg. 2022, 276, 776–783. [Google Scholar] [CrossRef] [PubMed]

| Characteristic | Total n = (552) | Training Cohort n = (409) | Validation Cohort n = (143) | p Value |
|---|---|---|---|---|
| Age | 0.55 | |||
| ≤40 (%) | 9 (1.6) | 8 (2.0) | 1 (0.7) | |
| 41–50 (%) | 42 (7.6) | 33 (8.1) | 9 (6.3) | |
| 51–60 (%) | 152 (27.5) | 118 (28.9) | 34 (23.8) | |
| 61–70 (%) | 178 (32.2) | 130 (31.8) | 48 (33.6) | |
| 71–80 (%) | 149 (27.0) | 105 (25.7) | 44 (30.8) | |
| >80 (%) | 22 (4.0) | 15 (3.7) | 7 (4.9) | |
| Mean (SD) | 64.3 (10.1) | 63.8 (10.1) | 65.8 (9.9) | |
| BMI | 0.82 | |||
| <18.5 (%) | 22 (4.0) | 17 (4.2) | 5 (3.5) | |
| 18.5–24.9 (%) | 216 (39.1) | 156 (38.1) | 60 (42.0) | |
| 25–29.9 (%) | 240 (43.5) | 182 (44.5) | 58 (40.6) | |
| ≥30 (%) | 74 (13.4) | 54 (13.2) | 20 (14.0) | |
| Mean (SD) | 25.8 (4.4) | 25.8 (4.5) | 25.7 (4.3) | |
| Sex | 0.44 | |||
| Male (%) | 439 (79.5) | 329 (80.4) | 110 (76.9) | |
| Female (%) | 113 (20.5) | 80 (19.6) | 33 (23.1) | |
| ECOG | 0.22 | |||
| 0 (%) | 354 (64.1) | 255 (62.3) | 99 (69.2) | |
| 1 (%) | 186 (33.7) | 146 (35.7) | 40 (28.0) | |
| 2 (%) | 12 (2.2) | 8 (2.0) | 4 (2.8) | |
| Histology | 0.07 | |||
| AC (%) | 360 (65.2) | 267 (65.3) | 93 (65.0) | |
| SCC (%) | 177 (32.1) | 128 (31.3) | 49 (34.4) | |
| Other (%) | 14 (2.5) | 14 (3.4) | 0 (0.) | |
| Neoadjuvant treatment | 0.3 | |||
| Chemotherapy (%) | 273 (49.5) | 212 (51.8) | 61 (42.7) | |
| Radiochemotherapy (%) | 183 (33.2) | 136 (33.3) | 47 (32.9) | |
| Radiotherapy alone (%) | 2 (0.4) | 2 (0.5) | 0 (0.0) | |
| Tpre | 0.25 | |||
| T0 (%) | 1 (0.2) | 1 (0.2) | 0 (0.0) | |
| T1 (%) | 42 (7.6) | 30 (7.3) | 12 (8.4) | |
| T2 (%) | 67 (12.1) | 54 (13.2) | 13 (9.1) | |
| T3 (%) | 300 (54.3) | 246 (60.1) | 54 (37.8) | |
| T4 (%) | 23 (4.2) | 19 (4.6) | 4 (2.8) | |
| Tis (%) | 1 (0.2) | 1 (0.2) | 0 (0.0) | |
| Tx (%) | 8 (1.4) | 4 (1.0) | 4 (2.8) | |
| Npre | <0.001 | |||
| N0 (%) | 124 (22.5) | 96 (23.5) | 28 (19.6) | |
| N1 (%) | 168 (30.4) | 129 (31.5) | 39 (27.3) | |
| N2 (%) | 74 (13.4) | 73 (17.8) | 1 (0.7) | |
| N3 (%) | 26 (4.7) | 24 (5.9) | 2 (1.4) | |
| Nx (%) | 10 (1.8) | 4 (1.0) | 6 (4.2) | |
| Comorbidities | ||||
| CCI mean (SD) | 7.4 (3.8) | 7.3 (3.8) | 7.6 (3.8) | 0.46 |
| Myocardial infarction (%) | 19 (3.4) | 13 (3.2) | 6 (4.2) | 0.76 |
| Peripheral vascular disease (%) | 24 (4.3) | 13 (3.2) | 11 (7.7) | 0.04 |
| Chronic pulmonary disease (%) | 125 (22.6) | 98 (24.0) | 27 (18.9) | 0.26 |
| Peptic ulcer disease (%) | 11 (2.0) | 6 (1.5) | 5 (3.5) | 0.25 |
| Liver disease mild (%) | 32 (5.8) | 19 (4.6) | 13 (9.1) | 0.08 |
| Diabetes without chronic complications (%) | 84 (15.2) | 59 (14.4) | 25 (17.5) | 0.46 |
| Hemiplegia or paraplegia (%) | 12 (2.2) | 7 (1.7) | 5 (3.5) | 0.35 |
| Liver disease moderate/severe (%) | 3 (0.5) | 3 (0.7) | 0 (0.0) | 0.71 |
| Renal disease (%) | 2 (0.4) | 2 (0.5) | 0 (0.0) | 0.98 |
| Metastatic solid tumor (%) | 154 (27.9) | 115 (28.1) | 39 (27.3) | 0.93 |
| 30-day mortality (%) | 15 (2.7) | 10 (2.4) | 5 (3.5) | 0.71 |
| 90-day mortality (%) | 32 (5.8) | 19 (4.6) | 13 (9.1) | 0.08 |
| Classifier | AUROC Mean, 95% CI (Low–High) | AUPRC Mean, 95% CI (Low–High) | MCC Mean, 95% CI (Low–High) | |
|---|---|---|---|---|
| 90-day mortality | Decision tree | 0.52 (0.51–0.53) | 0.21 (0.17–0.24) | 0.07 (0.05–0.09) |
| Gradient boosting | 0.64 (0.63–0.65) | 0.16 (0.15–0.16) | 0.12 (0.10–0.14) | |
| Linear support vector machine | 0.51 (0.50–0.52) | 0.10 (0.10–0.10) | 0.01 (0.00–0.01) | |
| Logistic regression | 0.64 (0.63–0.64) | 0.20 (0.19–0.21) | 0.26 (0.25–0.28) | |
| Neural network | 0.61 (0.60–0.63) | 0.25 (0.24–0.27) | 0.24 (0.21–0.27) | |
| Random forest | 0.64 (0.63–0.65) | 0.21 (0.20–0.22) | 0.27 (0.25–0.28) | |
| Support vector machine | 0.62 (0.61–0,63) | 0.23 (0.21–0.25) | 0.21 (0.18–0.24) | |
| IESG score stratification | 0.44 (0.32–0.56) | 0.11 (0.05–0.21) | 0.15 (0.03–0.27) | |
| 30-day mortality | Decision tree | 0.49 (0.47–0.50) | 0.17 (0.14–0.22) | 0.03 (0.02–0.05) |
| Gradient boosting | 0.58 (0.56–0.60) | 0.06 (0.06–0.07) | 0.03 (0.01–0.05) | |
| Linear support vector machine | 0.50 (0.49–0.51) | 0.04 (0.04–0.05) | 0.00 (0.00–0.00) | |
| Logistic regression | 0.45 (0.44–0.45) | 0.03 (0.03–0.04) | 0.04 (0.03–0.05) | |
| Neural network | 0.44 (0.41–0.46) | 0.07 (0.06–0.09) | 0.06 (0.03–0.08) | |
| Random forest | 0.47 (0.45–0.49) | 0.05 (0.03–0.06) | 0.07 (0.05–0.08) | |
| Support vector machine | 0.52 (0.51–0.54) | 0.08 (0.07–0.09) | 0.03 (0.01–0.04) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Winter, A.; van de Water, R.P.; Pfitzner, B.; Ibach, M.; Riepe, C.; Ahlborn, R.; Faraj, L.; Krenzien, F.; Dobrindt, E.M.; Raakow, J.; et al. Enhancing Preoperative Outcome Prediction: A Comparative Retrospective Case–Control Study on Machine Learning versus the International Esodata Study Group Risk Model for Predicting 90-Day Mortality in Oncologic Esophagectomy. Cancers 2024, 16, 3000. https://doi.org/10.3390/cancers16173000
Winter A, van de Water RP, Pfitzner B, Ibach M, Riepe C, Ahlborn R, Faraj L, Krenzien F, Dobrindt EM, Raakow J, et al. Enhancing Preoperative Outcome Prediction: A Comparative Retrospective Case–Control Study on Machine Learning versus the International Esodata Study Group Risk Model for Predicting 90-Day Mortality in Oncologic Esophagectomy. Cancers. 2024; 16(17):3000. https://doi.org/10.3390/cancers16173000
Chicago/Turabian StyleWinter, Axel, Robin P. van de Water, Bjarne Pfitzner, Marius Ibach, Christoph Riepe, Robert Ahlborn, Lara Faraj, Felix Krenzien, Eva M. Dobrindt, Jonas Raakow, and et al. 2024. "Enhancing Preoperative Outcome Prediction: A Comparative Retrospective Case–Control Study on Machine Learning versus the International Esodata Study Group Risk Model for Predicting 90-Day Mortality in Oncologic Esophagectomy" Cancers 16, no. 17: 3000. https://doi.org/10.3390/cancers16173000
APA StyleWinter, A., van de Water, R. P., Pfitzner, B., Ibach, M., Riepe, C., Ahlborn, R., Faraj, L., Krenzien, F., Dobrindt, E. M., Raakow, J., Sauer, I. M., Arnrich, B., Beyer, K., Denecke, C., Pratschke, J., & Maurer, M. M. (2024). Enhancing Preoperative Outcome Prediction: A Comparative Retrospective Case–Control Study on Machine Learning versus the International Esodata Study Group Risk Model for Predicting 90-Day Mortality in Oncologic Esophagectomy. Cancers, 16(17), 3000. https://doi.org/10.3390/cancers16173000








