Simple Summary
The rare ovarian cancer subtypes ovarian clear cell carcinoma (OCCC) and Small cell carcinoma of the ovary, hypercalcaemic type (SCCOHT), can have mutations in members of a complex known as SWI/SNF that regulates the accessibility of chromatin to factors involved in DNA repair and gene expression. Some mutations in a particular complex member also occur in endometrioid ovarian cancer (EnOC) and endometriosis. Patients with endometriosis have a greater risk of developing OCCC and EnOC, with endometriosis frequently present at the time of diagnosis of these malignancies. The OCCC and SCCOHT ovarian cancer subtypes are notoriously difficult to treat with chemotherapies based on platinum-drugs that are standard-of-care for most cases of ovarian cancer. Mutations in members of this chromatin-remodelling complex offer new opportunities for molecular therapeutics using drugs that inhibit different aspects of cellular processes, including DNA repair, epigenetic regulation, kinase activity, and immune checkpoints.
Abstract
SWI/SNF (SWItch/Sucrose Non-Fermentable) is the most frequently mutated chromatin-remodelling complex in human malignancy, with over 20% of tumours having a mutation in a SWI/SNF complex member. Mutations in specific SWI/SNF complex members are characteristic of rare chemoresistant ovarian cancer histopathological subtypes. Somatic mutations in ARID1A, encoding one of the mutually exclusive DNA-binding subunits of SWI/SNF, occur in 42–67% of ovarian clear cell carcinomas (OCCC). The concomitant somatic or germline mutation and epigenetic silencing of the mutually exclusive ATPase subunits SMARCA4 and SMARCA2, respectively, occurs in Small cell carcinoma of the ovary, hypercalcaemic type (SCCOHT), with SMARCA4 mutation reported in 69–100% of SCCOHT cases and SMARCA2 silencing seen 86–100% of the time. Somatic ARID1A mutations also occur in endometrioid ovarian cancer (EnOC), as well as in the chronic benign condition endometriosis, possibly as precursors to the development of the endometriosis-associated cancers OCCC and EnOC. Mutation of the ARID1A paralogue ARID1B can also occur in both OCCC and SCCOHT. Mutations in other SWI/SNF complex members, including SMARCA2, SMARCB1 and SMARCC1, occur rarely in either OCCC or SCCOHT. Abrogated SWI/SNF raises opportunities for pharmacological inhibition, including the use of DNA damage repair inhibitors, kinase and epigenetic inhibitors, as well as immune checkpoint blockade.
1. Introduction
Ovarian cancer spans a number of different histopathological subtypes, most of which have limited treatment options beyond surgical debulking and combination chemotherapy consisting of systemic platinum-based drugs including carboplatin and the taxane paclitaxel [1]. In the most common subtype, high-grade serous ovarian cancer (HGSOC), Poly (ADP-ribose) polymerase (PARP) inhibitors have been shown to increase both progression-free survival (PFS) and overall survival (OS) in the presence of molecular aberrations in genes such as BRCA1 and BRCA2 which encode proteins functioning in the homologous recombination repair (HRR) pathway [2,3,4,5,6]. Anti-angiogenics targeting vascular endothelial growth factor (VEGF), such as bevacizumab, are also in use, with clinical trials suggesting the benefit of combining bevacizumab with the PARP inhibitor olaparib [5,6]. Therapeutic targeting of mutations beyond those directly involved in the HRR pathway in ovarian cancer is yet to emerge into the clinic. This is an area attracting extensive interest, especially for treatment-resistant subtypes such as ovarian clear cell carcinoma (OCCC) and the rare Small cell carcinoma of the ovary, hypercalcaemic type (SCCOHT), both with a poorer prognosis compared to HGSOC [7,8].
The intersection of genetics and epigenomics to drive chromatin remodelling in malignancy provides further opportunities for both uncovering the fundamental mechanisms of gene regulation and identifying new therapeutic targets. The ATP-dependent chromatin-remodelling complex SWI/SNF (SWItch/Sucrose Non-Fermentable; also known as the BAF complex) is an important junction for these intersections, with aberrations in SWI/SNF complex members reported in around 20% of all human malignancies [9,10]. Mutations in SMARCA4, that encodes one of catalytic subunits of SWI/SNF, occur in around 5–7% of all human malignancies (reviewed in [11]). Similarly, mutations in ARID1A (AT-rich interactive domain-containing protein 1A) that encodes a DNA-binding subunit of SWI/SNF have been reported in ~6% of human malignancies [12].
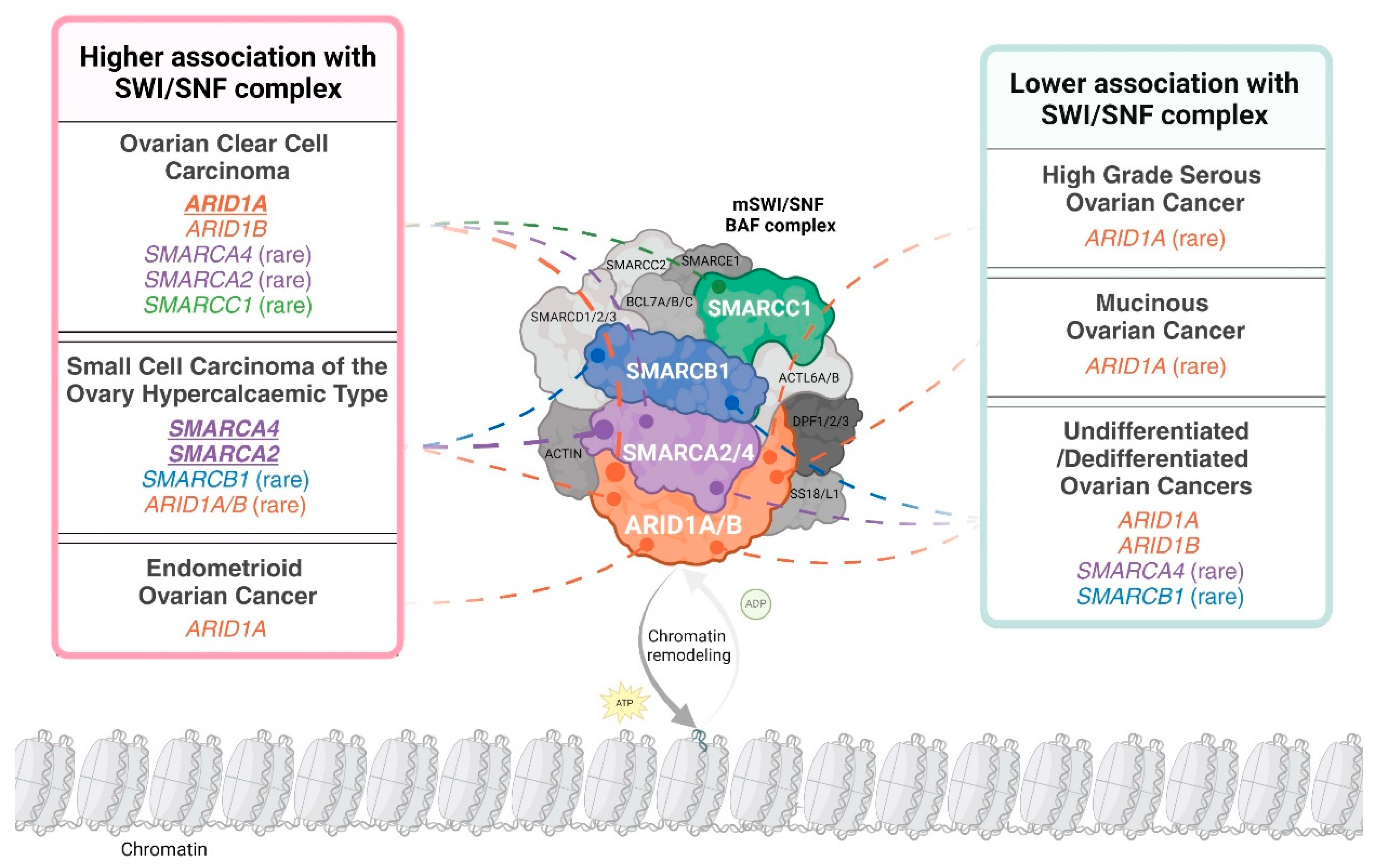
Subunits of the SWI/SNF complex are differentially mutated in distinct ovarian cancer subtypes. For example, ARID1A is frequently mutated in OCCC and endometrioid ovarian cancer (EnOC) but rarely mutated in HGSOC and mucinous ovarian cancer (MOC) [13,14,15,16]. ARID1B, the mutually exclusive paralogue of ARID1A, is mutated in OCCC [17]. Both ARID1A and ARID1B are rarely mutated in SCCOHT [18]. Interestingly, ARID1A is mutated in the benign condition endometriosis, with OCCC and EnOC described as endometriosis-associated ovarian cancers (EAOC) [14,19,20,21]. SMARCA4 is the predominant SWI/SNF complex member mutated in SCCOHT, with mutations reported infrequently in OCCC and HGSOC [17,22,23]. Other subunits of SWI/SNF, specifically SMARCA2, SMARCB1 and SMARCC1, are reported to be mutated at very low frequency in OCCC or SCCOHT [17,24,25]. The abrogation of SWI/SNF complex members is not only governed by genetic mutation, as SMARCA2 can be post-translationally silenced in SCCOHT [26] and more rarely in OCCC [26,27]. Mutations and epigenetic silencing of SWI/SNF complex members present therapeutic vulnerabilities, facilitating synthetic lethal approaches for the treatment of ovarian cancer [28,29]. Mutated or epigenetically silenced SWI/SNF subunits in ovarian cancer are depicted in Figure 1.
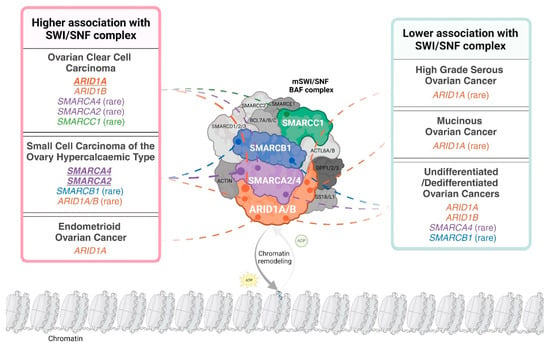
Figure 1.
The distribution of loss-of-function alterations in mammalian SWI/SNF (mSWI/SNF) chromatin-remodelling complex members across ovarian cancer histopathological subtypes. The schematic uses the canonical BAF (cBAF) formation of the SWI/SNF complex to depict subunit involvement. A loss-of-function alteration is defined as the presence of a pathogenic mutation in the encoding gene and/or the loss of corresponding protein expression. “Higher association with the SWI/SNF complex” is defined as subtypes where over 20% of cases have alterations in at least one complex member. An exception was made for undifferentiated/dedifferentiated ovarian cancers due to limited incidence reporting. Complex members identified as altered in over 40% of a specific subtype are indicated in bold and underline. An alteration is presented as ‘rare’ if less than 10 cases were reported in the published literature or large cohort analyses (N ≥ 100) report an incidence less than 10%. Distribution is based on data in Tables 2 and 3 and Tessier-Cloutier and colleagues [30]. Created with www.BioRender.com, Access Date: 9 August 2024.
In this review, we outline the extent of SWI/SNF complex member mutations and epigenetic regulation in a range of histopathological subtypes of ovarian cancer. We show that mutations of SWI/SNF complex members predominantly occur in the less frequent and very rare subtypes of ovarian cancer that generally respond poorly to current standard-of-care therapies. Lastly, we discuss the therapeutic opportunities that mutations in SWI/SNF complex members may provide for the treatment of patients with these rarer subtypes of ovarian cancer.
2. SWI/SNF Chromatin-Remodelling Complex
SWI/SNF is critical to maintaining healthy cellular functions, including during embryonic development and for stem cell pluripotency [31,32,33], in the development of male and female gametes [34,35], in cell cycle control [36], and in the DNA damage response where this complex is rapidly recruited to double-strand breaks (DSBs) [37,38]. There are three distinct forms of the mammalian SWI/SNF complex, specifically canonical BAF (cBAF), polybromo-associated BAF (PBAF) and non-canonical BAF (ncBAF), consisting of both common and distinct complex members encoded by 29 genes [39]. These distinct forms each have up to 15 members constituting mammalian chromatin-remodelling complexes [32].
The SWI/SNF complex has significant roles in transcription via its ability to modulate DNA accessibility. This complex is highly enriched at gene enhancers that bind transcription factors, where it is associated with the active chromatin mark histone H3 lysine 27 acetylation (H3K27ac) and has roles in directing lineage specificity [40,41]. SWI/SNF complexes are also located at promoters and transcription start sites (TSSs) where they function to regulate transcription [42]. Additionally, these complexes are known to preferentially associate with chromatin enriched with the active histone mark of monoubiquitylated histone H2B at lysine 120 (H2Bub1), which is reliant upon the histone writer Ring Finger Protein 20 (RNF20) [43,44]. Of note, SWI/SNF has antagonistic roles in transcription with proteins from the Polycomb group (PcG) such as EZH2 (Enhancer of Zeste Homolog 2) as part of the PRC2 (Polycomb repressor complex 2) that is associated with gene silencing via the catalysis of the repressive mark trimethylation of histone H3 lysine 127 (H3K127me3) [45,46,47].
Here, we focus on SWI/SNF complex member genes and their proteins, that are either mutated or themselves epigenetically regulated in rare subtypes of ovarian cancer. Specifically, we will review SMARCA4, SMARCA2, SMARCB1, SMARCC1, ARID1A and ARID1B. SMARCA4 (SWI/SNF-Related, Matrix-Associated, Actin-Dependent Regulator of Chromatin, Subfamily A, Member 4) and SMARCA2 (SWI/SNF-Related, Matrix-Associated, Actin-Dependent Regulator of Chromatin, Subfamily A, Member 2) are mutually exclusive ATP-dependent helicases that are present in all of cBAF, PBAF and ncBAF. These catalytic subunits are critical to chromatin remodelling, requiring ATP hydrolysis to enable nucleosome sliding and histone eviction, modulating access to DNA [39,48,49].
Non-catalytic SWI/SNF complex members also have roles to play in ovarian tumorigenesis. cBAF has 12 complex members and is the only BAF complex to include the mutually exclusive AT-rich interaction domain (ARID)-domain containing proteins ARID1A and ARID1B that bind to DNA (also known as BAF250A and BAF250B (BRG/BRM-associated factors A and B) [39,50]. SMARCB1 (SWI/SNF-Related, Matrix-Associated, Actin-Dependent Regulator of Chromatin, Subfamily B, Member 1) is an evolutionarily conserved subunit of SWI/SNF. The coiled-coil structural motif of the C-terminal domain of SMARCB1 contains a basic alpha-helix that directly binds to the acidic patch region of nucleosomes and is critical to the facilitation of chromatin remodelling [51]. SMARCC1 (SWI/SNF-Related, Matrix-Associated, Actin-Dependent Regulator of Chromatin Subfamily C Member 1), is a chromodomain-containing protein and core member of SWI/SNF that works as a scaffold to promote the stability of the complex, likely by preventing the proteasomal degradation of key complex members [39,52]. For ease of navigating the literature, the alternative nomenclature that is used for the SWI/SNF complex members discussed in this review is summarised in Table 1.

Table 1.
Nomenclature for SWI/SNF (BAF) complex members mutated or otherwise lost in ovarian cancer subtypes ^.
When viewed as an entire complex, SWI/SNF is the most frequently mutated chromatin-remodelling complex in human malignancy, with around 20% of human cancers harbouring a mutation in one of the SWI/SNF complex members (reviewed in [39,53]). Of note, component genes of SWI/SNF can also carry heterozygous mutations in neurodevelopmental and autism spectrum-like disorders including Coffin-Siris Syndrome [OMIM: #135900], Nicolaides–Baraitser syndrome [OMIM: #601358], congenital hydrocephalus and Hirschsprung’s disease [39,54]. The clinical association of mutated SWI/SNF complex members with developmental disorders supports key roles for complex members in determining cell fate [55]. SWI/SNF mutations could present viable therapeutic targets, increasing the response to immune checkpoint blockade (ICB) therapy, likely due to the differential expression of genes that stimulate the immune response. This would appear to be applicable even in tumours with low mutational burden, traditionally thought to respond poorly to immunotherapies [53].
3. SWI/SNF Complex Members Are Primarily Mutated in Poorer Prognosis Ovarian Cancer Subtypes and Endometriosis
Distinct subtypes of the group of ovarian cancers that harbour mutations of SWI/SNF complex members in catalytic, DNA-binding and core-stabilising subunits have some commonalities. In comparison to HGSOC that occurs in around 70% of all ovarian cancers, SWI/SNF-mutated ovarian cancers occur more rarely, present at an earlier age, and are more chemoresistant. With the exception of SCCOHT where both germline and somatic mutations can occur, the mutations of genes encoding SWI/SNF complex members in ovarian cancer occur somatically (see Table 2). While highly aggressive dedifferentiated and undifferentiated ovarian carcinomas are not discussed in depth in this review, these rare malignancies are also reported to have lost the expression of key SWI/SNF complex members [30]. The mutation of ARID1A has also been observed in endometriosis, with evidence suggesting that endometriotic lesions are precursors to the development of OCCC and EnOC, with disease progression driven by loss of this tumour suppressor [56]. In the following, we focus on the clinical nature and presentation of SWI/SNF-mutated ovarian cancers and discuss the extent of SWI/SNF complex member mutations in these tumours.
3.1. Ovarian Clear Cell Carcinoma (OCCC)
3.1.1. Clinical Presentation and Epidemiology of OCCC
The median age of diagnosis of patients with OCCC has been reported as 55 years, much lower than that for patients with the more frequently diagnosed serous ovarian cancers at 64 years [7]. OCCC is one of the rarer ovarian cancer histopathological subtypes, with reported differences in frequency based on race and geographical location. In North America and Europe, OCCC is reported to constitute between 1–12% of all ovarian cancers; however, in Asian countries, this frequency is higher [57]. In Korea, around 12.5% of patients with ovarian cancer have OCCC [58]. A study conducted at Qingdao University in China showed that 14.2% of women with ovarian cancer in a cohort of 697 patients from a single hospital had OCCC [59]. In Taiwan, around 20% of patients with ovarian cancer have been reported to have OCCC [60]. Japan demonstrates the highest frequency of OCCC patients, constituting almost 27% of all patients with ovarian cancer [61]. The Surveillance, Epidemiology, and End Results (SEER) study showed that 4.8%, 3.1%, and 11.1% of white, black, and Asian women, respectively, living with ovarian cancer in the USA, have the OCCC subtype [7]. The drivers of these global differences in frequency are currently unclear, although it is interesting to note that Asian women living in the USA have higher frequencies of OCCC compared to non-Asian women, suggesting that ethnicity does play a role in the development of this subtype.
OCCC exhibits distinctive histopathological characteristics and clinical behaviours compared to other ovarian cancer subtypes. Macroscopically, this tumour mostly presents as a unilateral mass that is either solid, a mix of solid and cystic, or a predominantly cystic mass. Microscopically, characteristic features often include a combination of solid, tubulo-cystic and papillary patterns, with cells having large nuclei and abundant cytoplasmic glycogen that give them a clear appearance [62,63,64,65,66]. Clear cell changes are also observed mixed with other ovarian cancer subtypes [66]. OCCC is frequently found in conjunction with endometriosis, with endometriotic lesions speculated to be the precursor lesion for this malignancy [56,67].
OCCC tends to be diagnosed at an earlier FIGO (International Federation of Gynaecology and Obstetrics) stage than HGSOC, with studies reporting between 48.5 and 56.3% of OCCC presenting at Stage I compared with 12–16.6% of serous ovarian cancers, and between 9.9–11% of OCCC at Stage II compared with 5.5–7.2% of serous tumours [7,68]. Conversely, when considering advanced stages, 20.9–30.7% of OCCC were reported as Stage III at diagnosis compared with 45.7–61.7% of serous tumours. Diagnosis with Stage IV disease occurred in only 10.9–11.8% of OCCC compared with 16.2–35.2% of serous tumours [7,68]. In a large meta-analysis, OCCC patients were shown to have a poorer prognosis relative to tumour stage than other ovarian cancer subtypes, especially when diagnosed with advanced disease [69]. Furthermore, it has been shown that when diagnosed at early stages, OCCC patients had better PFS times compared to patients with serous ovarian cancer; however, OS was significantly decreased for these patients compared to those with serous ovarian cancer [70]. Similar findings have been reported by others [59,71]. In the context of therapy, between 11–27% of women with OCCC respond to first-line platinum-based therapy, with only 1–2% of women responsive to treatment once relapse has occurred [68,72,73]. These studies clearly indicate that there would be benefits to diagnosing OCCC in patients at an earlier stage; however, as for all ovarian cancer, OCCC currently lacks early screening tests that would enable this goal.
3.1.2. SWI/SNF Complex Member Mutations in OCCC
Of the SWI/SNF complex members, ARID1A is the most frequently mutated in OCCC, with studies reporting between 42–67% of these tumours with somatic ARID1A mutation (Table 2) [13,14,15,17,74,75,76,77,78,79]. The immunohistochemical loss of ARID1A is seen in between 15–76% of OCCC (Table 3) [15,27,80,81,82]. ARID1A behaves as a tumour suppressor gene in OCCC, with both alleles presumed to be affected by concomitant variants, specifically mutations (deletions and insertions as well as nonsense mutations, all leading to premature STOP codons that would truncate the normal protein) and loss of heterozygosity, or the presence of presumably biallelic mutations [14,76]. In a study of 55 OCCC, the ARID1A paralogue ARID1B was reported to be mutated in 18% of tumours (Table 2) [17]. Tumours in which both ARID1A and ARID1B are mutated are reported to retain a wild-type allele of ARID1B, providing evidence that a certain level of wild-type ARID1B is essential to avoid synthetic lethality in ARID1A-mutant tumours [83]. An absence of functional ARID1A would seem to result in dependency on its mutually exclusive paralogue ARID1B, the depletion of which has been shown to destabilise SWI/SNF and inhibit cellular proliferation [84,85]. While this has yet to result in a new therapy for ARID1A-mutated tumours, the identification of a drug(s) that targets and abolishes ARID1B function in ARID1A-mutated tumours could represent a new therapeutic strategy for OCCC.

Table 2.
Mutations in genes encoding SWI/SNF complex members identified in primary ovarian cancers of different histotypes.
Table 2.
Mutations in genes encoding SWI/SNF complex members identified in primary ovarian cancers of different histotypes.
| Gene | Histotype | Mutated | Reference |
|---|---|---|---|
| ARID1A | OCCC | 66.7% (32 of 48) | Shibuya et al., 2017 [13] |
| OCCC | 62% (24 of 39) | Murakami et al. [75] | |
| OCCC | 57% (24 of 42) | Jones et al. [76] | |
| OCCC | 55% (17 of 31) | Wiegand et al. [79] | |
| OCCC | 49% (27 of 55) | Schnack et al. [78] | |
| OCCC | 46% (55 of 119) | Wiegand et al. [14] | |
| OCCC | 42% (23 of 55) | Itamochi et al. [17] | |
| OCCC | 41.5% (17 of 41) a | Kuroda et al. [15] | |
| OCCC | 7 of 9 | Su et al. [74] | |
| OCCC b | 1 of 1 | Kihara et al. [77] | |
| EnOC | 45% (9 of 20) a | Kuroda et al. [15] | |
| EnOC | 30% (10 of 33) | Wiegand et al. [14] | |
| EnOC | 21% (5 of 24) | Wiegand et al. [79] | |
| EnOC | 1 of 7 | Su et al. [74] | |
| HGSOC | 19% (6 of 32) | Vaicekauskaitė et al. [16] | |
| HGSOC | 0% (0 of 76) | Wiegand et al. [14] | |
| HGSOC | 0% (0 of 36) a | Kuroda et al. [15] | |
| MOC | 2 of 6 a | Kuroda et al. [15] | |
| SCCOHT | 1 of 6 | Auguste et al. [18] | |
| SCCOHT | 1 of 1 | Genestie et al. [86] | |
| SCCOHT | 1 of 1 | Sanders et al. [87] | |
| ARID1B | OCCC | 18% (10 of 55) | Itamochi et al. [17] |
| SCCOHT | 1 of 6 | Auguste et al. [18] | |
| SCCOHT | 1 of 1 | Genestie et al. [86] | |
| SMARCA4 | SCCOHT | 100% (12 of 12) c | Jelinic et al. [22] |
| SCCOHT | 100% (10 of 10) | Le Loarer et al. [88] | |
| SCCOHT | 92% (24 of 26) c | Witkowski et al., 2014 [89] | |
| SCCOHT | 91.9% (10 of 11) a | Jelinic et al. [90] | |
| SCCOHT | 83.3% (15 of 18) | Lin et al. [91] | |
| SCCOHT | 79% (19 of 24) a,c,d | Ramos et al. [25] | |
| SCCOHT | 69% (9 of 13) a,c,d | Ramos et al. [92] | |
| SCCOHT | 8 of 8 c | Moes-Sosnowska et al. [93] | |
| SCCOHT | 7 of 7 a | Mazibrada et al. [94] | |
| SCCOHT | 5 of 6 | Auguste et al. [18] | |
| SCCOHT | 2 of 2 | Kupryjańczyk et al. [95] | |
| SCCOHT | 2 of 2 a,c | Chandan et al. [96] | |
| SCCOHT | 1 of 1 c | Sanders et al. [87] | |
| SCCOHT | 1 of 1 | Li et al. [97] | |
| SCCOHT | 1 of 1 | Gao et al. [98] | |
| SCCOHT | 1 of 1 c | Pressey et al. [99] | |
| SCCOHT | 1 of 1 | Mathey et al. [100] | |
| SCCOHT | 1 of 1 c | Mehta et al. [101] | |
| SCCOHT | 1 of 1 c | Pastorczak et al. [102] | |
| SCCOHT | 1 of 1 c | Connor et al. [103] | |
| SCCOHT | 1 of 1 c | David et al. [104] | |
| SCCOHT | 1 of 1 | Bailey et al. [105] | |
| SCCOHT | 1 of 1 a c | Lavrut et al. [106] | |
| SCCOHT | 1 of 1 a | Fahiminiya et al. [107] | |
| SCCOHT | 1 of 1 a | Gao et al. [108] | |
| SCCOHT | 1 of 1 a | Aoyagi et al. [109] | |
| OCCC | 5% (3 of 55) | Itamochi et al. [17] | |
| HGSOC | 1 of 1 c | Muppala et al. [23] | |
| SMARCA2 | OCCC | 2% (1 of 55) | Itamochi et al. [17] |
| SMARCB1 | SCCOHT | 1 of 1 | Simões et al. [24] |
| SCCOHT | 1 of 1 | Ramos et al. [25] | |
| SMARCC1 | OCCC | 2% (1 of 55) | Itamochi et al. [17] |
Overlap between cohorts exists in some studies. Percentages are reported for cohort sizes ≥ 10. Abbreviations: OCCC, ovarian clear cell carcinoma; EnOC, endometrioid ovarian cancer; MOC, mucinous ovarian cancer; HGSOC, high-grade serous ovarian cancer; SCCOHT, Small cell carcinoma of the ovary, hypercalcemic type. Symbols: a Corresponding immunohistochemical data are reported in Table 2; b with immature teratoma component; c includes patients with confirmed germline mutation; d extensive overlap exists between cases in these reports.

Table 3.
Immunohistochemical analyses of specific SWI/SNF complex members identified in primary ovarian cancers of different histotypes.
Table 3.
Immunohistochemical analyses of specific SWI/SNF complex members identified in primary ovarian cancers of different histotypes.
| Complex | Histotype | Loss of Expression | Reference |
|---|---|---|---|
| Member | |||
| ARID1A | OCCC | 76% (31 of 41) a | Kuroda et al. [15] |
| OCCC | 55% (23 of 42) | Yamamoto et al. [80] | |
| OCCC | 39% (44 of 112) | Itamochi et al. [81] | |
| OCCC | 33% (30 of 92) | Bennett et al. 2021 [27] | |
| OCCC | 15% (9 of 60) | Katagiri et al. [82] | |
| EnOC | 60% (12 of 20) a | Kuroda et al. [15] | |
| HGSOC | 19% (7 of 36) a | Kuroda et al. [15] | |
| HGSOC | 0% (0 of 17) | Katagiri et al. [82] | |
| MOC | 0 of 6 a | Kuroda et al. [15] | |
| ARID1B | OCCC | 15% (8 of 53) | Sato et al. [85] |
| SMARCA4/ | SCCOHT | 100% (12 of 12) | Karianian-Phillipe et al. [110] |
| BRG1 | SCCOHT | 97% (34 of 25) a | Witkowski et al. [89] |
| SCCOHT | 96% (54 of 56) b | Clarke et al. [111] | |
| SCCOHT | 94% (16 of 17) | Conlon et al. [112] | |
| SCCOHT | 92% (46 of 50) c | Karnezis et al. [113] | |
| SCCOHT | 89% (16 of 18) | Zheng et al. [114] | |
| SCCOHT | 88% (39 of 44) | Genestie et al. [86] | |
| SCCOHT | 84% (16 of 19) a | Ramos et al. [25] | |
| SCCOHT | 82% (14 of 17) a | Ramos et al. [92] | |
| SCCOHT | 64% (7 of 11) a,d | Jelinic et al. [90] | |
| SCCOHT | 2 of 2 a | Chandan et al. [96] | |
| SCCOHT | 1 of 1 | Aggarwal et al. [115] | |
| SCCOHT | 1 of 1 e | Atwi et al. [116] | |
| SCCOHT | 1 of 1 a | Lavrut et al. [106] | |
| SCCOHT | 1 of 1 a | Fahiminiya et al. [107] | |
| SCCOHT | 1 of 1 | Altmann et al. [117] | |
| SCCOHT | 1 of 1 a | Gao et al. [108] | |
| SCCOHT | 1 of 1 | Saylany et al. [118] | |
| SCCOHT | 1 of 1 a | Aoyagi et al. [109] | |
| SCCOHT | 0 of 1 | Coşkun et al. [119] | |
| SCCOHT | 0 of 7 a | Mazibrada et al. [94] | |
| OCCC | 5% (1 of 20) | Jelinic et al. [26] | |
| OCCC | 4% (15 of 360) | Karnezis et al. [113] | |
| OCCC | 3% (1 of 37) | Conlon et al. [112] | |
| OCCC | 2% (2 of 93) | Ramos et al. [92] | |
| OCCC | 0% (0 of 105) | Bennett et al. [27] | |
| HGSOC | 0% (0 of 1198) | Karnezis et al. [113] | |
| HGSOC | 0% (0 of 204) | Ramos et al. [92] | |
| HGSOC | 0% (0 of 42) | Conlon et al. [112] | |
| HGSOC | 0% (0 of 33) | Karianian-Phillipe et al. [110] | |
| endometrioid f | 0% (0 of 268) | Karnezis et al. [113] | |
| EnOC | 0% (0 of 38) | Conlon et al. [112] | |
| endometrioid f | 0% (0 of 36) | Ramos et al. [92] | |
| mucinous g | 0% (0 of 110) | Karnezis et al. [113] | |
| mucinous g | 0% (0 of 14) | Ramos et al. [92] | |
| LGSOC | 0% (0 of 53) | Karnezis et al. [113] | |
| LGSOC | (0 of 9) | Ramos et al. [92] | |
| SMARCA2/ | SCCOHT | 100% (45 of 45) h | Karnezis et al. [113] |
| BRM | SCCOHT | 90% (9 of 10) | Jelinic et al. [26] |
| SCCOHT | 86% (31 of 36) | Genestie et al. [86] | |
| SCCOHT | 7 of 7 a | Mazibrada et al. [94] | |
| SCCOHT | 5 of 6 | Auguste et al. [18] | |
| SCCOHT | 1 of 1 | Sanders et al. [87] | |
| SCCOHT | 1 of 1 | Mehta et al. [101] | |
| SCCOHT | 1 of 1 | Simões et al. [24] | |
| SCCOHT | 1 of 1 | Altmann et al. [117] | |
| OCCC | 8% (8 of 104) | Bennett et al. [27] | |
| OCCC | 5% (1 of 20) | Jelinic et al. [26] | |
| SMARCB1/ | SCCOHT | 13% (2 of 16) | Ramos et al. [25] |
| INI-1 | SCCOHT | 6% (3 of 50) | Karnezis et al. [113] |
| SCCOHT | 0% (37 of 37) | Clarke et al. [111] | |
| SCCOHT | 0 of 1 | Coşkun et al. [119] | |
| SCCOHT | 0 of 1 | Mehta et al. [101] | |
| OCCC | 0% (0 of 150) | Bennett et al. [27] |
Percentages are reported for cohort sizes ≥10. There is extensive overlap in cohorts studied in Ramos et al. [92], Ramos et al. [25], and Karnezis et al. [113]. Abbreviations: OCCC, ovarian clear cell carcinoma; EnOC, endometrioid ovarian cancer; MOC, mucinous ovarian cancer; HGSOC, high-grade serous ovarian cancer; SCCOHT, Small cell carcinoma of the ovary hypercalcemic type. Symbols: a Corresponding gene mutation data are reported in Table 1; b 41 samples in this study previously published, including genetic and immunohistochemical data; c SCCOHT cohort consists of 46 primary tumours, 2 patient-derived xenografts and 2 cell lines; d three samples reported as “equivocal” for BRG1/SMARCA4 immunostaining and one with an in-frame deletion showed the retention of BRG1/SMARCA4; e subsequent germline testing revealed a SMARCA4 mutation; f all of endometrioid carcinoma, mixed carcinoma and borderline tumours; g both borderline and malignant mucinous tumours; h all tumours reported for loss of BRM/SMARCA2 immunostaining had a mutation in either SMARCA4 (43 of 45) or SMARCB1 (2 of 45).
A single study has reported additional SWI/SNF complex member mutations in a small number of OCCC cases, specifically in both of the mutually exclusive ATPase subunits SMARCA4 (3 of 55 tumours) and SMARCA2 (1 of 55 tumours), as well as in the core scaffold subunit SMARCC1 (1 of 55 tumours) (Table 2) [17]. Furthermore, the loss of SMARCA4 detected by immunohistochemistry has been reported in between 2–5% of OCCC [26,92,112,113]. The immunohistochemical loss of SMARCA2 has also been reported in OCCC in between 5–8% of cases [26,27]. The dual loss of SMARCA4 and SMARCA2 has not been reported in OCCC [26,27]. A single immunohistochemical study of SMARCB1 in 105 OCCCs showed the retention of this protein [27]. It will be interesting to determine whether additional mutations or otherwise dysregulated SWI/SNF complex members will be identified in new studies of primary OCCCs.
3.2. Small Cell Carcinoma of the Ovary, Hypercalcaemic Type (SCCOHT)
3.2.1. Clinical Presentation and Epidemiology of SCCOHT
SCCOHT is an exceedingly rare and aggressive subtype of ovarian cancer, believed to account for less than 0.01% of all ovarian malignancies [120]. Cells constituting primary SCCOHT tumours have been described as small and round with hyperchromatic nuclei and minimal cytoplasm [121]. The cell of origin has been under debate, although aspects of its clinical presentation that include positive staining for the germ-cell markers SALL4, OCT3/4, alpha-fetoprotein (AFP) and glypican 3 suggest that SCCOHT is most likely of germ-cell origin [122]. Similarities between SCCOHT and malignant rhabdoid tumours have been drawn, with one study suggesting that SCCOHT be renamed as ‘malignant rhabdoid tumour of the ovary’ [123].
In contrast to other subtypes of ovarian cancer that generally, but not always, occur in post-menopausal women, SCCOHT is predominantly diagnosed in the vastly different demographic of infants, children and women of child-bearing age, with a median age of onset of 25 years [8,124]. The youngest diagnosis of SCCOHT reported to date is in a 12-month-old child, while the oldest is a woman of 56 years [116,125]. A recent case report described a 28-year-old woman diagnosed in the last trimester of pregnancy [118]. Patients have reported with general symptoms including nausea, weight loss, constipation and fatigue, as well as abdominal pain and/or swelling. Between 50–70% of patients with SCCOHT have associated hypercalcaemia [8,126,127]. Immunohistochemical analyses of primary tumours have observed elevated parathyroid hormone-related protein (PTHrp) in some cases, suggesting that PTHrp may be the underlying cause of hypercalcaemia in SCCOHT [128]. In a systematic review of 67 studies describing 306 SCCOHT patients, elevation of the serum glycoprotein CA-125 was reported in around 80% [8].
The five-year OS for patients diagnosed at the earliest stage (FIGO Stage I) has been reported as 51%, with patients diagnosed after this time (FIGO Stages II-IV) having a reduced OS of only 24% [8]. Both familial and sporadic cases of SCCOHT have been reported [124,125]. The international SCCOHT consortium has proposed consensus guidelines for the diagnosis and care of SCCOHT patients, including radical surgery, adjuvant chemotherapy and radiotherapy [121]. Despite this, the risk of spread beyond the ovary remains high and outcomes are poor [127]. While SCCOHT tumours are initially chemosensitive, the time to recurrence can be short, with relapsed tumours displaying a reduced response to chemotherapy [121,127].
3.2.2. SWI/SNF Complex Member Mutations in SCCOHT
SCCOHT tumours have a low tumour mutational burden (TMB), with studies reporting less than six mutations/Mb on a background of genomic stability and low copy number changes [18,91]. The mutation of one of the SWI/SNF ATPases, SMARCA4, is the predominant mutation in SCCOHT, reported in between 69–100% of cases in studies of 10 or more patients [22,25,88,89,90,91,92]. Similar results have been reported in studies of less than 10 patients, as well as in many case studies [18,87,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109]. Numerous studies have reported the presence of a germline SMARCA4 mutation in SCCOHT [22,25,87,89,92,93,96,99,101,102,103,104,106]. This would indicate that at least in these tumours, SMARCA4 mutation is occurring as an early event across the timeline of tumorigenesis. SMARCA4 mutation has also been implied by immunohistochemical studies of SMARCA4 depletion in the absence of genetic analyses [86,110,111,112,113,114,115,116,117,118,119]. Reports of SMARCA4 mutations and protein loss in SCCOHT are summarised in Table 2 and Table 3.
The loss or depletion of SMARCA2, but not SMARCA2 mutation, is reported in SCCOHT concomitant with SMARCA4 mutations, implying epigenetic mechanisms of SMARCA2 silencing [18,24,26,86,87,94,101,113,117] (Table 2 and Table 3). The loss of both the SWI/SNF ATPases is exceedingly rare in human malignancy, as cancer cells normally need at least one of these ATPases to be functioning in order to survive, proliferate and metastasise [113]. Rare variants of non-ovarian cases of concomitant SMARCA2 and SMARCA4 loss have been reported for cancer types including lung [88,129], endometrial [130], gastrointestinal tract [131], sinonasal undifferentiated carcinoma [132], and rhabdoid tumours [133]. The key survival mechanisms of this intriguing dual loss of the mutually exclusive SWI/SNF ATPases SMARCA4 and SMARCA2 remain to be determined. It has been reported that after the dual loss of SMARCA4 and SMARCA2, the residual SWI/SNF complex can still bind to accessible chromatin but results in disrupted transcriptional regulation that impacts upon the expression of genes involved in cellular processes and behaviours, including differentiation, epithelial–mesenchymal transition (EMT), metastasis, DNA repair, apoptosis, adhesion, immunity, metabolism, drug metabolism, proliferation and angiogenesis [134,135].
The mutation of SMARCB1, one of the most conserved subunits of SWI/SNF, is rarely reported in SCCOHT. A 19-year-old woman presented with a highly aggressive case of SCCOHT that was found to have a somatic nonsense point mutation in SMARCB1 with loss of the wild-type allele and a low TMB of less than two mutations/Mb [24]. The patient received high-dose chemotherapy and stem cell transplantation; however, disease progression was rapid and she died 11 months after the presentation of initial symptoms. A homozygous frameshift SMARCB1 mutation in a SCCOHT tumour has also been reported [25] (Table 2). The immunohistochemical loss of SMARCB1 has been reported in 6% (3 of 50) [113] and 13% (2 of 16) [25] of SCCOHT tumours studied (Table 3). Mutation of ARID1A and ARID1B has also been infrequently reported in SCCOHT [18,86,87] (Table 2).
3.3. Endometrioid Ovarian Cancer (EnOC) and Other Ovarian Cancer Subtypes—Links to Abrogated SWI/SNF
Similar to OCCC, EnOC has also been associated with the presence of endometriosis, with these two subtypes together referred to as EAOCs [14,19,20,21,56]. A large systematic review and meta-analysis found a 2.3-fold higher risk of EnOC in women with endometriosis [136]. EnOC is a distinct ovarian cancer subtype accounting for up to ~15% of all ovarian cancers [137]. It tends to be diagnosed at an earlier stage (FIGO Stages I–II), and as a result, most patients have a more favourable prognosis than other ovarian cancer subtypes [138]. In addition to its association with endometriosis, EnOC is also associated with Lynch syndrome [139] and can occur in patients who have endometrial cancer [138,140]. Relative to SCCOHT and OCCC, there are fewer reports investigating SWI/SNF complex members in EnOC. ARID1A mutations have been reported in between 21–45% of cases of EnOC [14,15,74,79] (Table 2). Furthermore, the immunohistochemical loss of ARID1A has been reported in 60% (12 of 20) of EnOC cases [15]. In studies of EnOC, mixed carcinoma and borderline tumours did not identify the immunohistochemical loss of SMARCA4 (Table 3) [92,112,113].
Table 2 and Table 3 also summarise the involvement of SWI/SNF complex members in ovarian cancer subtypes other than OCCC, EnOC and SCCOHT. ARID1A mutations have been reported in a single study in 2 of 6 cases of MOC [15]; however, the same study did not report a corresponding loss of ARID1A following immunohistochemical analyses. Further, correlations observed between ARID1A mutations and CD8 and PD-L1 (programmed death ligand 1) levels in other ovarian cancer subtypes were not observed for MOC in this study [15]. Immunohistochemical studies of borderline and malignant MOC have not shown a loss of SMARCA4 [92,113]. Two immunohistochemical studies in low-grade serous ovarian cancer (LGSOC) reported a retention of SMARCA4 [92,113]. Of note, mutation of ARID1A has been reported infrequently in HGSOC [14,15,16], with a single case report of a SMARCA4 mutation in HGSOC [23]. This patient presented at 57 years of age, and her SMARCA4 mutation was found to be germline. It is important to note that this patient also had a BRCA2 variant of unknown significance that could not be excluded as a driver mutation of her HGSOC. Further, SMARCC1, referred to as BAF155, is proposed to be methylated by the arginine methyltransferase CARM1 (Coactivator-Associated Arginine Methyltransferase 1) in HGSOC, typically without mutations in BRCA1 or BRCA2 [141]. While interesting, additional studies of CARM1 amplification and BAF155 methylation leading to down-regulation in primary ovarian tumours would be required to draw further conclusions.
3.4. Endometriosis and the SWI/SNF Complex
Globally, endometriosis is believed to affect 5–10% of women of reproductive age [142], however, this figure increases for certain groups of patients, such as those who are symptomatic [143]. Despite these high frequencies, the exact cause of this chronic condition has not been determined, with theories including retrograde menstruation and endometriotic lesions being of stem cell origin [144]. Numerous studies have reported an elevated risk of developing ovarian cancer in patients with endometriosis [64,66,67,145,146,147,148,149,150,151]. The most recent and one of the largest of these studies by Barnard and colleagues investigated the Utah Population Database and reported a 4.2-fold higher ovarian cancer risk in women with endometriosis compared to those without, and a 9.7-fold greater risk for women with ovarian endometriomas and/or deep-infiltrating endometriosis [145]. Similar results were also seen in an earlier study, whereby a higher proportion of these ovarian cancers were OCCC or EnOC [136]. Endometriosis has been identified in between 21–51% of women with OCCC and 23–43% of women with EnOC [146,152,153].
Identical mutations in ARID1A have been identified in OCCC and synchronous endometriotic lesions, providing a strong argument for malignant OCCC arising from these apparently benign lesions [67]. Should this be the case, it would suggest that at least in OCCC associated with endometriosis, the loss of ARID1A is an early event in the timeline of tumorigenesis. Endometriotic lesions occurring without EAOCs or distant from an ARID1A-deficient EAOC have shown diffuse immunoreactivity for ARID1A [80]. The immunohistochemical loss of ARID1A in endometriotic lesions has been suggested as a putative prognostic biomarker for ovarian cancer risk [149]. The potential involvement of other abrogated SWI/SNF complex members in endometriosis is currently unknown. The underlying factors driving the malignant transformation of endometriotic lesions remain to be fully elucidated, with suggestions including exposure to oestrogen or an imbalance of oestrogen receptors, oxidative stress, inflammatory processes, local nutrient availability and metabolic reprogramming [19]. Whether ARID1A defects present an actionable target in endometriosis also remains to be determined, with the types of drugs discovered to treat malignancy needing to be considered in a different context for potential management of a predominantly benign chronic condition.
4. SWI/SNF Abrogation in Ovarian Cancer Represents Therapeutic Vulnerabilities
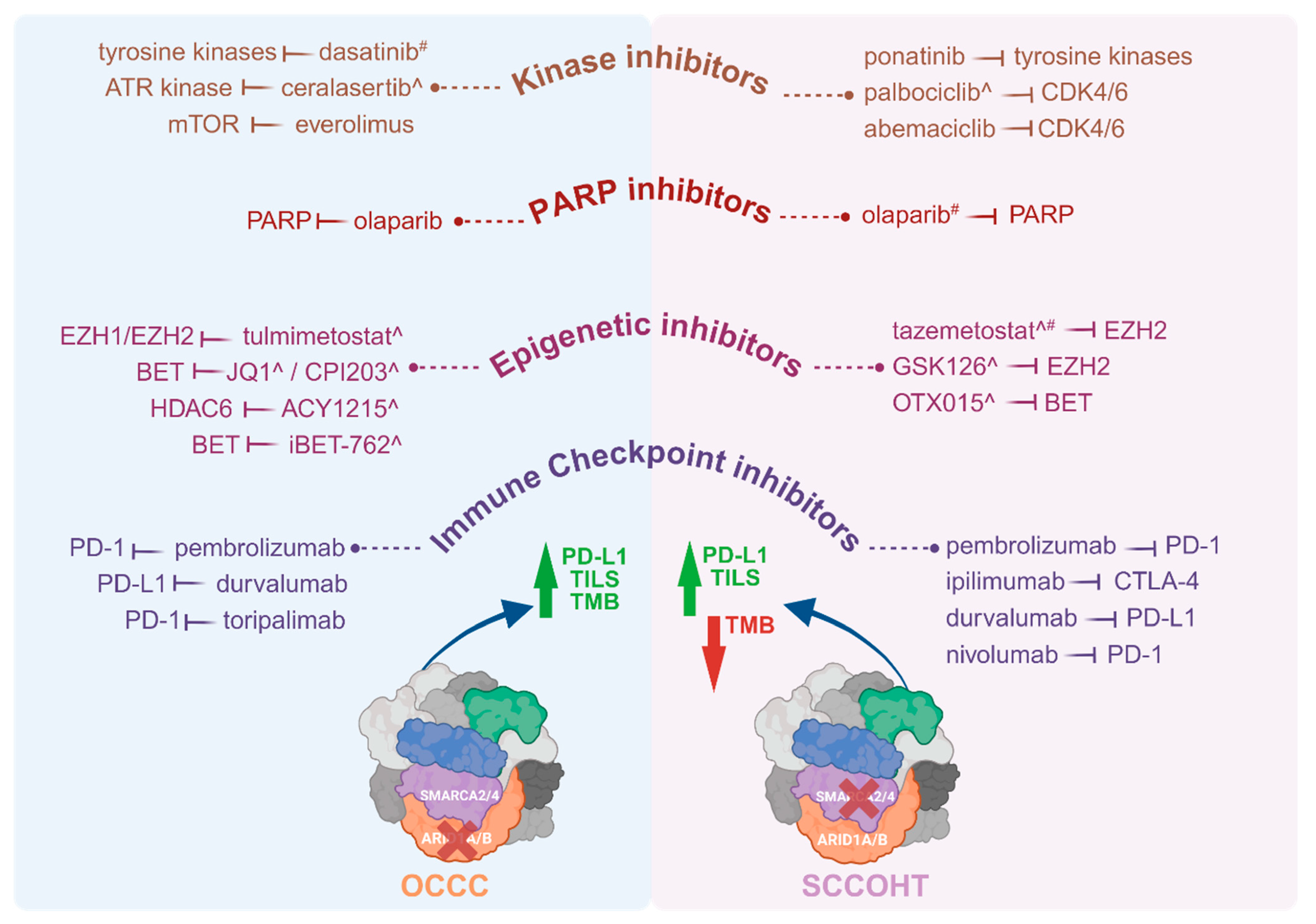
Pharmacological targeting of the effects of mutant SWI/SNF complex members, with a specific focus on mutant ARID1A and SMARCA4 for the purpose of treating patients with OCCC or SCCOHT, span a number of drug classes. These include immunotherapeutics, kinase inhibitors, inhibitors of the DNA damage response including PARP inhibitors, and epigenetic inhibitors (Figure 2).
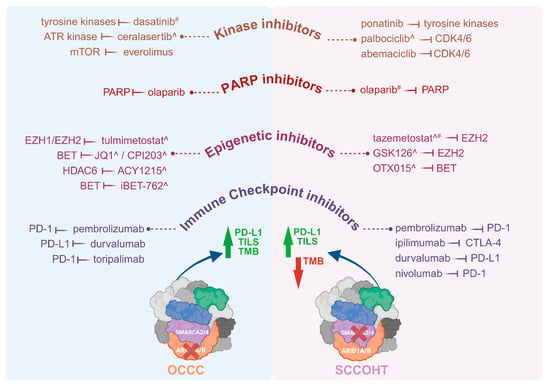
Figure 2.
Therapeutic drugs investigated in patients with OCCC or SCCOHT and pre-clinical models of these tumours. Molecular targeted therapies including immune checkpoint inhibitors, epigenetic inhibitors, PARP inhibitors and kinase inhibitors have been trialled in patients with OCCC or SCCOHT, as well as in vitro and in vivo pre-clinical models ^ of these malignancies. Where patients did not respond to, or tolerate, a drug, this is indicated by #. Drugs listed were administered to patients or tested in pre-clinical models either as monotherapies or in conjunction with other drug(s). A higher TMB is reported for OCCC, indicated by a green arrow, while SCCOHT has a low TMB, indicated by a red arrow. Both OCCC and SCCOHT have TILs. Both tumour types have high levels of PD-L1. Therapeutic drugs tested in OCCC patients, include pembrolizumab [154], durvalumab [155], toripalimab [156], olaparib [157], everolimus [156], and dasatinib [158], and in pre-clinical models include iBET-762 [159], ACY1215 [160], CPI203 [159], ceralasertib [161] and tulmimetostat [162]. Therapeutic drugs tested in SCCOHT patients include nivolumab [163], ipilimumab [163], pembrolizumab [103,164], durvalumab [108], olaparib [108,163], tazemetostat [165], abemaciclib [163], palbociclib [108] and ponatinib [163], and in pre-clinical models include GSK126 [166], OTX015 [167], tazemetostat [166,168] and palbociclib [169]. Drugs trialled in patients were on occasion administered either sequentially, informed by patient response, or together. Drug combinations of this nature in OCCC patients included pembrolizumab (combined with bevacizumab and cyclophosphamide) [154], pembrolizumab (combined with bevacizumab and olaparib) [157], and toripalimab (combined with everolimus) [156]. Drug combinations trialled in SCCOHT patients included pembrolizumab (following cycles of cisplatin/etoposide and carboplatin/paclitaxel) [103], nivolumab and ipilimumab (followed by ponatinib, abemaciclib and olaparib) [163]. In a single case report, a SCCOHT patient was administered six lines of chemotherapy of multiple drugs that included durvalumab, olaparib and palbociclib [108]. Abbreviations: ATR, Ataxia-telangiectasia-mutated (ATM) and RAD3-related; BET, bromo- and extra-terminal domain family; CDK4/6, cyclin-dependent kinases 4 and 6; CTLA-4, cytotoxic T-lymphocyte-associated protein 4; EZH1/2, Enhancer Of Zeste 1/2 Polycomb repressive complex 2 subunit; HDAC6, histone deacetylase 6; mTOR, mammalian target of rapamycin; OCCC, ovarian clear cell carcinoma; PARP, Poly (ADP-ribose) polymerase; PD-1, programmed cell death protein 1; PD-L1, programmed death-ligand 1; SCCOHT, Small cell carcinoma of the ovary, hypercalcaemic type; TILS, Tumour-infiltrating lymphocytes; TMB, tumour mutational burden. Both tumour types also have high levels of PD-L1. Created with www.BioRender.com, Access Date: 9 August 2024.
4.1. Immunotherapy
Most ovarian cancer subtypes, and especially the most frequently diagnosed HGSOC, at best show only modest responses to immunotherapy, with clinical trials suggesting responses as low as 8% of patients [170,171,172,173]. This does not appear to be the case for OCCC and SCCOHT, with ICB being trialled on patients with SCCOHT and OCCC. The knockout of ARID1A has been shown to increase levels of CD274 and the protein it encodes—PD-L1, the predominant ligand of programmed death 1 (PD-1)—both in vitro and in vivo [160,174]. Wild-type ARID1A and ARID1B are located at the promoter of CD274 in OCCC cells (shown in the human OVCA429 and RMG1 cell lines, as well as mouse ovarian ID8-Defb29/Vegf cancer cells), although ARID1B was not able to influence the levels of CD274 in ARID1A knockout (KO) cells [160]. CRISPR-Cas9 KO of ARID1A in ID8 cells investigated in both an intraperitoneal and an orthotopic model showed increased tumour-infiltrating lymphocytes and PD-L1 levels and a greater response to an anti-PD-L1 antibody compared to WT ARID1A tumours, with lower tumour burden and prolonged survival [174]. All of these factors suggest that a response to ICB is likely in the presence of ARID1A mutation.
ARID1A is a binding partner of the mismatch repair (MMR) protein MSH2, although it does not appear to regulate MSH2, or other MMR genes (MLH1, MSH3, MSH6, PMS1 and PMS2), at the transcriptional level [174]. OCCC has a higher TMB than other ovarian cancer subtypes, although this is not seen in all cases [15,175]. Like SCCOHT, OCCC has a relatively low level of copy number abnormalities, possibly due to the loss of ARID1A causing defects in telomere cohesion leading to the removal of defective chromosomal changes during mitosis [176]. The anti-PD-L1 monoclonal antibody durvalumab that blocks the binding of PD-L1 and PD-1, as well as CD80, has been trialled in OCCC (www.ClinicalTrials.gov, NCT03405454) [155]. A combination of pembrolizumab (anti-PD-1 monoclonal antibody), bevacizumab (anti-angiogenic anti-VEGF monoclonal antibody) and cyclophosphamide has been trialled in a small number of patients with OCCC, with durable responses observed, but also treatment-limiting toxicities [154]. ICB has been the focus of other studies in OCCC with mixed results in individual patients [156,177]. Investigation of an OCCC molecular signature that maximises the likelihood that a patient will respond to ICB is warranted [178].
In contrast to OCCC, SCCOHT tumours have low TMB and are genomically stable, but like OCCC have low copy number variations [18,91]. This molecular profile would suggest that SCCOHT may not be responsive to immunotherapy; however, many tumours have been shown to have high PD-L1 expression in both tumour and stromal cells accompanied by robust associated T-cell infiltration [90]. It is possible that the loss of SMARCA4 in SCCOHT reprograms the transcriptional landscape to influence tumour immunogenicity, creating an environment that is more permissive to ICB [90].
Anti-PD-1 ICB with pembrolizumab has been trialled in SCCOHT patients with encouraging results [90,164], although not all SCCOHT patients respond to ICB [103,164]. A 34-year-old patient diagnosed with FIGO Stage II SCCOHT with a somatic SMARCA4 mutation was reported to have a remarkable response of over 5 years of survival post recurrence, treated with ICB plus anti-angiogenic therapy and CDK4/6 (cyclin-dependent kinase 4 and 6) inhibitors [108]. A 21-year old patient diagnosed with SCCOHT and a germline SMARCA4 mutation underwent high-dose chemotherapy followed by an autologous stem cell transplant and a combination of drugs, including nivolumab (anti-PD-1 monoclonal antibody) and ipilimumab (anti-CTLA-4 (Cytotoxic T-Lymphocyte Associated Protein 4) monoclonal antibody), as well as the multi-tyrosine kinase inhibitor ponatinib and abemaciclib (selective CDK4/6 inhibitor), surviving at least three years post diagnosis [163]. Clinical trial activity focused on ICB for SCCOHT patients, including in combination with other therapies, have been undertaken or are currently active [108,121]. As for OCCC, decisions regarding ICB in SCCOHT patients will benefit from determining molecular signatures that are congruent with response to ICB drugs.
4.2. Kinase Inhibitors
Drugs in the receptor tyrosine kinase family have been shown to have selective benefit in SCCOHT cell lines with the dual loss of SMARCA4 and SMARCA2 [179]. Unbiased synthetic lethal screens using a short hairpin RNA (shRNA) targeting the human kinome in a SMARCA4/SMARCA2-deficient model of SCCOHT led to the identification of CDK4/6 inhibitors as a molecular target therapy for this malignancy [169]. The molecular mechanism underpinning this sensitivity is low levels of cyclin D1 in SMARCA4-deficient cells, given that SMARCA4 directly regulates the transcription of the cyclin D1 gene CCND1 [169]. As noted above, treatment responses in SCCOHT have been observed with ponatinib and abemaciclib in conjunction with other therapies, including ICB [163]. Dasatinib, a second-generation multi-tyrosine kinase inhibitor administered as a monotherapy, did not show treatment benefits for OCCC [158]. The loss of ARID1A has been linked to the activation of MAPK signalling [180], sensitivity to PI3K/AKT/mTOR inhibitors and ATR inhibitors [161,181], as well as aurora kinase inhibition [182], warranting further investigation for the development of molecularly targeted therapies against ARID1A-mutated ovarian cancers.
4.3. PARP Inhibitors
The SWI/SNF complex plays an active role in modelling the accessibility of chromatin to accommodate complexities of the DNA damage response [38,183]. In cells with intact SWI/SNF, ARID1A is recruited to DSBs through its association with the DNA damage checkpoint kinase ATR [174]. In cells lacking ARID1A, there is a delay in the recruitment of repair factors to sites of DNA damage. Further, in ARID1A null cells, increased PARP activity has been reported, making these cells susceptible to PARP inhibition with olaparib, and especially in conjunction with ionising radiation therapy [181]. This increased sensitivity of ARID1A null cells to PARP inhibition is seen both in vitro and in vivo [174]. Some OCCC cell lines are sensitive to PARP inhibitors in vitro [184]. The alkylating agent temozolomide (TMZ) combined with PARP inhibition has also been shown to induce replication fork instability and apoptosis in ARID1A mutant ovarian cancer xenografts [185]. An OCCC patient with an ARID1A mutated tumour achieved a partial and sustained response to the combination of olaparib, pembrolizumab and bevacizumab [157]. A compelling case for the use of PARP inhibitors in SCCOHT is yet to be made, given that olaparib is not always well tolerated in these patients [108,163].
4.4. Epigenetic Inhibitors
The interaction of the SWI/SNF complex with other epigenetic complexes and regulators offers opportunities for therapeutic interventions based on synthetic lethal relationships with mutant SWI/SNF complex members. To date, the exploration of epigenetic inhibitors to treat either OCCC or SCCOHT has been predominantly confined to pre-clinical models only. In this vein, the antagonistic relationship of SWI/SNF and the PRC2 has raised the pharmacological inhibition of the PRC2 catalytic subunit EZH2, a histone methyltransferase, in ARID1A mutant cells, including OCCC, as a potential therapy [186,187,188]. The second-generation EZH2 inhibitor tulmimetostat has shown promise in pre-clinical models of ARID1A mutated malignancies, including OCCC [162]. EZH2 inhibition is also under investigation for the treatment of SCCOHT. The EZH2 inhibitor tazemetostat (EPZ-6438) has displayed efficacy in pre-clinical models of SCCOHT with SMARCA4/SMARCA2 loss [166,168], as has the EZH2 inhibitor GSK126 [166]. It remains to be determined whether strong responses in SCCOHT patients will be generally achieved [189], however, a Phase I/II trial for tazemetostat (NCT02601950) failed stage 2 futility that needed a confirmed partial response or complete response in at least five patients based on the RECIST 1.1 criteria [165].
Preclinical models of SCCOHT have also demonstrated responses to pan-HDAC inhibitors, including quisinostat, and in combination with EZH2 inhibitors at sub-lethal doses have worked synergistically to induce apoptosis both in vitro and in vivo [189]. Using the same strategies of targeting mutant SWI/SNF, the HDAC6 inhibitor ACY1215 combined with anti-PD-L1 antibody reduced tumour burden and eliminated ascites in an in vivo model of ARID1A-inactivated OCCC [160]. Lastly, the BET (bromodomain and extra-terminal domain) family of proteins classified as epigenetic readers is being considered for inhibition in both OCCC and SCCOHT. BET inhibitors JQ1 and iBET-762 have shown efficacy in in vitro and in vivo models of ARID1A mutant OCCC [159]. Combination drug treatments with the BET inhibitor CPI203 and select PI3K-AKT inhibitors showed efficacy in preclinical models of OCCC, although this appeared to be independent of ARID1A mutation status [190]. Combinations of the BET inhibitor OTX015 that targets BET family member BRD2 (bromodomain containing 2) and MEK (mitogen-activated protein kinase kinase) inhibitors have also shown efficacy in preclinical models of SCCOHT, although this does not appear to be specific for tumours with concomitant SMARCA4 and SMARCA2 loss [167].
5. Conclusions
Mutation and epigenetic silencing of SWI/SNF complex members are associated with the poorer prognosis, chemoresistant subtypes of ovarian cancer occurring in younger patients. In the case of SCCOHT, this disease can occur in infants. ARID1A mutation is present in endometriosis, and given that OCCC and EnOC are identified as EAOCs, this points to the involvement of abrogated SWI/SNF as an early driver of tumorigenesis. Studies investigating the mechanistic behaviour of mutant SWI/SNF complex members are revealing potential therapeutic opportunities for the treatment of patients with OCCC and SCCOHT that have already demonstrated clinical benefit for some patients. A deeper understanding of mutant SWI/SNF in ovarian and other cancers, as well as in endometriosis, may reveal new opportunities for the pharmacological inhibition of the key molecular targets associated with these diseases.
Author Contributions
Conceptualisation, D.J.M., Y.M. and N.R.F.; data curation, N.R.F., Y.M., K.-A.D., T.X. and D.J.M.; writing—original draft preparation, D.J.M.; writing—review and editing, D.J.M., Y.M., N.R.F., T.X., S.B., E.G.K., T.S.S., A.A., F.A.S., N.T., N.A.B. and K.-A.D.; visualisation, N.R.F., D.J.M., A.A., F.A.S. and T.S.S.; supervision, D.J.M. and N.T.; project administration, D.J.M.; funding acquisition, D.J.M. and N.A.B. All authors have read and agreed to the published version of the manuscript.
Funding
Y.M. and T.X. are supported by a Chinese Scholarship Council scholarship. N.R.F. and T.S.S. are supported by Australian Government Research Training Program (RTP) scholarships. F.A.S. is supported by an Australian Medical Research Future Fund grant (APP1199620) to N.A.B. and D.J.M., and a Cancer Council NSW grant (2019/TPG1002) to D.J.M. N.A.B. is supported by the Hunter Medical Research Institute Vanessa McGuigan Memorial Fellowship. This work was supported by a National Health and Medical Research Council (NHMRC) Australia grant [2019296] (to D.J.M. and N.A.B.) supporting A.A.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Figures were generated using Biorender (www.Biorender.com, Access Date: 9 August 2024).
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Kehoe, S.; Hook, J.; Nankivell, M.; Jayson, G.C.; Kitchener, H.; Lopes, T.; Luesley, D.; Perren, T.; Bannoo, S.; Mascarenhas, M.; et al. Primary chemotherapy versus primary surgery for newly diagnosed advanced ovarian cancer (CHORUS): An open-label, randomised, controlled, non-inferiority trial. Lancet 2015, 386, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Dickson, K.A.; Yee, C.; Ma, Y.; Ford, C.E.; Bowden, N.A.; Marsh, D.J. Targeting Homologous Recombination Deficiency in Ovarian Cancer with PARP Inhibitors: Synthetic Lethal Strategies That Impact Overall Survival. Cancers 2022, 14, 4621. [Google Scholar] [CrossRef]
- Poveda, A.; Floquet, A.; Ledermann, J.A.; Asher, R.; Penson, R.T.; Oza, A.M.; Korach, J.; Huzarski, T.; Pignata, S.; Friedlander, M.; et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): A final analysis of a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2021, 22, 620–631. [Google Scholar] [CrossRef] [PubMed]
- Maiorano, B.A.; Maiorano, M.F.P.; Maiello, E. Olaparib and advanced ovarian cancer: Summary of the past and looking into the future. Front. Pharmacol. 2023, 14, 1162665. [Google Scholar] [CrossRef]
- Ray-Coquard, I.; Leary, A.; Pignata, S.; Cropet, C.; González-Martín, A.; Marth, C.; Nagao, S.; Vergote, I.; Colombo, N.; Mäenpää, J.; et al. Olaparib plus bevacizumab first-line maintenance in ovarian cancer: Final overall survival results from the PAOLA-1/ENGOT-ov25 trial. Ann. Oncol. 2023, 34, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Ray-Coquard, I.; Pautier, P.; Pignata, S.; Pérol, D.; González-Martín, A.; Berger, R.; Fujiwara, K.; Vergote, I.; Colombo, N.; Mäenpää, J.; et al. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2416–2428. [Google Scholar] [CrossRef]
- Chan, J.K.; Teoh, D.; Hu, J.M.; Shin, J.Y.; Osann, K.; Kapp, D.S. Do clear cell ovarian carcinomas have poorer prognosis compared to other epithelial cell types? A study of 1411 clear cell ovarian cancers. Gynecol. Oncol. 2008, 109, 370–376. [Google Scholar] [CrossRef]
- Wens, F.; Hulsker, C.C.C.; Fiocco, M.; Zsiros, J.; Smetsers, S.E.; de Krijger, R.R.; van der Steeg, A.F.W.; Zweemer, R.P.; Baas, I.O.; Roes, E.M.; et al. Small Cell Carcinoma of the Ovary, Hypercalcemic Type (SCCOHT): Patient Characteristics, Treatment, and Outcome-A Systematic Review. Cancers 2023, 15, 3794. [Google Scholar] [CrossRef]
- Kadoch, C.; Hargreaves, D.C.; Hodges, C.; Elias, L.; Ho, L.; Ranish, J.; Crabtree, G.R. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013, 45, 592–601. [Google Scholar] [CrossRef]
- Shain, A.H.; Pollack, J.R. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS ONE 2013, 8, e55119. [Google Scholar] [CrossRef]
- Mardinian, K.; Adashek, J.J.; Botta, G.P.; Kato, S.; Kurzrock, R. SMARCA4: Implications of an Altered Chromatin-Remodeling Gene for Cancer Development and Therapy. Mol. Cancer Ther. 2021, 20, 2341–2351. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Chen, X.; Su, C.; Ren, S.; Zhou, C. Pan-cancer analysis of ARID1A Alterations as Biomarkers for Immunotherapy Outcomes. J. Cancer 2020, 11, 776–780. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, Y.; Tokunaga, H.; Saito, S.; Shimokawa, K.; Katsuoka, F.; Bin, L.; Kojima, K.; Nagasaki, M.; Yamamoto, M.; Yaegashi, N.; et al. Identification of somatic genetic alterations in ovarian clear cell carcinoma with next generation sequencing. Genes Chromosomes Cancer 2018, 57, 51–60. [Google Scholar] [CrossRef]
- Wiegand, K.C.; Shah, S.P.; Al-Agha, O.M.; Zhao, Y.; Tse, K.; Zeng, T.; Senz, J.; McConechy, M.K.; Anglesio, M.S.; Kalloger, S.E.; et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 2010, 363, 1532–1543. [Google Scholar] [CrossRef]
- Kuroda, Y.; Chiyoda, T.; Kawaida, M.; Nakamura, K.; Aimono, E.; Yoshimura, T.; Takahashi, M.; Saotome, K.; Yoshihama, T.; Iwasa, N.; et al. ARID1A mutation/ARID1A loss is associated with a high immunogenic profile in clear cell ovarian cancer. Gynecol. Oncol. 2021, 162, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Vaicekauskaitė, I.; Dabkevičienė, D.; Šimienė, J.; Žilovič, D.; Čiurlienė, R.; Jarmalaitė, S.; Sabaliauskaitė, R. ARID1A, NOTCH and WNT Signature in Gynaecological Tumours. Int. J. Mol. Sci. 2023, 24, 5854. [Google Scholar] [CrossRef]
- Itamochi, H.; Oishi, T.; Oumi, N.; Takeuchi, S.; Yoshihara, K.; Mikami, M.; Yaegashi, N.; Terao, Y.; Takehara, K.; Ushijima, K.; et al. Whole-genome sequencing revealed novel prognostic biomarkers and promising targets for therapy of ovarian clear cell carcinoma. Br. J. Cancer 2017, 117, 717–724. [Google Scholar] [CrossRef]
- Auguste, A.; Blanc-Durand, F.; Deloger, M.; Le Formal, A.; Bareja, R.; Wilkes, D.C.; Richon, C.; Brunn, B.; Caron, O.; Devouassoux-Shisheboran, M.; et al. Small Cell Carcinoma of the Ovary, Hypercalcemic Type (SCCOHT) beyond SMARCA4 Mutations: A Comprehensive Genomic Analysis. Cells 2020, 9, 1496. [Google Scholar] [CrossRef]
- Steinbuch, S.C.; Lüß, A.M.; Eltrop, S.; Götte, M.; Kiesel, L. Endometriosis-Associated Ovarian Cancer: From Molecular Pathologies to Clinical Relevance. Int. J. Mol. Sci. 2024, 25, 4306. [Google Scholar] [CrossRef]
- Centini, G.; Schettini, G.; Pieri, E.; Giorgi, M.; Lazzeri, L.; Martire, F.G.; Mancini, V.; Raimondo, D.; Seracchioli, R.; Habib, N.; et al. Endometriosis-Related Ovarian Cancer: Where Are We Now? A Narrative Review towards a Pragmatic Approach. J. Clin. Med. 2024, 13, 1933. [Google Scholar] [CrossRef]
- Samartzis, E.P.; Noske, A.; Dedes, K.J.; Fink, D.; Imesch, P. ARID1A mutations and PI3K/AKT pathway alterations in endometriosis and endometriosis-associated ovarian carcinomas. Int. J. Mol. Sci. 2013, 14, 18824–18849. [Google Scholar] [CrossRef]
- Jelinic, P.; Mueller, J.J.; Olvera, N.; Dao, F.; Scott, S.N.; Shah, R.; Gao, J.; Schultz, N.; Gonen, M.; Soslow, R.A.; et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat. Genet. 2014, 46, 424–426. [Google Scholar] [CrossRef] [PubMed]
- Muppala, R.; Donenberg, T.; Huang, M.S.; Schlumbrecht, M.P. SMARCA4 germline gene mutation in a patient with epithelial ovarian: A case report. Gynecol. Oncol. Rep. 2017, 22, 45–47. [Google Scholar] [CrossRef]
- Simões, M.F.E.; da Costa, A.; Silva, T.N.; Fernandes, L.; Bovolim, G.; Torrezan, G.T.; Carraro, D.M.; Baiocchi, G.; Menezes, A.N.O.; Santana Dos Santos, E.; et al. Case Report of Small Cell Carcinoma of the Ovary, Hypercalcemic Type (Ovarian Rhabdoid Tumor) with SMARCB1 Mutation: A Literature Review of a Rare and Aggressive Condition. Curr. Oncol. 2022, 29, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Ramos, P.; Karnezis, A.N.; Hendricks, W.P.; Wang, Y.; Tembe, W.; Zismann, V.L.; Legendre, C.; Liang, W.S.; Russell, M.L.; Craig, D.W.; et al. Loss of the tumor suppressor SMARCA4 in small cell carcinoma of the ovary, hypercalcemic type (SCCOHT). Rare Dis. 2014, 2, e967148. [Google Scholar] [CrossRef] [PubMed]
- Jelinic, P.; Schlappe, B.A.; Conlon, N.; Tseng, J.; Olvera, N.; Dao, F.; Mueller, J.J.; Hussein, Y.; Soslow, R.A.; Levine, D.A. Concomitant loss of SMARCA2 and SMARCA4 expression in small cell carcinoma of the ovary, hypercalcemic type. Mod. Pathol. 2016, 29, 60–66. [Google Scholar] [CrossRef]
- Bennett, J.A.; Safdar, N.; Segal, J.P.; Lastra, R.R.; Oliva, E. Evaluation of SWI/SNF Protein Expression by Immunohistochemistry in Ovarian Clear Cell Carcinoma. Int. J. Gynecol. Pathol. 2021, 40, 156–164. [Google Scholar] [CrossRef]
- Shi, Y.; Shin, D.S. Dysregulation of SWI/SNF Chromatin Remodelers in NSCLC: Its Influence on Cancer Therapies including Immunotherapy. Biomolecules 2023, 13, 984. [Google Scholar] [CrossRef]
- Mandal, J.; Mandal, P.; Wang, T.L.; Shih, I.M. Treating ARID1A mutated cancers by harnessing synthetic lethality and DNA damage response. J. Biomed. Sci. 2022, 29, 71. [Google Scholar] [CrossRef]
- Tessier-Cloutier, B.; Kommoss, F.K.F.; Kolin, D.L.; Němejcová, K.; Smith, D.; Pors, J.; Stewart, C.J.R.; McCluggage, W.G.; Foulkes, W.D.; von Deimling, A.; et al. Dedifferentiated and Undifferentiated Ovarian Carcinoma: An Aggressive and Molecularly Distinct Ovarian Tumor Characterized by Frequent SWI/SNF Complex Inactivation. Mod. Pathol. 2024, 37, 100374. [Google Scholar] [CrossRef]
- Saha, D.; Animireddy, S.; Bartholomew, B. The SWI/SNF ATP-dependent chromatin remodeling complex in cell lineage priming and early development. Biochem. Soc. Trans. 2024, 52, 603–616. [Google Scholar] [CrossRef] [PubMed]
- Alfert, A.; Moreno, N.; Kerl, K. The BAF complex in development and disease. Epigenetics Chromatin 2019, 12, 19. [Google Scholar] [CrossRef]
- Sokpor, G.; Xie, Y.; Rosenbusch, J.; Tuoc, T. Chromatin Remodeling BAF (SWI/SNF) Complexes in Neural Development and Disorders. Front. Mol. Neurosci. 2017, 10, 243. [Google Scholar] [CrossRef] [PubMed]
- Abedini, A.; Landry, D.A.; Macaulay, A.D.; Vaishnav, H.; Parbhakar, A.; Ibrahim, D.; Salehi, R.; Maranda, V.; Macdonald, E.; Vanderhyden, B.C. SWI/SNF chromatin remodeling subunit Smarca4/BRG1 is essential for female fertility. Biol. Reprod. 2023, 108, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Fedoriw, A.M.; Magnuson, T. An essential role for a mammalian SWI/SNF chromatin-remodeling complex during male meiosis. Development 2012, 139, 1133–1140. [Google Scholar] [CrossRef]
- Smith, J.J.; Xiao, Y.; Parsan, N.; Medwig-Kinney, T.N.; Martinez, M.A.Q.; Moore, F.E.Q.; Palmisano, N.J.; Kohrman, A.Q.; Chandhok Delos Reyes, M.; Adikes, R.C.; et al. The SWI/SNF chromatin remodeling assemblies BAF and PBAF differentially regulate cell cycle exit and cellular invasion in vivo. PLoS Genet. 2022, 18, e1009981. [Google Scholar] [CrossRef]
- Harrod, A.; Lane, K.A.; Downs, J.A. The role of the SWI/SNF chromatin remodelling complex in the response to DNA double strand breaks. DNA Repair. 2020, 93, 102919. [Google Scholar] [CrossRef]
- Brownlee, P.M.; Meisenberg, C.; Downs, J.A. The SWI/SNF chromatin remodelling complex: Its role in maintaining genome stability and preventing tumourigenesis. DNA Repair. 2015, 32, 127–133. [Google Scholar] [CrossRef]
- Centore, R.C.; Sandoval, G.J.; Soares, L.M.M.; Kadoch, C.; Chan, H.M. Mammalian SWI/SNF Chromatin Remodeling Complexes: Emerging Mechanisms and Therapeutic Strategies. Trends Genet. 2020, 36, 936–950. [Google Scholar] [CrossRef]
- Alver, B.H.; Kim, K.H.; Lu, P.; Wang, X.; Manchester, H.E.; Wang, W.; Haswell, J.R.; Park, P.J.; Roberts, C.W. The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat. Commun. 2017, 8, 14648. [Google Scholar] [CrossRef]
- Wang, X.; Lee, R.S.; Alver, B.H.; Haswell, J.R.; Wang, S.; Mieczkowski, J.; Drier, Y.; Gillespie, S.M.; Archer, T.C.; Wu, J.N.; et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat. Genet. 2017, 49, 289–295. [Google Scholar] [CrossRef]
- Tolstorukov, M.Y.; Sansam, C.G.; Lu, P.; Koellhoffer, E.C.; Helming, K.C.; Alver, B.H.; Tillman, E.J.; Evans, J.A.; Wilson, B.G.; Park, P.J.; et al. Swi/Snf chromatin remodeling/tumor suppressor complex establishes nucleosome occupancy at target promoters. Proc. Natl. Acad. Sci. USA 2013, 110, 10165–10170. [Google Scholar] [CrossRef] [PubMed]
- Shema-Yaacoby, E.; Nikolov, M.; Haj-Yahya, M.; Siman, P.; Allemand, E.; Yamaguchi, Y.; Muchardt, C.; Urlaub, H.; Brik, A.; Oren, M.; et al. Systematic identification of proteins binding to chromatin-embedded ubiquitylated H2B reveals recruitment of SWI/SNF to regulate transcription. Cell Rep. 2013, 4, 601–608. [Google Scholar] [CrossRef] [PubMed]
- Marsh, D.J.; Ma, Y.; Dickson, K.A. Histone Monoubiquitination in Chromatin Remodelling: Focus on the Histone H2B Interactome and Cancer. Cancers 2020, 12, 3462. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.G.; Wang, X.; Shen, X.; McKenna, E.S.; Lemieux, M.E.; Cho, Y.J.; Koellhoffer, E.C.; Pomeroy, S.L.; Orkin, S.H.; Roberts, C.W. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 2010, 18, 316–328. [Google Scholar] [CrossRef]
- Wilson, M.R.; Reske, J.J.; Koeman, J.; Adams, M.; Joshi, N.R.; Fazleabas, A.T.; Chandler, R.L. SWI/SNF Antagonism of PRC2 Mediates Estrogen-Induced Progesterone Receptor Expression. Cells 2022, 11, 1000. [Google Scholar] [CrossRef]
- Marsh, D.J.; Shah, J.S.; Cole, A.J. Histones and their modifications in ovarian cancer—Drivers of disease and therapeutic targets. Front. Oncol. 2014, 4, 144. [Google Scholar] [CrossRef]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef]
- Schwabish, M.A.; Struhl, K. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol. Cell Biol. 2007, 27, 6987–6995. [Google Scholar] [CrossRef]
- Reisman, D.; Glaros, S.; Thompson, E.A. The SWI/SNF complex and cancer. Oncogene 2009, 28, 1653–1668. [Google Scholar] [CrossRef]
- Valencia, A.M.; Collings, C.K.; Dao, H.T.; St Pierre, R.; Cheng, Y.C.; Huang, J.; Sun, Z.Y.; Seo, H.S.; Mashtalir, N.; Comstock, D.E.; et al. Recurrent SMARCB1 Mutations Reveal a Nucleosome Acidic Patch Interaction Site That Potentiates mSWI/SNF Complex Chromatin Remodeling. Cell 2019, 179, 1342–1356.e23. [Google Scholar] [CrossRef] [PubMed]
- Sohn, D.H.; Lee, K.Y.; Lee, C.; Oh, J.; Chung, H.; Jeon, S.H.; Seong, R.H. SRG3 interacts directly with the major components of the SWI/SNF chromatin remodeling complex and protects them from proteasomal degradation. J. Biol. Chem. 2007, 282, 10614–10624. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Yuan, J.; Deng, Y.; Fan, X.; Shen, J. Emerging role of SWI/SNF complex deficiency as a target of immune checkpoint blockade in human cancers. Oncogenesis 2021, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Hourvitz, N.; Kurolap, A.; Mory, A.; Haratz, K.K.; Kidron, D.; Malinger, G.; Baris Feldman, H.; Yaron, Y. SMARCC1 is a susceptibility gene for congenital hydrocephalus with an autosomal dominant inheritance mode and incomplete penetrance. Prenat. Diagn. 2023, 43, 1374–1377. [Google Scholar] [CrossRef]
- Pagliaroli, L.; Trizzino, M. The Evolutionary Conserved SWI/SNF Subunits ARID1A and ARID1B Are Key Modulators of Pluripotency and Cell-Fate Determination. Front. Cell Dev. Biol. 2021, 9, 643361. [Google Scholar] [CrossRef]
- Samartzis, E.P.; Labidi-Galy, S.I.; Moschetta, M.; Uccello, M.; Kalaitzopoulos, D.R.; Perez-Fidalgo, J.A.; Boussios, S. Endometriosis-associated ovarian carcinomas: Insights into pathogenesis, diagnostics, and therapeutic targets-a narrative review. Ann. Transl. Med. 2020, 8, 1712. [Google Scholar] [CrossRef]
- del Carmen, M.G.; Birrer, M.; Schorge, J.O. Carcinosarcoma of the ovary: A review of the literature. Gynecol. Oncol. 2012, 125, 271–277. [Google Scholar] [CrossRef]
- Kim, S.I.; Ha, H.I.; Eoh, K.J.; Lim, J.; Won, Y.J.; Lim, M.C. Trends in the Incidence and Survival Rates of Primary Ovarian Clear Cell Carcinoma Compared to Ovarian Serous Carcinoma in Korea. Front. Oncol. 2022, 12, 874037. [Google Scholar] [CrossRef]
- Dong, S.; Yu, F.; Liu, Y.; Yu, X.; Sun, X.; Wang, W.; Wang, Y. Comparison of the clinical characteristics and prognosis between clear cell carcinomas and high-grade serous ovarian carcinomas. Ginekol. Pol. 2023, 94, 792–798. [Google Scholar] [CrossRef]
- Ku, F.C.; Wu, R.C.; Yang, L.Y.; Tang, Y.H.; Chang, W.Y.; Yang, J.E.; Wang, C.C.; Jung, S.M.; Lin, C.T.; Chang, T.C.; et al. Clear cell carcinomas of the ovary have poorer outcomes compared with serous carcinomas: Results from a single-center Taiwanese study. J. Formos. Med. Assoc. 2018, 117, 117–125. [Google Scholar] [CrossRef]
- Machida, H.; Matsuo, K.; Yamagami, W.; Ebina, Y.; Kobayashi, Y.; Tabata, T.; Kanauchi, M.; Nagase, S.; Enomoto, T.; Mikami, M. Trends and characteristics of epithelial ovarian cancer in Japan between 2002 and 2015: A JSGO-JSOG joint study. Gynecol. Oncol. 2019, 153, 589–596. [Google Scholar] [CrossRef]
- Pozzati, F.; Moro, F.; Pasciuto, T.; Gallo, C.; Ciccarone, F.; Franchi, D.; Mancari, R.; Giunchi, S.; Timmerman, D.; Landolfo, C.; et al. Imaging in gynecological disease (14): Clinical and ultrasound characteristics of ovarian clear cell carcinoma. Ultrasound Obstet. Gynecol. 2018, 52, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Ohkawa, K.; Amasaki, H.; Terashima, Y.; Aizawa, S.; Ishikawa, E. Clear cell carcinoma of the ovary: Light and electron microscopic studies. Cancer 1977, 40, 3019–3029. [Google Scholar] [CrossRef] [PubMed]
- Prat, J. Ovarian carcinomas: Five distinct diseases with different origins, genetic alterations, and clinicopathological features. Virchows Arch. 2012, 460, 237–249. [Google Scholar] [CrossRef]
- Gounaris, I.; Brenton, J.D. Molecular pathogenesis of ovarian clear cell carcinoma. Future Oncol. 2015, 11, 1389–1405. [Google Scholar] [CrossRef]
- Iida, Y.; Okamoto, A.; Hollis, R.L.; Gourley, C.; Herrington, C.S. Clear cell carcinoma of the ovary: A clinical and molecular perspective. Int. J. Gynecol. Cancer 2021, 31, 605–616. [Google Scholar] [CrossRef]
- Anglesio, M.S.; Bashashati, A.; Wang, Y.K.; Senz, J.; Ha, G.; Yang, W.; Aniba, M.R.; Prentice, L.M.; Farahani, H.; Li Chang, H.; et al. Multifocal endometriotic lesions associated with cancer are clonal and carry a high mutation burden. J. Pathol. 2015, 236, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, T.; Kamura, T.; Kigawa, J.; Terakawa, N.; Kikuchi, Y.; Kita, T.; Suzuki, M.; Sato, I.; Taguchi, K. Clinical characteristics of clear cell carcinoma of the ovary: A distinct histologic type with poor prognosis and resistance to platinum-based chemotherapy. Cancer 2000, 88, 2584–2589. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.Y.; Kim, T.J.; Kim, M.J.; Kim, H.J.; Song, T.; Kim, M.K.; Choi, C.H.; Lee, J.W.; Bae, D.S.; Kim, B.G. Prognosis of ovarian clear cell carcinoma compared to other histological subtypes: A meta-analysis. Gynecol. Oncol. 2011, 122, 541–547. [Google Scholar] [CrossRef]
- Oliver, K.E.; Brady, W.E.; Birrer, M.; Gershenson, D.M.; Fleming, G.; Copeland, L.J.; Tewari, K.; Argenta, P.A.; Mannel, R.S.; Secord, A.A.; et al. An evaluation of progression free survival and overall survival of ovarian cancer patients with clear cell carcinoma versus serous carcinoma treated with platinum therapy: An NRG Oncology/Gynecologic Oncology Group experience. Gynecol. Oncol. 2017, 147, 243–249. [Google Scholar] [CrossRef]
- Liu, H.; Xu, Y.; Ji, J.; Dong, R.; Qiu, H.; Dai, X. Prognosis of ovarian clear cell cancer compared with other epithelial cancer types: A population-based analysis. Oncol. Lett. 2020, 19, 1947–1957. [Google Scholar] [CrossRef] [PubMed]
- Mabuchi, S.; Sugiyama, T.; Kimura, T. Clear cell carcinoma of the ovary: Molecular insights and future therapeutic perspectives. J. Gynecol. Oncol. 2016, 27, e31. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.Z.; Ye, J.; Yee, C.V.; Lim, D.; Ngoi, N.Y.L.; Tan, D.S.P.; Huang, R.Y. Analysis of gene expression signatures identifies prognostic and functionally distinct ovarian clear cell carcinoma subtypes. EBioMedicine 2019, 50, 203–210. [Google Scholar] [CrossRef]
- Su, Y.F.; Tsai, E.M.; Chen, C.C.; Wu, C.C.; Er, T.K. Targeted sequencing of a specific gene panel detects a high frequency of ARID1A and PIK3CA mutations in ovarian clear cell carcinoma. Clin. Chim. Acta 2019, 494, 1–7. [Google Scholar] [CrossRef]
- Murakami, R.; Matsumura, N.; Brown, J.B.; Higasa, K.; Tsutsumi, T.; Kamada, M.; Abou-Taleb, H.; Hosoe, Y.; Kitamura, S.; Yamaguchi, K.; et al. Exome Sequencing Landscape Analysis in Ovarian Clear Cell Carcinoma Shed Light on Key Chromosomal Regions and Mutation Gene Networks. Am. J. Pathol. 2017, 187, 2246–2258. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Wang, T.L.; Shih Ie, M.; Mao, T.L.; Nakayama, K.; Roden, R.; Glas, R.; Slamon, D.; Diaz, L.A., Jr.; Vogelstein, B.; et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 2010, 330, 228–231. [Google Scholar] [CrossRef]
- Kihara, A.; Iizuka, T.; Endo, S.; Horie, K.; Kanda, H.; Niki, T. Ovarian clear cell carcinoma with an immature teratoma component showing ARID1A deficiency and an identical PIK3CA mutation. J. Obstet. Gynaecol. Res. 2021, 47, 3401–3407. [Google Scholar] [CrossRef]
- Schnack, T.H.; Oliveira, D.N.P.; Christiansen, A.P.; Høgdall, C.; Høgdall, E. Prognostic impact of molecular profiles and molecular signatures in clear cell ovarian cancer. Cancer Genet. 2023, 278–279, 9–16. [Google Scholar] [CrossRef]
- Wiegand, K.C.; Hennessy, B.T.; Leung, S.; Wang, Y.; Ju, Z.; McGahren, M.; Kalloger, S.E.; Finlayson, S.; Stemke-Hale, K.; Lu, Y.; et al. A functional proteogenomic analysis of endometrioid and clear cell carcinomas using reverse phase protein array and mutation analysis: Protein expression is histotype-specific and loss of ARID1A/BAF250a is associated with AKT phosphorylation. BMC Cancer 2014, 14, 120. [Google Scholar] [CrossRef]
- Yamamoto, S.; Tsuda, H.; Takano, M.; Tamai, S.; Matsubara, O. Loss of ARID1A protein expression occurs as an early event in ovarian clear-cell carcinoma development and frequently coexists with PIK3CA mutations. Mod. Pathol. 2012, 25, 615–624. [Google Scholar] [CrossRef]
- Itamochi, H.; Oumi, N.; Oishi, T.; Shoji, T.; Fujiwara, H.; Sugiyama, T.; Suzuki, M.; Kigawa, J.; Harada, T. Loss of ARID1A expression is associated with poor prognosis in patients with stage I/II clear cell carcinoma of the ovary. Int. J. Clin. Oncol. 2015, 20, 967–973. [Google Scholar] [CrossRef]
- Katagiri, A.; Nakayama, K.; Rahman, M.T.; Rahman, M.; Katagiri, H.; Nakayama, N.; Ishikawa, M.; Ishibashi, T.; Iida, K.; Kobayashi, H.; et al. Loss of ARID1A expression is related to shorter progression-free survival and chemoresistance in ovarian clear cell carcinoma. Mod. Pathol. 2012, 25, 282–288. [Google Scholar] [CrossRef]
- Helming, K.C.; Wang, X.; Roberts, C.W.M. Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer Cell 2014, 26, 309–317. [Google Scholar] [CrossRef]
- Helming, K.C.; Wang, X.; Wilson, B.G.; Vazquez, F.; Haswell, J.R.; Manchester, H.E.; Kim, Y.; Kryukov, G.V.; Ghandi, M.; Aguirre, A.J.; et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat. Med. 2014, 20, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Sato, E.; Nakayama, K.; Razia, S.; Nakamura, K.; Ishikawa, M.; Minamoto, T.; Ishibashi, T.; Yamashita, H.; Iida, K.; Kyo, S. ARID1B as a Potential Therapeutic Target for ARID1A-Mutant Ovarian Clear Cell Carcinoma. Int. J. Mol. Sci. 2018, 19, 1710. [Google Scholar] [CrossRef] [PubMed]
- Genestie, C.; Blanc-Durand, F.; Auguste, A.; Pautier, P.; Dunant, A.; Scoazec, J.Y.; Gouy, S.; Morice, P.; Bentivegna, E.; Maulard, A.; et al. Clinical utility of SMARCA4 testing by immunohistochemistry in rare ovarian tumours. Br. J. Cancer 2020, 122, 564–568. [Google Scholar] [CrossRef]
- Sanders, B.E.; Wolsky, R.; Doughty, E.S.; Wells, K.L.; Ghosh, D.; Ku, L.; Pressey, J.G.; Bitler, B.B.; Brubaker, L.W. Small cell carcinoma of the ovary hypercalcemic type (SCCOHT): A review and novel case with dual germline SMARCA4 and BRCA2 mutations. Gynecol. Oncol. Rep. 2022, 44, 101077. [Google Scholar] [CrossRef] [PubMed]
- Le Loarer, F.; Watson, S.; Pierron, G.; de Montpreville, V.T.; Ballet, S.; Firmin, N.; Auguste, A.; Pissaloux, D.; Boyault, S.; Paindavoine, S.; et al. SMARCA4 inactivation defines a group of undifferentiated thoracic malignancies transcriptionally related to BAF-deficient sarcomas. Nat. Genet. 2015, 47, 1200–1205. [Google Scholar] [CrossRef]
- Witkowski, L.; Carrot-Zhang, J.; Albrecht, S.; Fahiminiya, S.; Hamel, N.; Tomiak, E.; Grynspan, D.; Saloustros, E.; Nadaf, J.; Rivera, B.; et al. Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat. Genet. 2014, 46, 438–443. [Google Scholar] [CrossRef]
- Jelinic, P.; Ricca, J.; Van Oudenhove, E.; Olvera, N.; Merghoub, T.; Levine, D.A.; Zamarin, D. Immune-Active Microenvironment in Small Cell Carcinoma of the Ovary, Hypercalcemic Type: Rationale for Immune Checkpoint Blockade. J. Natl. Cancer Inst. 2018, 110, 787–790. [Google Scholar] [CrossRef]
- Lin, D.I.; Chudnovsky, Y.; Duggan, B.; Zajchowski, D.; Greenbowe, J.; Ross, J.S.; Gay, L.M.; Ali, S.M.; Elvin, J.A. Comprehensive genomic profiling reveals inactivating SMARCA4 mutations and low tumor mutational burden in small cell carcinoma of the ovary, hypercalcemic-type. Gynecol. Oncol. 2017, 147, 626–633. [Google Scholar] [CrossRef]
- Ramos, P.; Karnezis, A.N.; Craig, D.W.; Sekulic, A.; Russell, M.L.; Hendricks, W.P.; Corneveaux, J.J.; Barrett, M.T.; Shumansky, K.; Yang, Y.; et al. Small cell carcinoma of the ovary, hypercalcemic type, displays frequent inactivating germline and somatic mutations in SMARCA4. Nat. Genet. 2014, 46, 427–429. [Google Scholar] [CrossRef]
- Moes-Sosnowska, J.; Szafron, L.; Nowakowska, D.; Dansonka-Mieszkowska, A.; Budzilowska, A.; Konopka, B.; Plisiecka-Halasa, J.; Podgorska, A.; Rzepecka, I.K.; Kupryjanczyk, J. Germline SMARCA4 mutations in patients with ovarian small cell carcinoma of hypercalcemic type. Orphanet J. Rare Dis. 2015, 10, 32. [Google Scholar] [CrossRef]
- Mazibrada, J.; Jayatunge, N.; Domecq, C.; Witkowski, L.; Croce, S.; Foulkes, W.D.; McCluggage, W.G. Unusual Aspects of Small Cell Carcinoma of the Ovary of Hypercalcaemic Type: Retained SMARCA4 Immunohistochemical Staining and Positive Staining With TLE1. Am. J. Surg. Pathol. 2023, 47, 1261–1266. [Google Scholar] [CrossRef]
- Kupryjańczyk, J.; Dansonka-Mieszkowska, A.; Moes-Sosnowska, J.; Plisiecka-Hałasa, J.; Szafron, L.; Podgórska, A.; Rzepecka, I.K.; Konopka, B.; Budziłowska, A.; Rembiszewska, A.; et al. Ovarian small cell carcinoma of hypercalcemic type—Evidence of germline origin and SMARCA4 gene inactivation. a pilot study. Pol. J. Pathol. 2013, 64, 238–246. [Google Scholar] [CrossRef]
- Chandan, C.S.; Mir, A.W.; Mir, A.W.; Dar, F.A.; Yasin, S.B. Hypercalcemic Ovarian Carcinoma (Small Cell Carcinoma of the Ovary, Hypercalcemic Type (SCCOHT)): A case series and review of literature of a rare malignancy. Indian J. Surg. Oncol. 2024. [Google Scholar] [CrossRef]
- Li, G.; Jiang, Y. Case Report: A Durable Response to Camrelizumab and Apatinib Combination Therapy in a Heavily Treated Small Cell Carcinoma of the Ovary, Hypercalcemic Type. Front. Oncol. 2022, 12, 916790. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zheng, K.; Kang, M.; Xu, J.; Ning, Y.; Hu, W.; Li, K.; Kang, Y.; Xu, C. Establishment and characterization of a novel cell line (SCCOHT-CH-1) and PDX models derived from Chinese patients of small cell ovarian carcinoma of the hypercalcemic type. Hum. Cell 2023, 36, 2214–2227. [Google Scholar] [CrossRef] [PubMed]
- Pressey, J.G.; Dandoy, C.E.; Pater, L.E.; Sroga Rios, J.; Sisson, R.; Dasgupta, R.; Szabo, S. Small cell carcinoma of the ovary hypercalcemic type (SCCOHT): Comprehensive management of a newly diagnosed young adult. Gynecol. Oncol. 2020, 158, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Mathey, M.P.; de Jolinière, J.B.; Major, A.; Conrad, B.; Khomsi, F.; Betticher, D.; Devouassoux, M.; Feki, A. Rare case of remission of a patient with small cell carcinoma of the ovary, hypercalcaemic type (SCCOHT) stage IV: Case report. Int. J. Surg. Case Rep. 2020, 66, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Diwan, H.; Karki, D.; Bansal, D.; Kamboj, M.; Sharma, A.; Nathany, S.; Mattoo, S.; Kumar, D. Novel germline SMARCA4 mutation in Small Cell Carcinoma of the Ovary, Hypercalcemic Type. Curr. Probl. Cancer Case Rep. 2022, 8, 100205. [Google Scholar] [CrossRef]
- Pastorczak, A.; Krajewska, K.; Urbanska, Z.; Szmyd, B.; Salacinska-Los, E.; Kobos, J.; Mlynarski, W.; Trelinska, J. Ovarian carcinoma in children with constitutional mutation of SMARCA4: Single-family report and literature review. Fam. Cancer 2021, 20, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Connor, Y.D.; Miao, D.; Lin, D.I.; Hayne, C.; Howitt, B.E.; Dalrymple, J.L.; DeLeonardis, K.R.; Hacker, M.R.; Esselen, K.M.; Shea, M. Germline mutations of SMARCA4 in small cell carcinoma of the ovary, hypercalcemic type and in SMARCA4-deficient undifferentiated uterine sarcoma: Clinical features of a single family and comparison of large cohorts. Gynecol. Oncol. 2020, 157, 106–114. [Google Scholar] [CrossRef] [PubMed]
- David, M.P.; Venkatramani, R.; Lopez-Terrada, D.H.; Roy, A.; Patil, N.; Fisher, K.E. Multimodal molecular analysis of an atypical small cell carcinoma of the ovary, hypercalcemic type. Cold Spring Harb. Mol. Case Stud. 2018, 4, a002956. [Google Scholar] [CrossRef]
- Bailey, S.; Murray, M.J.; Witkowski, L.; Hook, E.; Hasselblatt, M.; Crawford, R.; Foulkes, W.D.; Tischkowitz, M.; Nicholson, J.C. Biallelic somatic SMARCA4 mutations in small cell carcinoma of the ovary, hypercalcemic type (SCCOHT). Pediatr. Blood Cancer 2015, 62, 728–730. [Google Scholar] [CrossRef]
- Lavrut, P.M.; Le Loarer, F.; Normand, C.; Grosos, C.; Dubois, R.; Buenerd, A.; Conter, C.; Dijoud, F.; Blay, J.Y.; Collardeau-Frachon, S. Small Cell Carcinoma of the Ovary, Hypercalcemic Type: Report of a Bilateral Case in a Teenager Associated with SMARCA4 Germline Mutation. Pediatr. Dev. Pathol. 2016, 19, 56–60. [Google Scholar] [CrossRef]
- Fahiminiya, S.; Sabbaghian, N.; Albrecht, S.; Nadaf, J.; Callegaro-Filho, D.; Foulkes, W.D. Ovarian small cell carcinoma in one of a pair of monozygous twins. Fam. Cancer 2019, 18, 161–163. [Google Scholar] [CrossRef]
- Gao, Y.; Zang, L.; Ye, Y.; Ma, F.; Kang, M.; Zheng, K.; Kang, Y.; Wang, H.; Xu, C. Immunotherapy combined with targeted therapy in advanced small cell carcinoma of the ovary of hypercalcemic type: A case of overall survival lasting for over 5 years. Eur. J. Obstet. Gynecol. Reprod. Biol. 2024, 297, 270–274. [Google Scholar] [CrossRef]
- Aoyagi, Y.; Kai, K.; Nishida, H.; Aso, S.; Kobayashi, E. Multiple Somatic Mutations of SMARCA4 in Small Cell Carcinoma of the Ovary, Hypercalcemic Type: A Case Report. Cureus 2024, 16, e60802. [Google Scholar] [CrossRef]
- Karanian-Philippe, M.; Velasco, V.; Longy, M.; Floquet, A.; Arnould, L.; Coindre, J.M.; Le Naoures-Méar, C.; Averous, G.; Guyon, F.; MacGrogan, G.; et al. SMARCA4 (BRG1) loss of expression is a useful marker for the diagnosis of ovarian small cell carcinoma of the hypercalcemic type (ovarian rhabdoid tumor): A comprehensive analysis of 116 rare gynecologic tumors, 9 soft tissue tumors, and 9 melanomas. Am. J. Surg. Pathol. 2015, 39, 1197–1205. [Google Scholar] [CrossRef]
- Clarke, B.A.; Witkowski, L.; Ton Nu, T.N.; Shaw, P.A.; Gilks, C.B.; Huntsman, D.; Karnezis, A.N.; Sebire, N.; Lamovec, J.; Roth, L.M.; et al. Loss of SMARCA4 (BRG1) protein expression as determined by immunohistochemistry in small-cell carcinoma of the ovary, hypercalcaemic type distinguishes these tumours from their mimics. Histopathology 2016, 69, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Conlon, N.; Silva, A.; Guerra, E.; Jelinic, P.; Schlappe, B.A.; Olvera, N.; Mueller, J.J.; Tornos, C.; Jungbluth, A.A.; Young, R.H.; et al. Loss of SMARCA4 Expression Is Both Sensitive and Specific for the Diagnosis of Small Cell Carcinoma of Ovary, Hypercalcemic Type. Am. J. Surg. Pathol. 2016, 40, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Karnezis, A.N.; Wang, Y.; Ramos, P.; Hendricks, W.P.; Oliva, E.; D′Angelo, E.; Prat, J.; Nucci, M.R.; Nielsen, T.O.; Chow, C.; et al. Dual loss of the SWI/SNF complex ATPases SMARCA4/BRG1 and SMARCA2/BRM is highly sensitive and specific for small cell carcinoma of the ovary, hypercalcaemic type. J. Pathol. 2016, 238, 389–400. [Google Scholar] [CrossRef]
- Zheng, K.; Gao, Y.; Xu, C.; Kang, Y. Clinical characteristics and status of treatment of small-cell carcinoma of the ovary, hypercalcemic type in the Chinese population: A meta-analysis. J. Gynecol. Oncol. 2024, 35, e96. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, D.; Gupta, P.; Chhabra, P.; Peters, N.J.; Bansal, D.; Srinivasan, R.; Kakkar, N. Small Cell Carcinoma of Ovary, Hypercalcemic Type: Cytologic, Histopathologic, and Immunohistochemical Landscapes of a Rare Case. J. Pediatr. Adolesc. Gynecol. 2021, 34, 98–102. [Google Scholar] [CrossRef]
- Atwi, D.; Quinton, M.R.; Kiser, R.M.; Pokala, H.R.; Rooms, L.M.; Yu, Z. Small Cell Carcinoma of the Ovary, Hypercalcemic Type, in a 12-Month-Old Girl. Pediatr. Dev. Pathol. 2021, 24, 493–497. [Google Scholar] [CrossRef]
- Altmann, J.; Schmitt, W.; Bashian, N.; Sehouli, J. A dramatic response to checkpoint inhibitor in a woman with small cell carcinoma of the hypercalcemic type of the ovary. Gynecol. Oncol. 2024, 181, 99–101. [Google Scholar] [CrossRef]
- Saylany, L.; Ellepola, H.; Broom, E. A Rare Case of Advanced-Stage Small Cell Carcinoma of the Ovary Identified During the Third Trimester of Pregnancy. Cureus 2024, 16, e55696. [Google Scholar] [CrossRef]
- Coşkun, Ç.; Kurucu, N.; Usubutun, A.; Soyer, T.; Ozcan, H.N.; Çelik Ertaş, N.B.; Kutluk, T. Small Cell Carcinoma of Ovary, Hypercalcemic Type: A Rare Case Report. J. Pediatr. Adolesc. Gynecol. 2023, 36, 112–115. [Google Scholar] [CrossRef]
- Young, R.H.; Goodman, A.; Penson, R.T.; Russell, A.H.; Uppot, R.N.; Tambouret, R.H. Case records of the Massachusetts General Hospital. Case 8-2010. A 22-year-old woman with hypercalcemia and a pelvic mass. N. Engl. J. Med. 2010, 362, 1031–1040. [Google Scholar] [CrossRef]
- Tischkowitz, M.; Huang, S.; Banerjee, S.; Hague, J.; Hendricks, W.P.D.; Huntsman, D.G.; Lang, J.D.; Orlando, K.A.; Oza, A.M.; Pautier, P.; et al. Small-Cell Carcinoma of the Ovary, Hypercalcemic Type-Genetics, New Treatment Targets, and Current Management Guidelines. Clin. Cancer Res. 2020, 26, 3908–3917. [Google Scholar] [CrossRef]
- McCluggage, W.G.; Witkowski, L.; Clarke, B.A.; Foulkes, W.D. Clinical, morphological and immunohistochemical evidence that small-cell carcinoma of the ovary of hypercalcaemic type (SCCOHT) may be a primitive germ-cell neoplasm. Histopathology 2017, 70, 1147–1154. [Google Scholar] [CrossRef] [PubMed]
- Foulkes, W.D.; Clarke, B.A.; Hasselblatt, M.; Majewski, J.; Albrecht, S.; McCluggage, W.G. No small surprise—Small cell carcinoma of the ovary, hypercalcaemic type, is a malignant rhabdoid tumour. J. Pathol. 2014, 233, 209–214. [Google Scholar] [CrossRef]
- Witkowski, L.; Goudie, C.; Ramos, P.; Boshari, T.; Brunet, J.S.; Karnezis, A.N.; Longy, M.; Knost, J.A.; Saloustros, E.; McCluggage, W.G.; et al. The influence of clinical and genetic factors on patient outcome in small cell carcinoma of the ovary, hypercalcemic type. Gynecol. Oncol. 2016, 141, 454–460. [Google Scholar] [CrossRef]
- Witkowski, L.; Donini, N.; Byler-Dann, R.; Knost, J.A.; Albrecht, S.; Berchuck, A.; McCluggage, W.G.; Hasselblatt, M.; Foulkes, W.D. The hereditary nature of small cell carcinoma of the ovary, hypercalcemic type: Two new familial cases. Fam. Cancer 2017, 16, 395–399. [Google Scholar] [CrossRef]
- As Maintenance Therapy, Olaparib′s Benefits Continue. Cancer Discov. 2022, 12, 6–7. [CrossRef] [PubMed]
- Ray-Coquard, I.; Morice, P.; Lorusso, D.; Prat, J.; Oaknin, A.; Pautier, P.; Colombo, N. Non-epithelial ovarian cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018, 29, iv1–iv18. [Google Scholar] [CrossRef] [PubMed]
- Matias-Guiu, X.; Prat, J.; Young, R.H.; Capen, C.C.; Rosol, T.J.; Delellis, R.A.; Scully, R.E. Human parathyroid hormone-related protein in ovarian small cell carcinoma. An immunohistochemical study. Cancer 1994, 73, 1878–1881. [Google Scholar] [CrossRef] [PubMed]
- Rekhtman, N.; Montecalvo, J.; Chang, J.C.; Alex, D.; Ptashkin, R.N.; Ai, N.; Sauter, J.L.; Kezlarian, B.; Jungbluth, A.; Desmeules, P.; et al. SMARCA4-Deficient Thoracic Sarcomatoid Tumors Represent Primarily Smoking-Related Undifferentiated Carcinomas Rather Than Primary Thoracic Sarcomas. J. Thorac. Oncol. 2020, 15, 231–247. [Google Scholar] [CrossRef]
- Karnezis, A.N.; Hoang, L.N.; Coatham, M.; Ravn, S.; Almadani, N.; Tessier-Cloutier, B.; Irving, J.; Meng, B.; Li, X.; Chow, C.; et al. Loss of switch/sucrose non-fermenting complex protein expression is associated with dedifferentiation in endometrial carcinomas. Mod. Pathol. 2016, 29, 302–314. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Q.; Sun, S.; Li, Z.; Cui, Z.; Liu, Q.; Zhang, Y.; Xiong, S.; Zhang, S. Expression of SMARCA2 and SMARCA4 in gastric adenocarcinoma and construction of a nomogram prognostic model. Int. J. Clin. Oncol. 2023, 28, 1487–1500. [Google Scholar] [CrossRef] [PubMed]
- Libera, L.; Ottini, G.; Sahnane, N.; Pettenon, F.; Turri-Zanoni, M.; Lambertoni, A.; Chiaravalli, A.M.; Leone, F.; Battaglia, P.; Castelnuovo, P.; et al. Methylation Drivers and Prognostic Implications in Sinonasal Poorly Differentiated Carcinomas. Cancers 2021, 13, 5030. [Google Scholar] [CrossRef] [PubMed]
- Andrianteranagna, M.; Cyrta, J.; Masliah-Planchon, J.; Nemes, K.; Corsia, A.; Leruste, A.; Holdhof, D.; Kordes, U.; Orbach, D.; Corradini, N.; et al. SMARCA4-deficient rhabdoid tumours show intermediate molecular features between SMARCB1-deficient rhabdoid tumours and small cell carcinomas of the ovary, hypercalcaemic type. J. Pathol. 2021, 255, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Marquez-Vilendrer, S.B.; Rai, S.K.; Gramling, S.J.; Lu, L.; Reisman, D.N. Loss of the SWI/SNF ATPase subunits BRM and BRG1 drives lung cancer development. Oncoscience 2016, 3, 322–336. [Google Scholar] [CrossRef]
- Orlando, K.A.; Douglas, A.K.; Abudu, A.; Wang, Y.; Tessier-Cloutier, B.; Su, W.; Peters, A.; Sherman, L.S.; Moore, R.; Nguyen, V.; et al. Re-expression of SMARCA4/BRG1 in small cell carcinoma of ovary, hypercalcemic type (SCCOHT) promotes an epithelial-like gene signature through an AP-1-dependent mechanism. Elife 2020, 9, e59073. [Google Scholar] [CrossRef]
- Kvaskoff, M.; Mahamat-Saleh, Y.; Farland, L.V.; Shigesi, N.; Terry, K.L.; Harris, H.R.; Roman, H.; Becker, C.M.; As-Sanie, S.; Zondervan, K.T.; et al. Endometriosis and cancer: A systematic review and meta-analysis. Hum. Reprod. Update 2021, 27, 393–420. [Google Scholar] [CrossRef]
- Chen, S.; Li, Y.; Qian, L.; Deng, S.; Liu, L.; Xiao, W.; Zhou, Y. A Review of the Clinical Characteristics and Novel Molecular Subtypes of Endometrioid Ovarian Cancer. Front. Oncol. 2021, 11, 668151. [Google Scholar] [CrossRef]
- Chiang, Y.C.; Chen, C.A.; Huang, C.Y.; Hsieh, C.Y.; Cheng, W.F. Synchronous primary cancers of the endometrium and ovary. Int. J. Gynecol. Cancer 2008, 18, 159–164. [Google Scholar] [CrossRef]
- Mitric, C.; Salman, L.; Abrahamyan, L.; Kim, S.R.; Pechlivanoglou, P.; Chan, K.K.W.; Gien, L.T.; Ferguson, S.E. Mismatch-repair deficiency, microsatellite instability, and lynch syndrome in ovarian cancer: A systematic review and meta-analysis. Gynecol. Oncol. 2023, 170, 133–142. [Google Scholar] [CrossRef]
- van Niekerk, C.C.; Bulten, J.; Vooijs, G.P.; Verbeek, A.L. The Association between Primary Endometrioid Carcinoma of the Ovary and Synchronous Malignancy of the Endometrium. Obstet. Gynecol. Int. 2010, 2010, 465162. [Google Scholar] [CrossRef]
- Karakashev, S.; Zhu, H.; Wu, S.; Yokoyama, Y.; Bitler, B.G.; Park, P.H.; Lee, J.H.; Kossenkov, A.V.; Gaonkar, K.S.; Yan, H.; et al. CARM1-expressing ovarian cancer depends on the histone methyltransferase EZH2 activity. Nat. Commun. 2018, 9, 631. [Google Scholar] [CrossRef]
- Zondervan, K.T.; Becker, C.M.; Koga, K.; Missmer, S.A.; Taylor, R.N.; Viganò, P. Endometriosis. Nat. Rev. Dis. Primers 2018, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Harder, C.; Velho, R.V.; Brandes, I.; Sehouli, J.; Mechsner, S. Assessing the true prevalence of endometriosis: A narrative review of literature data. Int. J. Gynaecol. Obstet. 2024; online ahead of print. [Google Scholar] [CrossRef]
- Adilbayeva, A.; Kunz, J. Pathogenesis of Endometriosis and Endometriosis-Associated Cancers. Int. J. Mol. Sci. 2024, 25, 7624. [Google Scholar] [CrossRef] [PubMed]
- Barnard, M.E.; Farland, L.V.; Yan, B.; Wang, J.; Trabert, B.; Doherty, J.A.; Meeks, H.D.; Madsen, M.; Guinto, E.; Collin, L.J.; et al. Endometriosis Typology and Ovarian Cancer Risk. Jama 2024, 332, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Mortlock, S.; Corona, R.I.; Kho, P.F.; Pharoah, P.; Seo, J.H.; Freedman, M.L.; Gayther, S.A.; Siedhoff, M.T.; Rogers, P.A.W.; Leuchter, R.; et al. A multi-level investigation of the genetic relationship between endometriosis and ovarian cancer histotypes. Cell Rep. Med. 2022, 3, 100542. [Google Scholar] [CrossRef]
- Zhu, C.; Zhu, J.; Qian, L.; Liu, H.; Shen, Z.; Wu, D.; Zhao, W.; Xiao, W.; Zhou, Y. Clinical characteristics and prognosis of ovarian clear cell carcinoma: A 10-year retrospective study. BMC Cancer 2021, 21, 322. [Google Scholar] [CrossRef]
- Chiaffarino, F.; Cipriani, S.; Ricci, E.; Esposito, G.; Parazzini, F.; Vercellini, P. Histologic Subtypes in Endometriosis-Associated Ovarian Cancer and Ovarian Cancer Arising in Endometriosis: A Systematic Review and Meta-Analysis. Reprod. Sci. 2024, 31, 1642–1650. [Google Scholar] [CrossRef]
- Stamp, J.P.; Gilks, C.B.; Wesseling, M.; Eshragh, S.; Ceballos, K.; Anglesio, M.S.; Kwon, J.S.; Tone, A.; Huntsman, D.G.; Carey, M.S. BAF250a Expression in Atypical Endometriosis and Endometriosis-Associated Ovarian Cancer. Int. J. Gynecol. Cancer 2016, 26, 825–832. [Google Scholar] [CrossRef]
- Capozzi, V.A.; Scarpelli, E.; dell′Omo, S.; Rolla, M.; Pezzani, A.; Morganelli, G.; Gaiano, M.; Ghi, T.; Berretta, R. Atypical Endometriosis: A Comprehensive Systematic Review of Pathological Patterns and Diagnostic Challenges. Biomedicines 2024, 12, 1209. [Google Scholar] [CrossRef]
- Ness, R.B. Endometriosis and ovarian cancer: Thoughts on shared pathophysiology. Am. J. Obstet. Gynecol. 2003, 189, 280–294. [Google Scholar] [CrossRef]
- Vercellini, P.; Parazzini, F.; Bolis, G.; Carinelli, S.; Dindelli, M.; Vendola, N.; Luchini, L.; Crosignani, P.G. Endometriosis and ovarian cancer. Am. J. Obstet. Gynecol. 1993, 169, 181–182. [Google Scholar] [CrossRef]
- Jimbo, H.; Yoshikawa, H.; Onda, T.; Yasugi, T.; Sakamoto, A.; Taketani, Y. Prevalence of ovarian endometriosis in epithelial ovarian cancer. Int. J. Gynaecol. Obstet. 1997, 59, 245–250. [Google Scholar] [CrossRef]
- Glynn, S.M.; Gaillard, S.; Stone, R.L.; Fader, A.N.; Beavis, A.L. Pembrolizumab with bevacizumab and cyclophosphamide for the treatment of recurrent ovarian clear cell carcinoma: A case series. Gynecol. Oncol. Rep. 2024, 53, 101374. [Google Scholar] [CrossRef] [PubMed]
- Ngoi, N.Y.; Heong, V.; Ow, S.; Chay, W.Y.; Kim, H.S.; Choi, C.H.; Goss, G.; Goh, J.C.; Tai, B.C.; Lim, D.G.; et al. A multicenter phase II randomized trial of durvalumab (MEDI-4736) versus physician′s choice chemotherapy in recurrent ovarian clear cell adenocarcinoma (MOCCA). Int. J. Gynecol. Cancer 2020, 30, 1239–1242. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, W.; Zhang, X.; Fang, Y.; Yue, X.; Zhang, X.; He, Q.; Fu, N.; Wang, S.; Ma, T.; et al. Effective Disease Control After Combinatorial Treatment with a PD-1 Antibody and an mTOR Inhibitor for Recurrent Ovarian Clear Cell Carcinomas: A Case Report and Literature Review. Onco Targets Ther. 2021, 14, 5429–5434. [Google Scholar] [CrossRef]
- Zhao, Y.; Jiang, Y. Remarkable Response to the Triplet Combination of Olaparib with Pembrolizumab and Bevacizumab in the Third-Line Treatment of an Ovarian Clear Cell Carcinoma Patient with an ARID1A Mutation: A Case Report. Onco Targets Ther. 2022, 15, 323–328. [Google Scholar] [CrossRef]
- O′Cearbhaill, R.E.; Miller, A.; Soslow, R.A.; Lankes, H.A.; DeLair, D.; Segura, S.; Chavan, S.; Zamarin, D.; DeBernardo, R.; Moore, K.; et al. A phase 2 study of dasatinib in recurrent clear cell carcinoma of the ovary, fallopian tube, peritoneum or endometrium: NRG oncology/gynecologic oncology group study 0283. Gynecol. Oncol. 2023, 176, 16–24. [Google Scholar] [CrossRef]
- Berns, K.; Caumanns, J.J.; Hijmans, E.M.; Gennissen, A.M.C.; Severson, T.M.; Evers, B.; Wisman, G.B.A.; Jan Meersma, G.; Lieftink, C.; Beijersbergen, R.L.; et al. ARID1A mutation sensitizes most ovarian clear cell carcinomas to BET inhibitors. Oncogene 2018, 37, 4611–4625. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, T.; Fatkhutdinov, N.; Zundell, J.A.; Tcyganov, E.N.; Nacarelli, T.; Karakashev, S.; Wu, S.; Liu, Q.; Gabrilovich, D.I.; Zhang, R. HDAC6 Inhibition Synergizes with Anti-PD-L1 Therapy in ARID1A-Inactivated Ovarian Cancer. Cancer Res. 2019, 79, 5482–5489. [Google Scholar] [CrossRef] [PubMed]
- Chien, W.; Tyner, J.W.; Gery, S.; Zheng, Y.; Li, L.Y.; Gopinatha Pillai, M.S.; Nam, C.; Bhowmick, N.A.; Lin, D.C.; Koeffler, H.P. Treatment for ovarian clear cell carcinoma with combined inhibition of WEE1 and ATR. J. Ovarian Res. 2023, 16, 80. [Google Scholar] [CrossRef]
- Keller, P.J.; Adams, E.J.; Wu, R.; Côté, A.; Arora, S.; Cantone, N.; Meyer, R.; Mertz, J.A.; Gehling, V.; Cui, J.; et al. Comprehensive Target Engagement by the EZH2 Inhibitor Tulmimetostat Allows for Targeting of ARID1A Mutant Cancers. Cancer Res. 2024, 84, 2501–2517. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.K.; Esselen, K.M.; Kolin, D.L.; Lee, L.J.; Matulonis, U.A.; Konstantinopoulos, P.A. Combined CDK4/6 and PD-1 Inhibition in Refractory SMARCA4-Deficient Small-Cell Carcinoma of the Ovary, Hypercalcemic Type. JCO Precis. Oncol. 2020, 4, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Blatnik, A.; Dragoš, V.; Blatnik, O.; Stegel, V.; Klančar, G.; Novaković, S.; Drev, P.; Žagar, T.; Merlo, S.; Škof, E.; et al. A Population-Based Study of Patients With Small Cell Carcinoma of the Ovary, Hypercalcemic Type, Encompassing a 30-Year Period. Arch. Pathol. Lab. Med. 2024, 148, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.L.; Blay, J.-Y.; Agulnik, M.; Chugh, R.; Mir, O.; Italiano, A.; Thomas, D.; Gupta, A.; Jahan, T.; Cote, G.; et al. A phase II, multicenter study of the EZH2 inhibitor tazemetostat in adults (rhabdoid tumor cohort) (NCT02601950). Ann. Oncol. 2018, 29, viii580–viii581. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, S.Y.; Karnezis, A.N.; Colborne, S.; Santos, N.D.; Lang, J.D.; Hendricks, W.P.; Orlando, K.A.; Yap, D.; Kommoss, F.; et al. The histone methyltransferase EZH2 is a therapeutic target in small cell carcinoma of the ovary, hypercalcaemic type. J. Pathol. 2017, 242, 371–383. [Google Scholar] [CrossRef]
- Shorstova, T.; Su, J.; Zhao, T.; Dahabieh, M.; Leibovitch, M.; De Sa Tavares Russo, M.; Avizonis, D.; Rajkumar, S.; Watson, I.R.; Del Rincón, S.V.; et al. Reprogramming of Nucleotide Metabolism Mediates Synergy between Epigenetic Therapy and MAP Kinase Inhibition. Mol. Cancer Ther. 2021, 20, 64–75. [Google Scholar] [CrossRef]
- Chan-Penebre, E.; Armstrong, K.; Drew, A.; Grassian, A.R.; Feldman, I.; Knutson, S.K.; Kuplast-Barr, K.; Roche, M.; Campbell, J.; Ho, P.; et al. Selective Killing of SMARCA2- and SMARCA4-deficient Small Cell Carcinoma of the Ovary, Hypercalcemic Type Cells by Inhibition of EZH2: In Vitro and In Vivo Preclinical Models. Mol. Cancer Ther. 2017, 16, 850–860. [Google Scholar] [CrossRef]
- Xue, Y.; Meehan, B.; Macdonald, E.; Venneti, S.; Wang, X.Q.D.; Witkowski, L.; Jelinic, P.; Kong, T.; Martinez, D.; Morin, G.; et al. CDK4/6 inhibitors target SMARCA4-determined cyclin D1 deficiency in hypercalcemic small cell carcinoma of the ovary. Nat. Commun. 2019, 10, 558. [Google Scholar] [CrossRef]
- Zhao, L.; Chen, X.; Wu, H.; He, Q.; Ding, L.; Yang, B. Strategies to synergize PD-1/PD-L1 targeted cancer immunotherapies to enhance antitumor responses in ovarian cancer. Biochem. Pharmacol. 2023, 215, 115724. [Google Scholar] [CrossRef]
- Yoon, W.H.; DeFazio, A.; Kasherman, L. Immune checkpoint inhibitors in ovarian cancer: Where do we go from here? Cancer Drug Resist. 2023, 6, 358–377. [Google Scholar] [CrossRef]
- Pawłowska, A.; Rekowska, A.; Kuryło, W.; Pańczyszyn, A.; Kotarski, J.; Wertel, I. Current Understanding on Why Ovarian Cancer Is Resistant to Immune Checkpoint Inhibitors. Int. J. Mol. Sci. 2023, 24, 10859. [Google Scholar] [CrossRef] [PubMed]
- Matulonis, U.A.; Shapira-Frommer, R.; Santin, A.D.; Lisyanskaya, A.S.; Pignata, S.; Vergote, I.; Raspagliesi, F.; Sonke, G.S.; Birrer, M.; Provencher, D.M.; et al. Antitumor activity and safety of pembrolizumab in patients with advanced recurrent ovarian cancer: Results from the phase II KEYNOTE-100 study. Ann. Oncol. 2019, 30, 1080–1087. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Ju, Z.; Zhao, W.; Wang, L.; Peng, Y.; Ge, Z.; Nagel, Z.D.; Zou, J.; Wang, C.; Kapoor, P.; et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat. Med. 2018, 24, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Ackroyd, S.A.; Arguello, D.; Ramos, P.; Mahdi, H.; ElNaggar, A.; Winer, I.; Holloway, R.; Krivak, T.; Jones, N.; Turner, V.G.; et al. Molecular portraits of clear cell ovarian and endometrial carcinoma with comparison to clear cell renal cell carcinoma. Gynecol. Oncol. 2023, 169, 164–171. [Google Scholar] [CrossRef]
- Zhao, B.; Lin, J.; Rong, L.; Wu, S.; Deng, Z.; Fatkhutdinov, N.; Zundell, J.; Fukumoto, T.; Liu, Q.; Kossenkov, A.; et al. ARID1A promotes genomic stability through protecting telomere cohesion. Nat. Commun. 2019, 10, 4067. [Google Scholar] [CrossRef]
- Sia, T.Y.; Manning-Geist, B.; Gordhandas, S.; Murali, R.; Marra, A.; Liu, Y.L.; Friedman, C.F.; Hollmann, T.J.; Zivanovic, O.; Chi, D.S.; et al. Treatment of ovarian clear cell carcinoma with immune checkpoint blockade: A case series. Int. J. Gynecol. Cancer 2022, 32, 1017–1024. [Google Scholar] [CrossRef]
- Pérez-Mies, B.; Carretero-Barrio, I.; Palacios, J. Immune-related gene expression signatures: A step forward in the stratification of patients with ovarian clear cell carcinoma(†). J. Pathol. 2022, 256, 366–368. [Google Scholar] [CrossRef]
- Lang, J.D.; Hendricks, W.P.D.; Orlando, K.A.; Yin, H.; Kiefer, J.; Ramos, P.; Sharma, R.; Pirrotte, P.; Raupach, E.A.; Sereduk, C.; et al. Ponatinib Shows Potent Antitumor Activity in Small Cell Carcinoma of the Ovary Hypercalcemic Type (SCCOHT) through Multikinase Inhibition. Clin. Cancer Res. 2018, 24, 1932–1943. [Google Scholar] [CrossRef]
- Mandal, J.; Yu, Z.C.; Shih, I.M.; Wang, T.L. ARID1A loss activates MAPK signaling via DUSP4 downregulation. J. Biomed. Sci. 2023, 30, 94. [Google Scholar] [CrossRef]
- Park, Y.; Chui, M.H.; Suryo Rahmanto, Y.; Yu, Z.C.; Shamanna, R.A.; Bellani, M.A.; Gaillard, S.; Ayhan, A.; Viswanathan, A.; Seidman, M.M.; et al. Loss of ARID1A in Tumor Cells Renders Selective Vulnerability to Combined Ionizing Radiation and PARP Inhibitor Therapy. Clin. Cancer Res. 2019, 25, 5584–5594. [Google Scholar] [CrossRef]
- Lheureux, S.; Tinker, A.; Clarke, B.; Ghatage, P.; Welch, S.; Weberpals, J.I.; Dhani, N.C.; Butler, M.O.; Tonkin, K.; Tan, Q.; et al. A Clinical and Molecular Phase II Trial of Oral ENMD-2076 in Ovarian Clear Cell Carcinoma (OCCC): A Study of the Princess Margaret Phase II Consortium. Clin. Cancer Res. 2018, 24, 6168–6174. [Google Scholar] [CrossRef] [PubMed]
- Lans, H.; Marteijn, J.A.; Vermeulen, W. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin 2012, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Wilkerson, P.M.; Dedes, K.J.; Samartzis, E.P.; Dedes, I.; Lambros, M.B.; Natrajan, R.; Gauthier, A.; Piscuoglio, S.; Töpfer, C.; Vukovic, V.; et al. Preclinical evaluation of the PARP inhibitor BMN-673 for the treatment of ovarian clear cell cancer. Oncotarget 2017, 8, 6057–6066. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.C.; Li, T.; Tully, E.; Huang, P.; Chen, C.N.; Oberdoerffer, P.; Gaillard, S.; Shih, I.M.; Wang, T.L. Temozolomide Sensitizes ARID1A-Mutated Cancers to PARP Inhibitors. Cancer Res. 2023, 83, 2750–2762. [Google Scholar] [CrossRef]
- Bitler, B.G.; Aird, K.M.; Garipov, A.; Li, H.; Amatangelo, M.; Kossenkov, A.V.; Schultz, D.C.; Liu, Q.; Shih Ie, M.; Conejo-Garcia, J.R.; et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat. Med. 2015, 21, 231–238. [Google Scholar] [CrossRef]
- Rehman, H.; Chandrashekar, D.S.; Balabhadrapatruni, C.; Nepal, S.; Balasubramanya, S.A.H.; Shelton, A.K.; Skinner, K.R.; Ma, A.H.; Rao, T.; Agarwal, S.; et al. ARID1A-deficient bladder cancer is dependent on PI3K signaling and sensitive to EZH2 and PI3K inhibitors. JCI Insight 2022, 7, e155899. [Google Scholar] [CrossRef]
- Bitler, B.G.; Aird, K.M.; Zhang, R. Epigenetic synthetic lethality in ovarian clear cell carcinoma: EZH2 and ARID1A mutations. Mol. Cell Oncol. 2016, 3, e1032476. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, S.Y.; Colborne, S.; Lambert, G.; Shin, C.Y.; Santos, N.D.; Orlando, K.A.; Lang, J.D.; Hendricks, W.P.D.; Bally, M.B.; et al. Histone Deacetylase Inhibitors Synergize with Catalytic Inhibitors of EZH2 to Exhibit Antitumor Activity in Small Cell Carcinoma of the Ovary, Hypercalcemic Type. Mol. Cancer Ther. 2018, 17, 2767–2779. [Google Scholar] [CrossRef]
- Shigeta, S.; Lui, G.Y.L.; Shaw, R.; Moser, R.; Gurley, K.E.; Durenberger, G.; Rosati, R.; Diaz, R.L.; Ince, T.A.; Swisher, E.M.; et al. Targeting BET Proteins BRD2 and BRD3 in Combination with PI3K-AKT Inhibition as a Therapeutic Strategy for Ovarian Clear Cell Carcinoma. Mol. Cancer Ther. 2021, 20, 691–703. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).