Future AI Will Most Likely Predict Antibody-Drug Conjugate Response in Oncology: A Review and Expert Opinion
Abstract
:Simple Summary
Abstract
1. Introduction
2. Prediction of Cancer Responsiveness and Resistance to ADCs
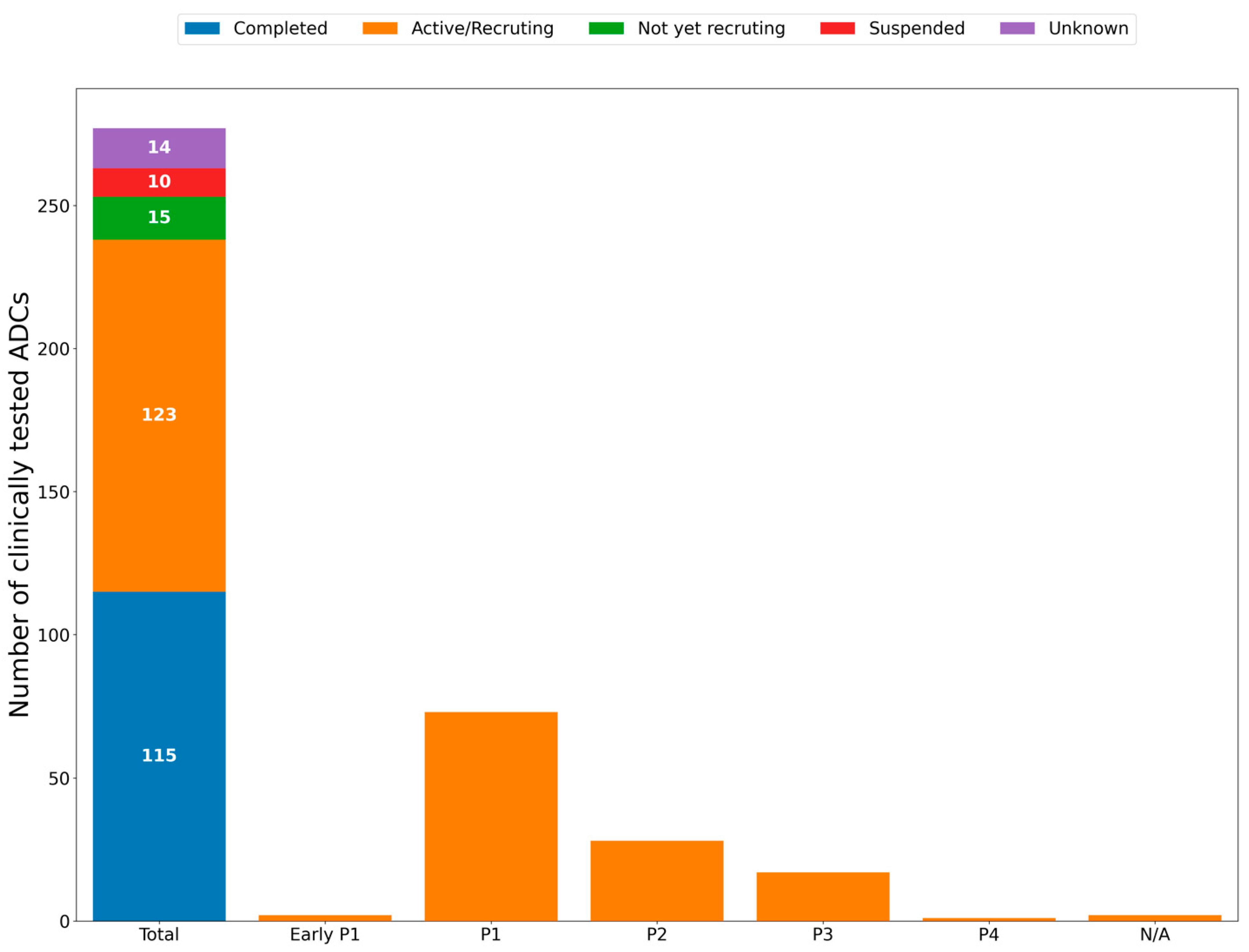
3. Anticancer ADCs That Have Entered Clinical Trials
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Paul, D.; Sanap, G.; Shenoy, S.; Kalyane, D.; Kalia, K.; Tekade, R.K. Artificial intelligence in drug discovery and development. Drug Discov. Today 2021, 26, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Luc Steels, R.B. The Artificial Life Route to Artificial Intelligence. 1995. Available online: https://www.routledge.com/The-Artificial-Life-Route-to-Artificial-Intelligence-Building-Embodied-Situated-Agents/Steels-Brooks/p/book/9781138545854 (accessed on 2 September 2024).
- Bielecki, A. Foundations of Artificial Neural Networks. In Models of Neurons and Perceptrons: Selected Problems and Challenges; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- AI’s potential to accelerate drug discovery needs a reality check. Nature 2023, 622, 217. [CrossRef] [PubMed]
- Schneider, G. Automating drug discovery. Nat. Rev. Drug Discov. 2018, 17, 97–113. [Google Scholar] [CrossRef] [PubMed]
- Sobhani, N.; Tardiel-Cyril, D.R.; Chai, D.; Generali, D.; Li, J.R.; Vazquez-Perez, J.; Lim, J.M.; Morris, R.; Bullock, Z.N.; Davtyan, A.; et al. Artificial intelligence-powered discovery of small molecules inhibiting CTLA-4 in cancer. BJC Rep. 2024, 2. [Google Scholar] [CrossRef]
- DiMasi, J.A.; Grabowski, H.G.; Hansen, R.W. Innovation in the pharmaceutical industry: New estimates of R&D costs. J. Health Econ. 2016, 47, 20–33. [Google Scholar] [CrossRef]
- Peters, C.; Brown, S. Antibody-drug conjugates as novel anti-cancer chemotherapeutics. Biosci. Rep. 2015, 35. [Google Scholar] [CrossRef]
- Sobhani, N. New and Most Powerful Molecules for the Treatment and Diagnosis of Neuroendocrine Cancers (Nets) and the Stem Cells of Nets. Patent IT201600108581A1, 27 April 2018. [Google Scholar]
- Liu, K.; Li, M.; Li, Y.; Li, Y.; Chen, Z.; Tang, Y.; Yang, M.; Deng, G.; Liu, H. A review of the clinical efficacy of FDA-approved antibody–drug conjugates in human cancers. Mol. Cancer 2024, 23, 62. [Google Scholar] [CrossRef]
- Chen, W.; Liu, X.; Zhang, S.; Chen, S. Artificial intelligence for drug discovery: Resources, methods, and applications. Mol. Ther. Nucleic Acids 2023, 31, 691–702. [Google Scholar] [CrossRef]
- Wallach, I.; Dzamba, M.; Heifets, A. AtomNet: A Deep Convolutional Neural Network for Bioactivity Prediction in Structure-based Drug Discovery. arXiv 2015, arXiv:1510.02855. [Google Scholar]
- Rajapakse, V.N.; Luna, A.; Yamade, M.; Loman, L.; Varma, S.; Sunshine, M.; Iorio, F.; Sousa, F.G.; Elloumi, F.; Aladjem, M.I.; et al. CellMinerCDB for Integrative Cross-Database Genomics and Pharmacogenomics Analyses of Cancer Cell Lines. iScience 2018, 10, 247–264. [Google Scholar] [CrossRef]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 19, A68–A77. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Sobhani, N.; Generali, D.; Zanconati, F.; Bortul, M.; Scaggiante, B. Cell-free DNA integrity for the monitoring of breast cancer: Future perspectives? World J. Clin. Oncol. 2018, 9, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Conca, V.; Ciracì, P.; Boccaccio, C.; Minelli, A.; Antoniotti, C.; Cremolini, C. Waiting for the “liquid revolution” in the adjuvant treatment of colon cancer patients: A review of ongoing trials. Cancer Treat. Rev. 2024, 126, 102735. [Google Scholar] [CrossRef]
- Sobhani, N.; Sirico, M.; Generali, D.; Zanconati, F.; Scaggiante, B. Circulating cell-free nucleic acids as prognostic and therapy predictive tools for metastatic castrate-resistant prostate cancer. World J. Clin. Oncol. 2020, 11, 450–463. [Google Scholar] [CrossRef]
- Gao, Q.; Zeng, Q.; Wang, Z.; Li, C.; Xu, Y.; Cui, P.; Zhu, X.; Lu, H.; Wang, G.; Cai, S.; et al. Circulating cell-free DNA for cancer early detection. Innovation 2022, 3, 100259. [Google Scholar] [CrossRef]
- Hsieh, C.; Laguna, A.; Ikeda, I.; Maxwell, A.W.P.; Chapiro, J.; Nadolski, G.; Jiao, Z.; Bai, H.X. Using Machine Learning to Predict Response to Image-guided Therapies for Hepatocellular Carcinoma. Radiology 2023, 309, e222891. [Google Scholar] [CrossRef]
- Bzdok, D.; Altman, N.; Krzywinski, M. Statistics versus machine learning. Nat. Methods 2018, 15, 233–234. [Google Scholar] [CrossRef]
- Morshid, A.; Elsayes, K.M.; Khalaf, A.M.; Elmohr, M.M.; Yu, J.; Kaseb, A.O.; Hassan, M.; Mahvash, A.; Wang, Z.; Hazle, J.D.; et al. A machine learning model to predict hepatocellular carcinoma response to transcatheter arterial chemoembolization. Radiol. Artif. Intell. 2019, 1. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, Z.; An, C.; Chen, H.; Li, X. Multi-Task Deep Learning Approach for Simultaneous Objective Response Prediction and Tumor Segmentation in HCC Patients with Transarterial Chemoembolization. J. Pers. Med. 2022, 12, 248. [Google Scholar] [CrossRef] [PubMed]
- Mazaheri, Y.; Thakur, S.B.; Bitencourt, A.G.; Lo Gullo, R.; Hötker, A.M.; Bates, D.D.B.; Akin, O. Evaluation of cancer outcome assessment using MRI: A review of deep-learning methods. BJR Open 2022, 4, 20210072. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jiang, Y.; Jin, Z.; Jiang, W.; Zhang, B.; Wang, C.; Wu, L.; Chen, L.; Chen, Q.; Liu, S.; et al. Real-time automatic prediction of treatment response to transcatheter arterial chemoembolization in patients with hepatocellular carcinoma using deep learning based on digital subtraction angiography videos. Cancer Imaging 2022, 22, 23. [Google Scholar] [CrossRef]
- Ma, Q.P.; He, X.L.; Li, K.; Wang, J.F.; Zeng, Q.J.; Xu, E.J.; He, X.Q.; Li, S.Y.; Kun, W.; Zheng, R.Q.; et al. Dynamic Contrast-Enhanced Ultrasound Radiomics for Hepatocellular Carcinoma Recurrence Prediction After Thermal Ablation. Mol. Imaging Biol. 2021, 23, 572–585. [Google Scholar] [CrossRef]
- Peng, J.; Lu, F.; Huang, J.; Zhang, J.; Gong, W.; Hu, Y.; Wang, J. Development and validation of a pyradiomics signature to predict initial treatment response and prognosis during transarterial chemoembolization in hepatocellular carcinoma. Front. Oncol. 2022, 12, 853254. [Google Scholar] [CrossRef]
- Lyshchik, A.; Kono, Y.; Dietrich, C.F.; Jang, H.J.; Kim, T.K.; Piscaglia, F.; Vezeridis, A.; Willmann, J.K.; Wilson, S.R. Contrast-enhanced ultrasound of the liver: Technical and lexicon recommendations from the ACR CEUS LI-RADS working group. Abdom Radiol. 2018, 43, 861–879. [Google Scholar] [CrossRef]
- Artificial Intelligence and Machine Learning in Software as a Medical Device. 2024. Available online: https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device (accessed on 2 September 2024).
- Puijk, R.S.; Ahmed, M.; Adam, A.; Arai, Y.; Arellano, R.; de Baère, T.; Bale, R.; Bellera, C.; Binkert, C.A.; Brace, C.L.; et al. Consensus Guidelines for the Definition of Time-to-Event End Points in Image-guided Tumor Ablation: Results of the SIO and DATECAN Initiative. Radiology 2021, 301, 533–540. [Google Scholar] [CrossRef]
- Iseke, S.; Zeevi, T.; Kucukkaya, A.S.; Raju, R.; Gross, M.; Haider, S.P.; Petukhova-Greenstein, A.; Kuhn, T.N.; Lin, M.; Nowak, M.; et al. Machine Learning Models for Prediction of Posttreatment Recurrence in Early-Stage Hepatocellular Carcinoma Using Pretreatment Clinical and MRI Features: A Proof-of-Concept Study. AJR Am. J. Roentgenol. 2023, 220, 245–255. [Google Scholar] [CrossRef]
- Yu, L.; Li, Y. Artificial Intelligence Decision-Making Transparency and Employees’ Trust: The Parallel Multiple Mediating Effect of Effectiveness and Discomfort. Behav. Sci. 2022, 12, 127. [Google Scholar] [CrossRef]
- Wang, F.; Casalino, L.P.; Khullar, D. Deep Learning in Medicine-Promise, Progress, and Challenges. JAMA Intern. Med. 2019, 179, 293–294. [Google Scholar] [CrossRef] [PubMed]
- Calderaro, J.; Seraphin, T.P.; Luedde, T.; Simon, T.G. Artificial intelligence for the prevention and clinical management of hepatocellular carcinoma. J. Hepatol. 2022, 76, 1348–1361. [Google Scholar] [CrossRef] [PubMed]
- Damen, J.A.; Hooft, L.; Schuit, E.; Debray, T.P.; Collins, G.S.; Tzoulaki, I.; Lassale, C.M.; Siontis, G.C.; Chiocchia, V.; Roberts, C.; et al. Prediction models for cardiovascular disease risk in the general population: Systematic review. bmj 2016, 353, i2416. [Google Scholar] [CrossRef] [PubMed]
- Malinchoc, M.; Kamath, P.S.; Gordon, F.D.; Peine, C.J.; Rank, J.; ter Borg, P.C. A model to predict poor survival in patients undergoing transjugular intrahepatic portosystemic shunts. Hepatology 2000, 31, 864–871. [Google Scholar] [CrossRef] [PubMed]
- van Gorp, M.J.; Steyerberg, E.W.; Van der Graaf, Y. Decision guidelines for prophylactic replacement of Björk-Shiley convexo-concave heart valves: Impact on clinical practice. Circulation 2004, 109, 2092–2096. [Google Scholar] [CrossRef]
- Wilson, P.W.; D’Agostino, R.B.; Levy, D.; Belanger, A.M.; Silbershatz, H.; Kannel, W.B. Prediction of coronary heart disease using risk factor categories. Circulation 1998, 97, 1837–1847. [Google Scholar] [CrossRef]
- Johnston, S.C.; Rothwell, P.M.; Nguyen-Huynh, M.N.; Giles, M.F.; Elkins, J.S.; Bernstein, A.L.; Sidney, S. Validation and refinement of scores to predict very early stroke risk after transient ischaemic attack. Lancet 2007, 369, 283–292. [Google Scholar] [CrossRef]
- Hippisley-Cox, J.; Coupland, C.; Brindle, P. Development and validation of QRISK3 risk prediction algorithms to estimate future risk of cardiovascular disease: Prospective cohort study. bmj 2017, 357, j2099. [Google Scholar] [CrossRef]
- Bouwmeester, W.; Zuithoff, N.P.; Mallett, S.; Geerlings, M.I.; Vergouwe, Y.; Steyerberg, E.W.; Altman, D.G.; Moons, K.G. Reporting and methods in clinical prediction research: A systematic review. PLoS Med. 2012, 9, e1001221. [Google Scholar] [CrossRef]
- Collins, G.S.; de Groot, J.A.; Dutton, S.; Omar, O.; Shanyinde, M.; Tajar, A.; Voysey, M.; Wharton, R.; Yu, L.M.; Moons, K.G.; et al. External validation of multivariable prediction models: A systematic review of methodological conduct and reporting. BMC Med. Res. Methodol. 2014, 14, 40. [Google Scholar] [CrossRef]
- Gary, S.; Collins, K.G.M.M. Reporting of artificial intelligence prediction models. Lancet 2019, 393, 1577–1579. [Google Scholar] [CrossRef]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD Statement. BMC Med. 2015, 13, 1. [Google Scholar] [CrossRef] [PubMed]
- Moons, K.G.; Altman, D.G.; Reitsma, J.B.; Ioannidis, J.P.; Macaskill, P.; Steyerberg, E.W.; Vickers, A.J.; Ransohoff, D.F.; Collins, G.S. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): Explanation and elaboration. Ann. Intern. Med. 2015, 162, W1–W73. [Google Scholar] [CrossRef]
- Ratner, M. FDA backs clinician-free AI imaging diagnostic tools. Nat. Biotechnol. 2018, 36, 673–674. [Google Scholar] [CrossRef] [PubMed]
- Collins, G.S.; Dhiman, P.; Andaur Navarro, C.L.; Ma, J.; Hooft, L.; Reitsma, J.B.; Logullo, P.; Beam, A.L.; Peng, L.; Van Calster, B.; et al. Protocol for development of a reporting guideline (TRIPOD-AI) and risk of bias tool (PROBAST-AI) for diagnostic and prognostic prediction model studies based on artificial intelligence. BMJ Open 2021, 11, e048008. [Google Scholar] [CrossRef] [PubMed]
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef]
- Li, T.; Li, Y.; Zhu, X.; He, Y.; Wu, Y.; Ying, T.; Xie, Z. Artificial intelligence in cancer immunotherapy: Applications in neoantigen recognition, antibody design and immunotherapy response prediction. Semin. Cancer Biol. 2023, 91, 50–69. [Google Scholar] [CrossRef]
- All about Cancer; American Cancer Society: Atlanta, GA, USA, 2024.
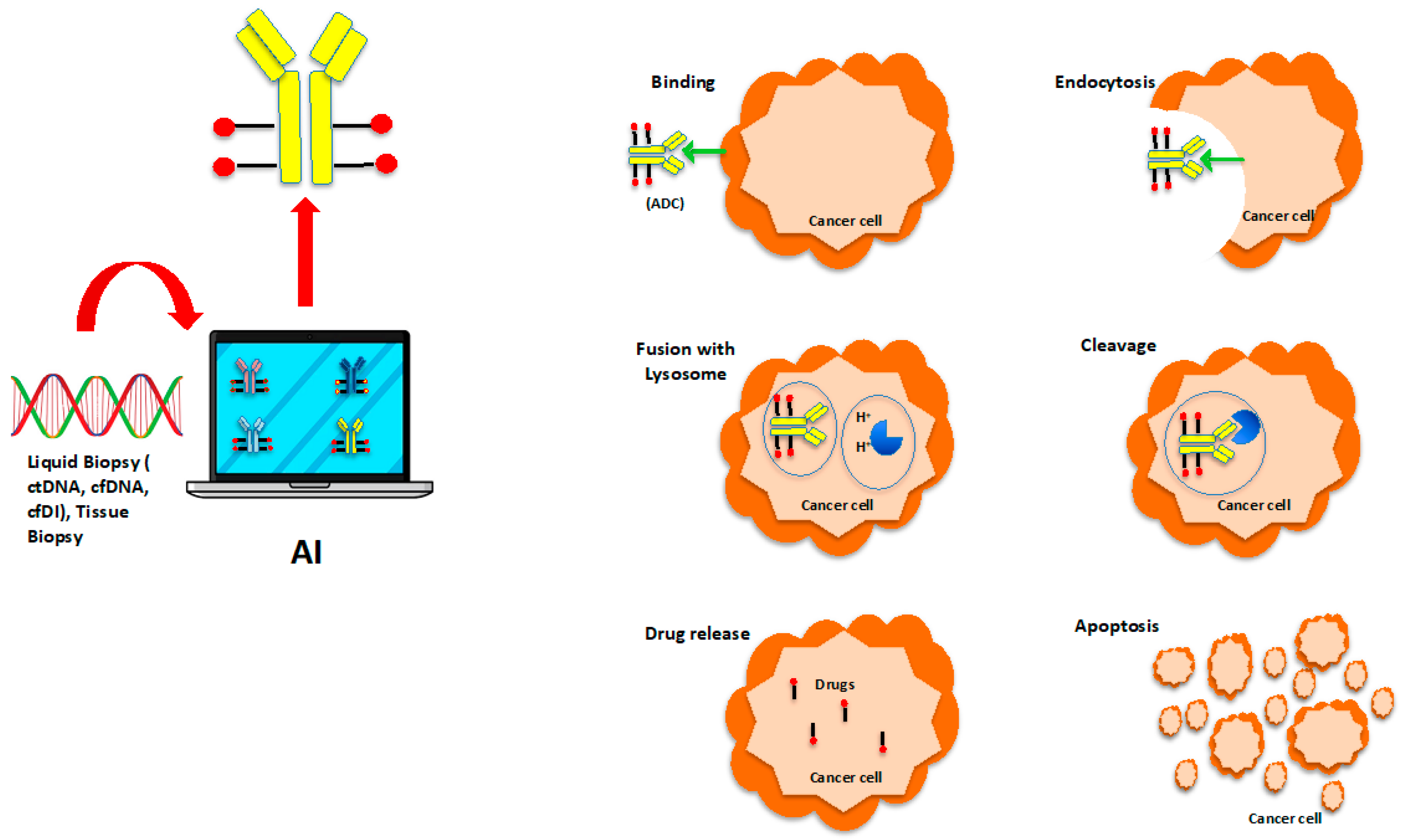
| Name | Main Features | Web Link |
|---|---|---|
| CGHub | Cancer genomics data repository | https://docs.gdc.cancer.gov/Encyclopedia/pages/Cancer_Genomics_Hub/ (accessed on 2 September 2024) |
| TCGA | Comprehensive database of cancer patients’ genomic, epigenomic, transcriptomic, and proteomic data. | https://www.cancer.gov/ccg/research/genome-sequencing/tcga (accessed on 2 September 2024) |
| CCLE | Comprehensive genetic database of cancer cell lines | https://sites.broadinstitute.org/ccle (accessed on 2 September 2024) |
| EGA | European genetic, phenotypic, and clinical data repository | https://ega-archive.org/ (accessed on 2 September 2024) |
| DepMap | High data quality visualization tool | https://depmap.org/portal/ (accessed on 2 September 2024) |
| SomamiR | Cancer somatic mutation and miRNA correlation | https://compbio.uthsc.edu/SomamiR/ (accessed on 2 September 2024) |
| COSMIC | Comprehensive somatic mutation database | https://cancer.sanger.ac.uk/cosmic (accessed on 2 September 2024) |
| MethyCancer | DNA methylations, cancer-related genes, and mutations in correlation with additional cancer information | http://methycancer.psych.ac.cn/ (accessed on 2 September 2024) |
| CTRP | connecting genetic, cellular features, lineage to cancer cell-lines sensitivity to small molecules | https://portals.broadinstitute.org/ctrp/ (accessed on 2 September 2024) |
| gCSI | Large number of transcriptomics data | https://pharmacodb.pmgenomics.ca/datasets/4 (accessed on 2 September 2024) |
| GDSC | Drug response, including genomics markers of drug sensitivity | https://www.cancerrxgene.org/ (accessed on 2 September 2024) |
| NCI60 | Large number of drug and genomics data | https://discover.nci.nih.gov/cellminer/loadDownload.do (accessed on 2 September 2024) https://dtp.cancer.gov/databases_tools/bulk_data.htm (accessed on 2 September 2024) |
| canSAR | Comprehensive drug discovery database | https://cansarblack.icr.ac.uk/ (accessed on 2 September 2024) |
| cBioPortal | Large database of cancer genomics data | https://www.cbioportal.org/datasets (accessed on 2 September 2024) |
| UCSC | Synthetical genomics information | https://genome.ucsc.edu/ (accessed on 2 September 2024) |
| dbNSFP | Non-synonymous single-nucleotide variants | https://sites.google.com/site/jpopgen/dbNSFP (accessed on 2 September 2024) |
| NONCODE | Non-coding RNAs database | http://www.noncode.org/ (accessed on 2 September 2024) |
| TCIA | Comprehensive immunogenomic data from the NGS of 20 solid tumors from TCGA | https://www.tcia.at/home (accessed on 2 September 2024) |
| ARCHS4 | Comprehensive RNA-Sequenced data from human and mouse | https://maayanlab.cloud/archs4/ (accessed on 2 September 2024) |
| NCT Number | Study Title | Study URL | Study Status | Conditions | ADC | Sponsor |
|---|---|---|---|---|---|---|
| NCT06340568 | A Clinical Study of the Anti-cancer Effects of an Investigational Therapy or Chemotherapy in Patients With Recurring Uterine Cancer | https://clinicaltrials.gov/study/NCT06340568 (accessed on 2 September 2024) | Not yet recruiting | Endometrial Cancer | DRUG: BNT323/DB-1303|DRUG: Doxorubicin|DRUG: Paclitaxel | BioNTech SE |
| NCT05609968 | Study of Pembrolizumab (MK-3475) Monotherapy Versus Sacituzumab Govitecan in Combination With Pembrolizumab for Participants With Metastatic Non-small Cell Lung Cancer (NSCLC) With Programmed Cell Death Ligand 1 (PD-L1) Tumor Proportion Score (TPS) ‚ ≥50% (MK-3475-D46) | https://clinicaltrials.gov/study/NCT05609968 (accessed on 2 September 2024) | Recruiting | Carcinoma|Non-Small Cell Lung Cancer | BIOLOGICAL: Sacituzumab govitecan|BIOLOGICAL: Pembrolizumab | Merck Sharp & Dohme LLC |
| NCT03529110 | DS-8201a Versus T-DM1 for Human Epidermal Growth Factor Receptor 2 (HER2)-Positive, Unresectable and/or Metastatic Breast Cancer Previously Treated With Trastuzumab and Taxane [DESTINY-Breast03] | https://clinicaltrials.gov/study/NCT03529110 (accessed on 2 September 2024) | Active—Not yet recruiting | Breast Cancer | DRUG: Trastuzumab deruxtecan (T-DXd)|DRUG: Ado-trastuzumab emtansine (T-DM1) | Daiichi Sankyo |
| NCT06203210 | A Study of Ifinatamab Deruxtecan Versus Treatment of Physician’s Choice in Subjects With Relapsed Small Cell Lung Cancer | https://clinicaltrials.gov/study/NCT06203210 (accessed on 2 September 2024) | Not yet recruiting | Small Cell Lung Cancer | DRUG: Ifinatamab deruxtecan|DRUG: Topotecan|DRUG: Amrubicin|DRUG: Lurbinectedin | Daiichi Sankyo |
| NCT02631876 | A Study of Mirvetuximab Soravtansine vs. Investigator’s Choice of Chemotherapy in Women With Folate Receptor (FR) Alpha Positive Advanced Epithelial Ovarian Cancer (EOC), Primary Peritoneal or Fallopian Tube Cancer | https://clinicaltrials.gov/study/NCT02631876 (accessed on 2 September 2024) | Completed | Epithelial Ovarian Cancer|Primary Peritoneal Carcinoma|Fallopian Tube Cancer|Ovarian Cancer | DRUG: Mirvetuximab soravtansine|DRUG: Paclitaxel|DRUG: Pegylated liposomal doxorubicin|DRUG: Topotecan | ImmunoGen, Inc. |
| NCT03734029 | Trastuzumab Deruxtecan (DS-8201a) Versus Investigator’s Choice for HER2-low Breast Cancer That Has Spread or Cannot be Surgically Removed [DESTINY-Breast04] | https://clinicaltrials.gov/study/NCT03734029 (accessed on 2 September 2024) | Active—Not yet recruiting | Breast Cancer | DRUG: Trastuzumab deruxtecan (DS-8201a)|DRUG: Capecitabine|DRUG: Eribulin|DRUG: Gemcitabine|DRUG: Paclitaxel|DRUG: Nab-paclitaxel | Daiichi Sankyo |
| NCT04494425 | Study of Trastuzumab Deruxtecan (T-DXd) vs. Investigator’s Choice Chemotherapy in HER2-low, Hormone Receptor Positive, Metastatic Breast Cancer | https://clinicaltrials.gov/study/NCT04494425 (accessed on 2 September 2024) | Active—Not yet recruiting | Advanced or Metastatic Breast Cancer | DRUG: Trastuzumab deruxtecan|DRUG: Capecitabine|DRUG: Paclitaxel|DRUG: Nab-Paclitaxel | AstraZeneca |
| NCT04595565 | Sacituzumab Govitecan in Primary HER2-negative Breast Cancer | https://clinicaltrials.gov/study/NCT04595565 (accessed on 2 September 2024) | Recruiting | HER2-negative Breast Cancer|Triple Negative Breast Cancer | DRUG: Capecitabine|DRUG: Carboplatin|DRUG: Cisplatin|DRUG: Sacituzumab govitecan | German Breast Group |
| NCT05687266 | Phase III, Open-label, First-line Study of Dato-DXd in Combination With Durvalumab and Carboplatin for Advanced NSCLC Without Actionable Genomic Alterations | https://clinicaltrials.gov/study/NCT05687266 (accessed on 2 September 2024) | Recruting | NSCLC | DRUG: Datopotamab deruxtecan|DRUG: Durvalumab|DRUG: Carboplatin|DRUG: Pembrolizumab|DRUG: Cisplatin|DRUG: Pemetrexed|DRUG: Paclitaxel | AstraZeneca |
| NCT05104866 | A Phase-3, Open-Label, Randomized Study of Dato-DXd Versus Investigator’s Choice of Chemotherapy (ICC) in Participants With Inoperable or Metastatic HR-Positive, HER2-Negative Breast Cancer Who Have Been Treated With One or Two Prior Lines of Systemic Chemotherapy (TROPION-Breast01) | https://clinicaltrials.gov/study/NCT05104866 (accessed on 2 September 2024) | Active—Not yet recruiting | Breast Cancer | DRUG: Dato-DXd|DRUG: Capecitabine|DRUG: Gemcitabine|DRUG: Eribulin|DRUG: Vinorelbine | AstraZeneca |
| NCT06161025 | A Study of Raludotatug Deruxtecan (R-DXd) in Subjects With Platinum-resistant, High-grade Ovarian, Primary Peritoneal, or Fallopian Tube Cancer | https://clinicaltrials.gov/study/NCT06161025 (accessed on 2 September 2024) | Recruiting | Solid Cancer | DRUG: R-DXd|DRUG: Gemcitabine|DRUG: Paclitaxel|DRUG: Topotecan|DRUG: PLD | Daiichi Sankyo |
| NCT04639986 | Asian Study of Sacituzumab Govitecan (IMMU-132) in HR+/HER2− Metastatic Breast Cancer (MBC) | https://clinicaltrials.gov/study/NCT04639986 (accessed on 2 September 2024) | Active—Not yet recruiting | Metastatic Breast Cancer | DRUG: Sacituzumab govitecan-hziy|DRUG: Eribulin mesylate injection|DRUG: Capecitabine oral product|DRUG: Gemcitabine injection|DRUG: Vinorelbine injection | Gilead Sciences |
| NCT04296890 | A Study of Mirvetuximab Soravtansine in Platinum-Resistant, Advanced High-Grade Epithelial Ovarian, Primary Peritoneal, or Fallopian Tube Cancers With High Folate Receptor-Alpha Expression | https://clinicaltrials.gov/study/NCT04296890 (accessed on 2 September 2024) | Completed | Epithelial Ovarian Cancer|Peritoneal Cancer|Fallopian Tube Cancer | DRUG: Mirvetuximab soravtansine | ImmunoGen, Inc. |
| NCT01100502 | A Phase 3 Study of Brentuximab Vedotin (SGN-35) in Patients at High Risk of Residual Hodgkin Lymphoma Following Stem Cell Transplant (The AETHERA Trial) | https://clinicaltrials.gov/study/NCT01100502 (accessed on 2 September 2024) | Completed | Disease, Hodgkin | DRUG: Brentuximab vedotin|DRUG: Placebo | Seagen Inc. |
| NCT06103864 | A Phase III Study of Dato-DXd With or Without Durvalumab Compared With Investigator’s Choice of Chemotherapy in Combination With Pembrolizumab in Patients With PD-L1 Positive Locally Recurrent Inoperable or Metastatic Triple-negative Breast Cancer | https://clinicaltrials.gov/study/NCT06103864 (accessed on 2 September 2024) | Recruiting | Breast Cancer | DRUG: Dato-DXd|DRUG: Durvalumab|DRUG: Paclitaxel|DRUG: Nab-paclitaxel|DRUG: Gemcitabine|DRUG: Carboplatin|DRUG: Pembrolizumab | AstraZeneca |
| NCT01712490 | A Frontline Therapy Trial in Participants With Advanced Classical Hodgkin Lymphoma | https://clinicaltrials.gov/study/NCT01712490 (accessed on 2 September 2024) | Active—Not yet recruiting | Hodgkin Lymphoma | DRUG: Brentuximab vedotin|DRUG: Doxorubicin|DRUG: Bleomycin|DRUG: Vinblastine|DRUG: Dacarbazine | Takeda |
| NCT05622890 | A Single-arm Clinical Trial of IMGN853 in Chinese Adult Patients With Platinum-resistant, Epithelial Ovarian Cancer | https://clinicaltrials.gov/study/NCT05622890 (accessed on 2 September 2024) | Recruiting | Epithelial Ovarian Cancer|Peritoneal Cancer|Fallopian Tube Cancer | DRUG: Mirvetuximab soravtansine | Hangzhou Zhongmei Huadong Pharmaceutical Co., Ltd. |
| NCT06112379 | A Phase III Randomised Study to Evaluate Dato-DXd and Durvalumab for Neoadjuvant/Adjuvant Treatment of Triple-Negative or Hormone Receptor-low/HER2-negative Breast Cancer | https://clinicaltrials.gov/study/NCT06112379 (accessed on 2 September 2024) | Recruiting | Breast Cancer | DRUG: Dato-DXd|DRUG: Durvalumab|DRUG: Pembrolizumab|DRUG: Doxorubicin|DRUG: Epirubicin|DRUG: Cyclophosphamide|DRUG: Paclitaxel|DRUG: Carboplatin|DRUG: Capecitabine|DRUG: Olaparib | AstraZeneca |
| NCT04209855 | A Study of Mirvetuximab Soravtansine vs. Investigator’s Choice of Chemotherapy in Platinum-Resistant, Advanced High-Grade Epithelial Ovarian, Primary Peritoneal, or Fallopian Tube Cancers With High Folate Receptor-Alpha Expression | https://clinicaltrials.gov/study/NCT04209855 (accessed on 2 September 2024) | Active—Not yet recruiting | Epithelial Ovarian Cancer|Peritoneal Cancer|Fallopian Tube Cancer | DRUG: Mirvetuximab soravtansine|DRUG: Paclitaxel|DRUG: Topotecan|DRUG: Pegylated liposomal doxorubicin | ImmunoGen, Inc. |
| NCT05751512 | A Study to Evaluate MRG003 vs. Cetuximab/Methotrexate in in the Treatment of Patients With RM-SCCHN | https://clinicaltrials.gov/study/NCT05751512 (accessed on 2 September 2024) | Not yet recruiting | Squamous Cell Carcinoma of the Head and Neck | DRUG: MRG003|DRUG: Cetuximab injection|DRUG: Methotrexate injection | Shanghai Miracogen Inc. |
| NCT05374512 | A Study of Dato-DXd Versus Investigator’s Choice Chemotherapy in Patients With Locally Recurrent Inoperable or Metastatic Triple-negative Breast Cancer, Who Are Not Candidates for PD-1/PD-L1 Inhibitor Therapy (TROPION-Breast02) | https://clinicaltrials.gov/study/NCT05374512 (accessed on 2 September 2024) | Recruiting | Breast Cancer | DRUG: Dato-DXd|DRUG: Paclitaxel|DRUG: Nab-paclitaxel|DRUG: Carboplatin|DRUG: Capecitabine|DRUG: Eribulin mesylate | AstraZeneca |
| NCT05629585 | A Study of Dato-DXd With or Without Durvalumab Versus Investigator’s Choice of Therapy in Patients With Stage I-III Triple-negative Breast Cancer Without Pathological Complete Response Following Neoadjuvant Therapy (TROPION-Breast03) | https://clinicaltrials.gov/study/NCT05629585 (accessed on 2 September 2024) | Recruiting | Breast Cancer | DRUG: Dato-DXd|DRUG: Durvalumab|DRUG: Capecitabine|DRUG: Pembrolizumab | AstraZeneca |
| NCT03523585 | DS-8201a in Pre-treated HER2 Breast Cancer That Cannot be Surgically Removed or Has Spread [DESTINY-Breast02] | https://clinicaltrials.gov/study/NCT03523585 (accessed on 2 September 2024) | Active—Not yet recruiting | Breast Cancer | DRUG: Trastuzumab deruxtecan|DRUG: Capecitabine|DRUG: Lapatinib|DRUG: Trastuzumab | Daiichi Sankyo |
| NCT01777152 | ECHELON-2: A Comparison of Brentuximab Vedotin and CHP With Standard-of-care CHOP in the Treatment of Patients With CD30-positive Mature T-cell Lymphomas | https://clinicaltrials.gov/study/NCT01777152 (accessed on 2 September 2024) | Completed | Anaplastic Large-Cell Lymphoma|Non-Hodgkin Lymphoma|T-Cell Lymphoma | DRUG: Brentuximab vedotin|DRUG: Doxorubicin|DRUG: Prednisone|DRUG: Vincristine|DRUG: Cyclophosphamide | Seagen Inc. |
| NCT06074588 | MK-2870 Versus Chemotherapy in Previously Treated Advanced or Metastatic Nonsquamous Non-small Cell Lung Cancer (NSCLC) With EGFR Mutations or Other Genomic Alterations (MK-2870-004) | https://clinicaltrials.gov/study/NCT06074588 (accessed on 2 September 2024) | Recruiting | Non-small Cell Lung Cancer (NSCLC) | BIOLOGICAL: MK-2870|DRUG: Docetaxel|DRUG: Pemetrexed | Merck Sharp & Dohme LLC |
| NCT03474107 | A Study to Evaluate Enfortumab Vedotin Versus (vs) Chemotherapy in Subjects With Previously Treated Locally Advanced or Metastatic Urothelial Cancer (EV-301) | https://clinicaltrials.gov/study/NCT03474107 (accessed on 2 September 2024) | Active—Not yet recruiting | Ureteral Cancer|Urothelial Cancer|Bladder Cancer | DRUG: Enfortumab Vedotin|DRUG: Docetaxel|DRUG: Vinflunine|DRUG: Paclitaxel | Astellas Pharma Global Development, Inc. |
| NCT05754853 | A Study of MRG002 Versus Investigator’s Choice of Chemotherapy in the Treatment of Patients With HER2-positive Unresectable Advanced or Metastatic Urothelial Cancer | https://clinicaltrials.gov/study/NCT05754853 (accessed on 2 September 2024) | Recruiting | Advanced or Metastatic Urothelium Cancer | DRUG: MRG002|DRUG: Docetaxel injection|DRUG: Paclitaxel injection|DRUG: Gemcitabine hydrochloride for injection|DRUG: Pemetrexed disodium injection | Shanghai Miracogen Inc. |
| NCT05445778 | Mirvetuximab Soravtansine With Bevacizumab Versus Bevacizumab as Maintenance in Platinum-sensitive Ovarian, Fallopian Tube, or Peritoneal Cancer (GLORIOSA) | https://clinicaltrials.gov/study/NCT05445778 (accessed on 2 September 2024) | Recruiting | Ovarian Cancer|Peritoneal Cancer|Fallopian Tube Cancer | DRUG: Mirvetuximab soravtansine plus bevacizumab|DRUG: Bevacizumab | ImmunoGen, Inc. |
| NCT02785900 | Vadastuximab Talirine (SGN-CD33A; 33A) Combined With Azacitidine or Decitabine in Older Patients With Newly Diagnosed Acute Myeloid Leukemia | https://clinicaltrials.gov/study/NCT02785900 (accessed on 2 September 2024) | Terminated | Acute Myeloid Leukemia | DRUG: 33A|DRUG: Placebo|DRUG: Azacitidine|DRUG: Decitabine | Seagen Inc. |
| NCT06132958 | MK-2870 in Post Platinum and Post Immunotherapy Endometrial Cancer (MK-2870-005) | https://clinicaltrials.gov/study/NCT06132958 (accessed on 2 September 2024) | Recruiting | Endometrial Cancer | BIOLOGICAL: MK-2870|DRUG: Doxorubicin|DRUG: Paclitaxel | Merck Sharp & Dohme LLC |
| NCT02573324 | A Study of ABT-414 in Participants With Newly Diagnosed Glioblastoma (GBM) With Epidermal Growth Factor Receptor (EGFR) Amplification | https://clinicaltrials.gov/study/NCT02573324 (accessed on 2 September 2024) | Completed | Glioblastoma|Gliosarcoma | DRUG: Temozolomide|DRUG: Depatuxizumab mafodotin|RADIATION: Radiation|DRUG: Placebo for ABT-414 | AbbVie |
| NCT03262935 | SYD985 vs. Physician’s Choice in Participants With HER2-positive Locally Advanced or Metastatic Breast Cancer | https://clinicaltrials.gov/study/NCT03262935 (accessed on 2 September 2024) | Completed | Metastatic Breast Cancer | DRUG: (Vic-)Trastuzumab duocarmazine|DRUG: Physician’s choice | Byondis B.V. |
| NCT04924699 | A Study of MRG002 in the Treatment of Patients With HER2-positive Unresectable Locally Advanced or Metastatic Breast Cancer | https://clinicaltrials.gov/study/NCT04924699 (accessed on 2 September 2024) | Recruiting | Advanced Breast Cancer|Metastatic Breast Cancer | DRUG: MRG002|DRUG: Trastuzumab emtansine for injection | Shanghai Miracogen Inc. |
| NCT05950945 | Trastuzumab Deruxtecan (T-DXd) in Patients Who Have Hormone Receptor-negative and Hormone Receptor-positive HER2-low or HER2 IHC 0 Metastatic Breast Cancer | https://clinicaltrials.gov/study/NCT05950945 (accessed on 2 September 2024) | Recruiting | Breast Cancer | DRUG: Trastuzumab deruxtecan | Daiichi Sankyo |
| NCT05329545 | Upifitamab Rilsodotin Maintenance in Platinum-Sensitive Recurrent Ovarian Cancer (UP-NEXT) | https://clinicaltrials.gov/study/NCT05329545 (accessed on 2 September 2024) | Terminated | High Grade Serous Ovarian Cancer|Fallopian Tube Cancer|Primary Peritoneal Cancer | DRUG: Upifitimab rilsodotin|OTHER: Placebo | Mersana Therapeutics |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sobhani, N.; D’Angelo, A.; Pittacolo, M.; Mondani, G.; Generali, D. Future AI Will Most Likely Predict Antibody-Drug Conjugate Response in Oncology: A Review and Expert Opinion. Cancers 2024, 16, 3089. https://doi.org/10.3390/cancers16173089
Sobhani N, D’Angelo A, Pittacolo M, Mondani G, Generali D. Future AI Will Most Likely Predict Antibody-Drug Conjugate Response in Oncology: A Review and Expert Opinion. Cancers. 2024; 16(17):3089. https://doi.org/10.3390/cancers16173089
Chicago/Turabian StyleSobhani, Navid, Alberto D’Angelo, Matteo Pittacolo, Giuseppina Mondani, and Daniele Generali. 2024. "Future AI Will Most Likely Predict Antibody-Drug Conjugate Response in Oncology: A Review and Expert Opinion" Cancers 16, no. 17: 3089. https://doi.org/10.3390/cancers16173089







