Cellular and Microbial In Vitro Modelling of Gastrointestinal Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
3. Results
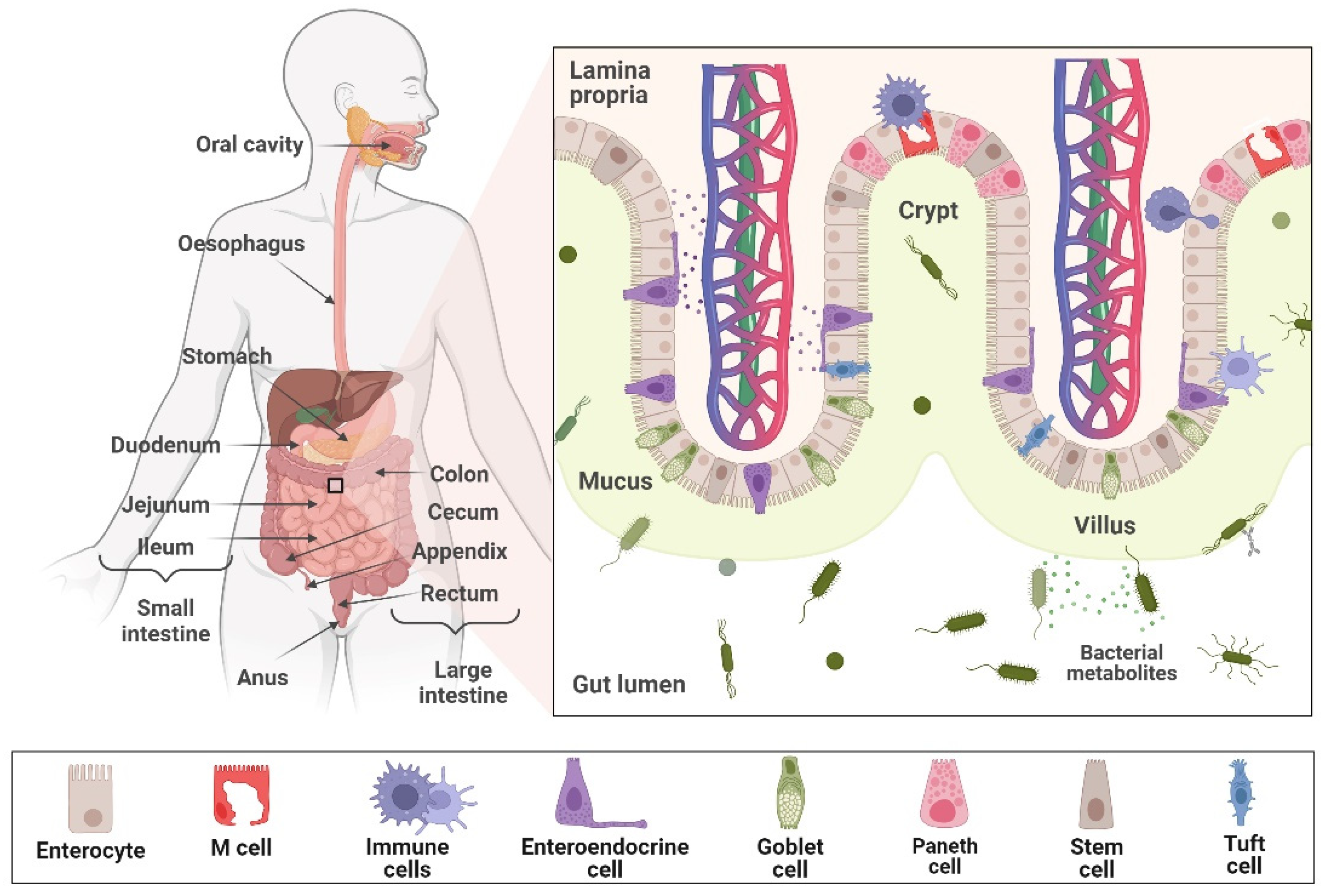
3.1. In Vitro Models of Gastrointestinal Tract
3.1.1. Cell Culture-Based In Vitro Models
- Primary Cells
- Immortalised Cells
- Stem Cell-Based Sources for the Gastrointestinal Tract Tissue Models
- The Transwell® System
3.1.2. 3D-Engineered Gastrointestinal Tissue Models
- Gastrointestinal Organoids
- Advanced 3D Intestinal Models
3.2. Bioreactor-Based In Vitro Models
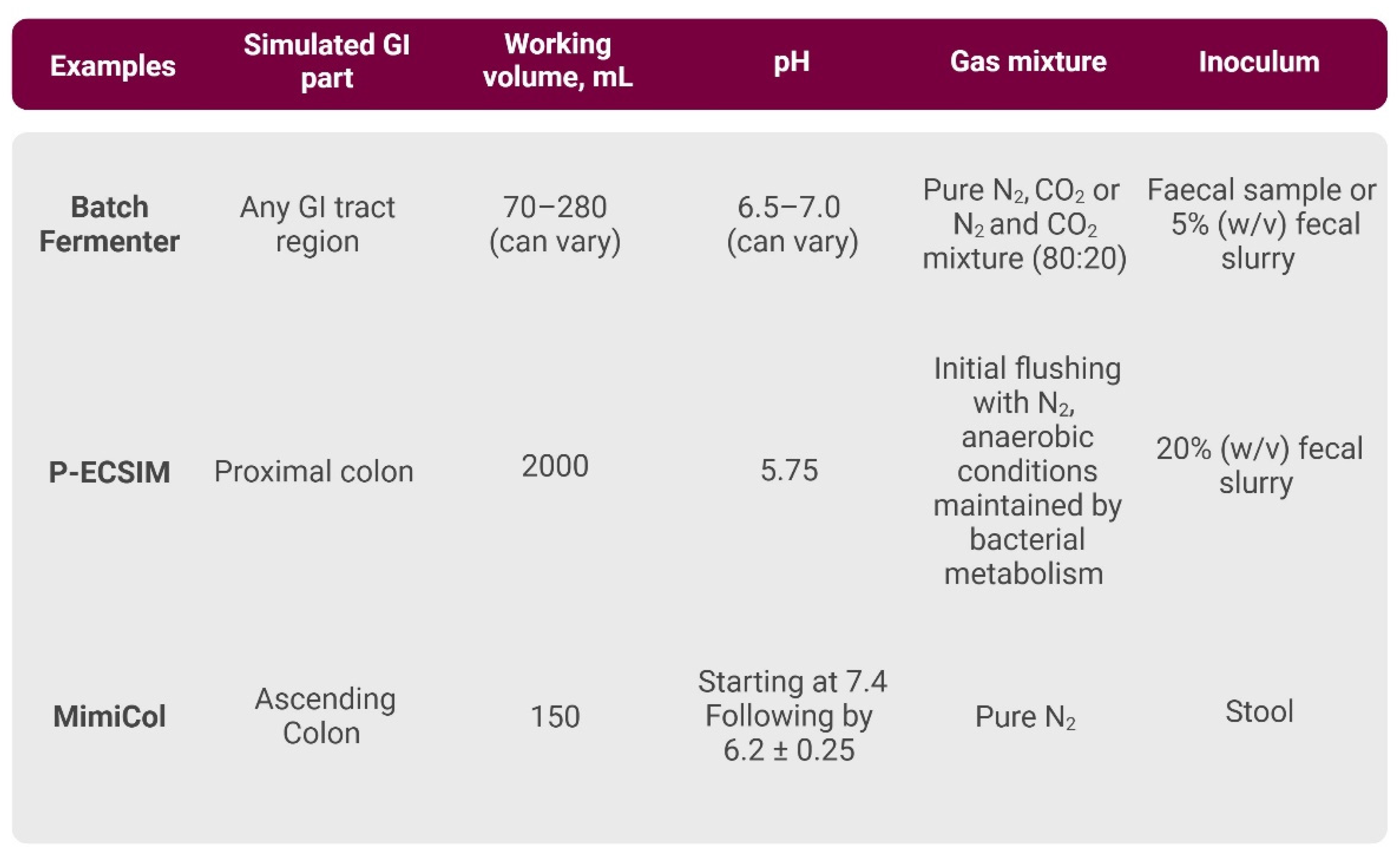
3.2.1. Single-Vessel In Vitro Models
- Batch Fermenter
- P-ECSIM
- MimiCol
3.2.2. Multi-Vessel In Vitro Models
- SHIME
- TIM-1 and TIM-2
- SIMGI
- 3S-ECSIM
- The Host–Microbiota Interaction Model
- The EnteroMix Model
- The Polyfermentor Intestinal Model
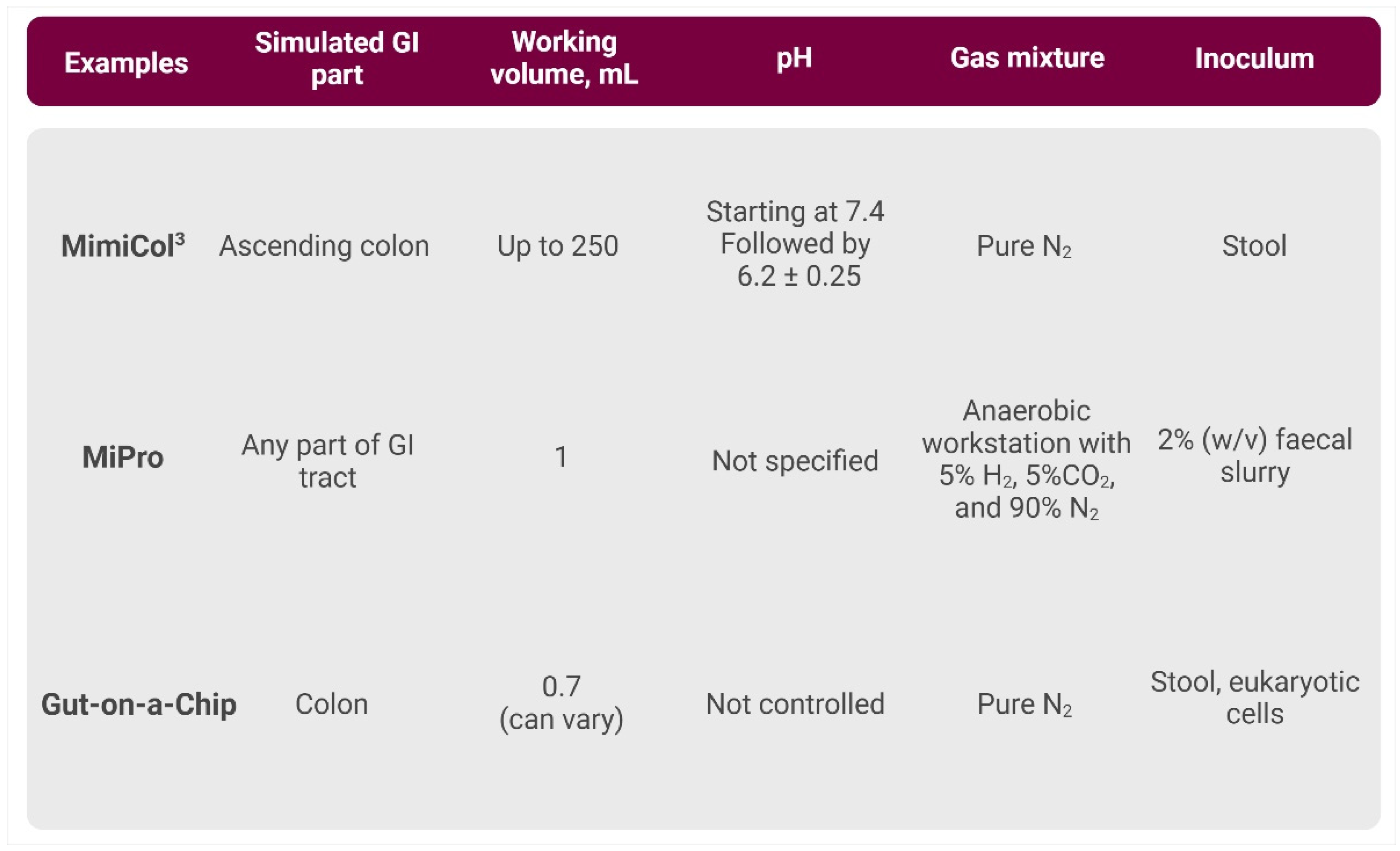
3.2.3. High-Throughput In Vitro Models
- MimiCol3
- 96-Well-Plate-Based Models
- Gut-on-a-Chip Models
3.3. Bioreactor-Based Approaches to Study Gastrointestinal Tract Cancer
3.4. Modelling the Side Effects of Cancer Treatment
Radiation-Induced Toxicity
- Chemotherapy and Antibiotics-Induced Dysbiosis
- Oralisation Process
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Eaton, S.E.; Kaczmarek, J.; Mahmood, D.; McDiarmid, A.M.; Norarfan, A.N.; Scott, E.G.; Then, C.K.; Tsui, H.Y.; Kiltie, A.E. Exploiting Dietary Fibre and the Gut Microbiota in Pelvic Radiotherapy Patients. Br. J. Cancer 2022, 127, 2087–2098. [Google Scholar] [CrossRef]
- Possenti, L.; Mecchi, L.; Rossoni, A.; Sangalli, V.; Bersini, S.; Cicchetti, A.; Costantino, M.L.; Candrian, C.; Arrigoni, C.; Rancati, T.; et al. Radiobiological Studies of Microvascular Damage through In Vitro Models: A Methodological Perspective. Cancers 2021, 13, 1182. [Google Scholar] [CrossRef] [PubMed]
- Bouges, E.; Segers, C.; Leys, N.; Lebeer, S.; Zhang, J.; Mastroleo, F. Human Intestinal Organoids and Microphysiological Systems for Modeling Radiotoxicity and Assessing Radioprotective Agents. Cancers 2023, 15, 5859. [Google Scholar] [CrossRef]
- Antfolk, M.; Jensen, K.B. A Bioengineering Perspective on Modelling the Intestinal Epithelial Physiology in Vitro. Nat. Commun. 2020, 11, 6244. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. The Intestinal Crypt, a Prototype Stem Cell Compartment. Cell 2013, 154, 274–284. [Google Scholar] [CrossRef]
- Clevers, H.; Conder, R.K.; Li, V.S.W.; Lutolf, M.P.; Vallier, L.; Chan, S.; Grikscheit, T.C.; Jensen, K.B.; De Coppi, P. Tissue-Engineering the Intestine: The Trials before the Trials. Cell Stem Cell 2019, 24, 855–859. [Google Scholar] [CrossRef]
- Min, S.; Kim, S.; Cho, S.-W. Gastrointestinal Tract Modeling Using Organoids Engineered with Cellular and Microbiota Niches. Exp. Mol. Med. 2020, 52, 227–237. [Google Scholar] [CrossRef]
- Creff, J.; Malaquin, L.; Besson, A. In Vitro Models of Intestinal Epithelium: Toward Bioengineered Systems. J. Tissue Eng. 2021, 12, 2041731420985202. [Google Scholar] [CrossRef] [PubMed]
- Snyder, J.; Wang, C.-M.; Zhang, A.Q.; Li, Y.; Luchan, J.; Hosic, S.; Koppes, R.; Carrier, R.L.; Koppes, A. Materials and Microenvironments for Engineering the Intestinal Epithelium. Ann. Biomed. Eng. 2020, 48, 1916–1940. [Google Scholar] [CrossRef]
- Huh, D.; Hamilton, G.A.; Ingber, D.E. From 3D Cell Culture to Organs-on-Chips. Trends Cell Biol. 2011, 21, 745–754. [Google Scholar] [CrossRef]
- Thorne, C.A.; Chen, I.W.; Sanman, L.E.; Cobb, M.H.; Wu, L.F.; Altschuler, S.J. Enteroid Monolayers Reveal an Autonomous WNT and BMP Circuit Controlling Intestinal Epithelial Growth and Organization. Dev. Cell 2018, 44, 624–633.e4. [Google Scholar] [CrossRef]
- Hirt, C.; Papadimitropoulos, A.; Muraro, M.G.; Mele, V.; Panopoulos, E.; Cremonesi, E.; Ivanek, R.; Schultz-Thater, E.; Droeser, R.A.; Mengus, C.; et al. Bioreactor-Engineered Cancer Tissue-like Structures Mimic Phenotypes, Gene Expression Profiles and Drug Resistance Patterns Observed “in Vivo”. Biomaterials 2015, 62, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning from 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- VanDussen, K.L.; Marinshaw, J.M.; Shaikh, N.; Miyoshi, H.; Moon, C.; Tarr, P.I.; Ciorba, M.A.; Stappenbeck, T.S. Development of an Enhanced Human Gastrointestinal Epithelial Culture System to Facilitate Patient-Based Assays. Gut 2015, 64, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Fogh, J.; Fogh, J.M.; Orfeo, T. One Hundred and Twenty-Seven Cultured Human Tumor Cell Lines Producing Tumors in Nude Mice. J. Natl. Cancer Inst. 1977, 59, 221–226. [Google Scholar] [CrossRef]
- Simon-Assmann, P.; Turck, N.; Sidhoum-Jenny, M.; Gradwohl, G.; Kedinger, M. In Vitro Models of Intestinal Epithelial Cell Differentiation. Cell Biol. Toxicol. 2007, 23, 241–256. [Google Scholar] [CrossRef]
- Costa, J.; Ahluwalia, A. Advances and Current Challenges in Intestinal in Vitro Model Engineering: A Digest. Front. Bioeng. Biotechnol. 2019, 7, 144. [Google Scholar] [CrossRef]
- Smetanová, L.; Štětinová, V.; Svoboda, Z.; Květina, J. Caco-2 Cells, Biopharmaceutics Classification System (BCS) and Biowaiter. Acta Medica 2011, 54, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Antunes, F.; Andrade, F.; Araújo, F.; Ferreira, D.; Sarmento, B. Establishment of a Triple Co-Culture in Vitro Cell Models to Study Intestinal Absorption of Peptide Drugs. Eur. J. Pharm. Biopharm. 2013, 83, 427–435. [Google Scholar] [CrossRef]
- Sambuy, Y.; De Angelis, I.; Ranaldi, G.; Scarino, M.L.; Stammati, A.; Zucco, F. The Caco-2 Cell Line as a Model of the Intestinal Barrier: Influence of Cell and Culture-Related Factors on Caco-2 Cell Functional Characteristics. Cell Biol. Toxicol. 2005, 21, 1–26. [Google Scholar] [CrossRef]
- Li, N.; Wang, D.; Sui, Z.; Qi, X.; Ji, L.; Wang, X.; Yang, L. Development of an Improved Three-Dimensional in Vitro Intestinal Mucosa Model for Drug Absorption Evaluation. Tissue Eng. Part C Methods 2013, 19, 708–719. [Google Scholar] [CrossRef] [PubMed]
- Lesuffleur, T.; Barbat, A.; Dussaulx, E.; Zweibaum, A. Growth Adaptation to Methotrexate of HT-29 Human Colon Carcinoma Cells Is Associated with Their Ability to Differentiate into Columnar Absorptive and Mucus-Secreting Cells. Cancer Res. 1990, 50, 6334–6343. [Google Scholar]
- Araújo, F.; Pereira, C.; Costa, J.; Barrias, C.; Granja, P.L.; Sarmento, B. In Vitro M-like Cells Genesis through a Tissue-Engineered Triple-Culture Intestinal Model. J. Biomed. Mater. Res. B Appl. Biomater. 2016, 104, 782–788. [Google Scholar] [CrossRef]
- Dharmsathaphorn, K.; McRoberts, J.A.; Mandel, K.G.; Tisdale, L.D.; Masui, H. A Human Colonic Tumor Cell Line That Maintains Vectorial Electrolyte Transport. Am. J. Physiol. 1984, 246, G204–G208. [Google Scholar] [CrossRef] [PubMed]
- McCool, D.J.; Marcon, M.A.; Forstner, J.F.; Forstner, G.G. The T84 Human Colonic Adenocarcinoma Cell Line Produces Mucin in Culture and Releases It in Response to Various Secretagogues. Biochem. J. 1990, 267, 491–500. [Google Scholar] [CrossRef] [PubMed]
- McKay, D.M.; Philpott, D.J.; Perdue, M.H. Review Article: In Vitro Models in Inflammatory Bowel Disease Research--a Critical Review. Aliment. Pharmacol. Ther. 1997, 11 (Suppl. S3), 70–80. [Google Scholar] [CrossRef]
- Ferrari, G.; Cusella-De Angelis, G.; Coletta, M.; Paolucci, E.; Stornaiuolo, A.; Cossu, G.; Mavilio, F. Muscle Regeneration by Bone Marrow-Derived Myogenic Progenitors. Science 1998, 279, 1528–1530. [Google Scholar] [CrossRef]
- Pittenger, M.F.; Mackay, A.M.; Beck, S.C.; Jaiswal, R.K.; Douglas, R.; Mosca, J.D.; Moorman, M.A.; Simonetti, D.W.; Craig, S.; Marshak, D.R. Multilineage Potential of Adult Human Mesenchymal Stem Cells. Science 1999, 284, 143–147. [Google Scholar] [CrossRef]
- Choi, R.S.; Vacanti, J.P. Preliminary Studies of Tissue-Engineered Intestine Using Isolated Epithelial Organoid Units on Tubular Synthetic Biodegradable Scaffolds. Transpl. Proc. 1997, 29, 848–851. [Google Scholar] [CrossRef]
- Gu, W.; Hong, X.; Le Bras, A.; Nowak, W.N.; Issa Bhaloo, S.; Deng, J.; Xie, Y.; Hu, Y.; Ruan, X.Z.; Xu, Q. Smooth Muscle Cells Differentiated from Mesenchymal Stem Cells Are Regulated by MicroRNAs and Suitable for Vascular Tissue Grafts. J. Biol. Chem. 2018, 293, 8089–8102. [Google Scholar] [CrossRef]
- Dalerba, P.; Dylla, S.J.; Park, I.-K.; Liu, R.; Wang, X.; Cho, R.W.; Hoey, T.; Gurney, A.; Huang, E.H.; Simeone, D.M.; et al. Phenotypic Characterization of Human Colorectal Cancer Stem Cells. Proc. Natl. Acad. Sci. USA 2007, 104, 10158–10163. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Costa, J.; Sarmento, B.; Araújo, F. 3.3—Cell-Based in Vitro Models for Intestinal Permeability Studies. In Concepts and Models for Drug Permeability Studies; Sarmento, B., Ed.; Woodhead Publishing: Sawston, UK, 2016; pp. 57–81. ISBN 978-0-08-100094-6. [Google Scholar]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 Stem Cells Build Crypt-Villus Structures in Vitro without a Mesenchymal Niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Ootani, A.; Li, X.; Sangiorgi, E.; Ho, Q.T.; Ueno, H.; Toda, S.; Sugihara, H.; Fujimoto, K.; Weissman, I.L.; Capecchi, M.R.; et al. Sustained in Vitro Intestinal Epithelial Culture within a Wnt-Dependent Stem Cell Niche. Nat. Med. 2009, 15, 701–706. [Google Scholar] [CrossRef]
- Preksha, G.; Yesheswini, R.; Srikanth, C.V. Cell Culture Techniques in Gastrointestinal Research: Methods, Possibilities and Challenges. Indian J. Pathol. Microbiol. 2021, 64, S52–S57. [Google Scholar] [CrossRef] [PubMed]
- Bartfeld, S.; Bayram, T.; van de Wetering, M.; Huch, M.; Begthel, H.; Kujala, P.; Vries, R.; Peters, P.J.; Clevers, H. In Vitro Expansion of Human Gastric Epithelial Stem Cells and Their Responses to Bacterial Infection. Gastroenterology 2015, 148, 126–136.e6. [Google Scholar] [CrossRef] [PubMed]
- Schlaermann, P.; Toelle, B.; Berger, H.; Schmidt, S.C.; Glanemann, M.; Ordemann, J.; Bartfeld, S.; Mollenkopf, H.J.; Meyer, T.F. A Novel Human Gastric Primary Cell Culture System for Modelling Helicobacter pylori Infection in Vitro. Gut 2016, 65, 202–213. [Google Scholar] [CrossRef]
- Wilson, S.S.; Tocchi, A.; Holly, M.K.; Parks, W.C.; Smith, J.G. A Small Intestinal Organoid Model of Non-Invasive Enteric Pathogen–Epithelial Cell Interactions. Mucosal Immunol. 2015, 8, 352–361. [Google Scholar] [CrossRef]
- Hibiya, S.; Tsuchiya, K.; Hayashi, R.; Fukushima, K.; Horita, N.; Watanabe, S.; Shirasaki, T.; Nishimura, R.; Kimura, N.; Nishimura, T.; et al. Long-Term Inflammation Transforms Intestinal Epithelial Cells of Colonic Organoids. J. Crohns Colitis 2017, 11, 621–630. [Google Scholar] [CrossRef]
- Peuker, K.; Muff, S.; Wang, J.; Künzel, S.; Bosse, E.; Zeissig, Y.; Luzzi, G.; Basic, M.; Strigli, A.; Ulbricht, A.; et al. Epithelial Calcineurin Controls Microbiota-Dependent Intestinal Tumor Development. Nat. Med. 2016, 22, 506–515. [Google Scholar] [CrossRef]
- Perrone, F.; Zilbauer, M. Biobanking of Human Gut Organoids for Translational Research. Exp. Mol. Med. 2021, 53, 1451–1458. [Google Scholar] [CrossRef]
- Zhou, Z.; Cong, L.; Cong, X. Patient-Derived Organoids in Precision Medicine: Drug Screening, Organoid-on-a-Chip and Living Organoid Biobank. Front. Oncol. 2021, 11, 762184. [Google Scholar] [CrossRef]
- Leonard, F.; Collnot, E.-M.; Lehr, C.-M. A Three-Dimensional Coculture of Enterocytes, Monocytes and Dendritic Cells to Model Inflamed Intestinal Mucosa in Vitro. Mol. Pharm. 2010, 7, 2103–2119. [Google Scholar] [CrossRef]
- Ehrbar, M.; Sala, A.; Lienemann, P.; Ranga, A.; Mosiewicz, K.; Bittermann, A.; Rizzi, S.C.; Weber, F.E.; Lutolf, M.P. Elucidating the Role of Matrix Stiffness in 3D Cell Migration and Remodeling. Biophys. J. 2011, 100, 284–293. [Google Scholar] [CrossRef]
- Ayvaz, I.; Sunay, D.; Sariyar, E.; Erdal, E.; Karagonlar, Z.F. Three-Dimensional Cell Culture Models of Hepatocellular Carcinoma—A Review. J. Gastrointest. Cancer 2021, 52, 1294–1308. [Google Scholar] [CrossRef] [PubMed]
- Dosh, R.H.; Essa, A.; Jordan-Mahy, N.; Sammon, C.; Le Maitre, C.L. Use of Hydrogel Scaffolds to Develop an in Vitro 3D Culture Model of Human Intestinal Epithelium. Acta Biomater. 2017, 62, 128–143. [Google Scholar] [CrossRef] [PubMed]
- Franck, D.; Chung, Y.G.; Coburn, J.; Kaplan, D.L.; Estrada, C.R.; Mauney, J.R. In Vitro Evaluation of Bi-Layer Silk Fibroin Scaffolds for Gastrointestinal Tissue Engineering. J. Tissue Eng. 2014, 5, 2041731414556849. [Google Scholar] [CrossRef]
- Zakhem, E.; Raghavan, S.; Suhar, R.A.; Bitar, K.N. Bioengineering and Regeneration of Gastrointestinal Tissue: Where Are We Now and What Comes Next? Expert Opin. Biol. Ther. 2019, 19, 527–537. [Google Scholar] [CrossRef]
- Biju, T.S.; Priya, V.V.; Francis, A.P. Role of Three-Dimensional Cell Culture in Therapeutics and Diagnostics: An Updated Review. Drug Deliv. Transl. Res. 2023, 13, 2239–2253. [Google Scholar] [CrossRef] [PubMed]
- Dynamic Tissue Engineering Scaffolds with Stimuli-Responsive Macroporosity Formation—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/23489920/ (accessed on 19 June 2024).
- Li, Y.; Kilian, K.A. Bridging the Gap: From 2D Cell Culture to 3D Microengineered Extracellular Matrices. Adv. Healthc. Mater. 2015, 4, 2780–2796. [Google Scholar] [CrossRef]
- Ağagündüz, D.; Cocozza, E.; Cemali, Ö.; Bayazıt, A.D.; Nanì, M.F.; Cerqua, I.; Morgillo, F.; Saygılı, S.K.; Berni Canani, R.; Amero, P.; et al. Understanding the Role of the Gut Microbiome in Gastrointestinal Cancer: A Review. Front. Pharmacol. 2023, 14, 1130562. [Google Scholar] [CrossRef]
- Wroblewski, L.E.; Peek, R.M.; Coburn, L.A. The Role of the Microbiome in GI Cancer. Gastroenterol. Clin. N. Am. 2016, 45, 543–556. [Google Scholar] [CrossRef] [PubMed]
- Nienow, A.W. Stirring and Stirred-Tank Reactors. Chem. Ing. Tech. 2014, 86, 2063–2074. [Google Scholar] [CrossRef]
- Comprehensive Biotechnology. Available online: https://www.sciencedirect.com/referencework/9780080885049/comprehensive-biotechnology (accessed on 27 May 2024).
- Wu, W.-K.; Chen, C.-C.; Panyod, S.; Chen, R.-A.; Wu, M.-S.; Sheen, L.-Y.; Chang, S.-C. Optimization of Fecal Sample Processing for Microbiome Study—The Journey from Bathroom to Bench. J. Formos. Med. Assoc. 2019, 118, 545–555. [Google Scholar] [CrossRef]
- Sardelli, L.; Perottoni, S.; Tunesi, M.; Boeri, L.; Fusco, F.; Petrini, P.; Albani, D.; Giordano, C. Technological tools and strategies for culturing human gut microbiota in engineered in vitro models. Biotechnol. Bioeng. 2021, 118, 2886–2905. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Zheng, X.; Hou, W.; Zhuang, Y.; Pi, X.; Yang, J. The Study of a Remote-Controlled Gastrointestinal Drug Delivery and Sampling System. Telemed. J. E Health 2008, 14, 715–719. [Google Scholar] [CrossRef] [PubMed]
- El Houari, A.; Ecale, F.; Mercier, A.; Crapart, S.; Laparre, J.; Soulard, B.; Ramnath, M.; Berjeaud, J.-M.; Rodier, M.-H.; Crépin, A. Development of an in Vitro Model of Human Gut Microbiota for Screening the Reciprocal Interactions with Antibiotics, Drugs, and Xenobiotics. Front. Microbiol. 2022, 13, 828359. [Google Scholar] [CrossRef]
- Takagi, R.; Sasaki, K.; Sasaki, D.; Fukuda, I.; Tanaka, K.; Yoshida, K.-I.; Kondo, A.; Osawa, R. A Single-Batch Fermentation System to Simulate Human Colonic Microbiota for High-Throughput Evaluation of Prebiotics. PLoS ONE 2016, 11, e0160533. [Google Scholar] [CrossRef]
- Wang, X.; Gibson, G.R. Effects of the in Vitro Fermentation of Oligofructose and Inulin by Bacteria Growing in the Human Large Intestine. J. Appl. Bacteriol. 1993, 75, 373–380. [Google Scholar] [CrossRef]
- Dai, J.; Yoon, S.H.; Sim, H.Y.; Yang, Y.S.; Oh, T.K.; Kim, J.F.; Hong, J.W. Charting Microbial Phenotypes in Multiplex Nanoliter Batch Bioreactors. Anal. Chem. 2013, 85, 5892–5899. [Google Scholar] [CrossRef]
- Roupar, D.; Berni, P.; Martins, J.T.; Caetano, A.C.; Teixeira, J.A.; Nobre, C. Bioengineering Approaches to Simulate Human Colon Microbiome Ecosystem. Trends Food Sci. Technol. 2021, 112, 808–822. [Google Scholar] [CrossRef]
- Hegner, R.; Koch, C.; Riechert, V.; Harnisch, F. Microbiome-Based Carboxylic Acids Production: From Serum Bottles to Bioreactors. RSC Adv. 2017, 7, 15362–15371. [Google Scholar] [CrossRef]
- Carcano, S. A Model for Cell Growth in Batch Bioreactors; Politecnico di Milano: Milan, Italy, 2010. [Google Scholar]
- Feria-Gervasio, D.; Denis, S.; Alric, M.; Brugère, J.-F. In Vitro Maintenance of a Human Proximal Colon Microbiota Using the Continuous Fermentation System P-ECSIM. Appl. Microbiol. Biotechnol. 2011, 91, 1425–1433. [Google Scholar] [CrossRef] [PubMed]
- Brugère, J.-F.; Féria-Gervasio, D.; Popse, Z.; Tottey, W.; Alric, M. The ECSIM Concept (Environmental Control System for Intestinal Microbiota) and Its Derivative Versions to Help Better Understand Human Gut Biology. Appl. Biomed. Eng. 2011, 4, 63–82. [Google Scholar]
- Beeck, R.; Dols, A.; Schneider, F.; Seradj, D.-S.; Krause, J.; Schick, P.; Weitschies, W. An Advanced Bioreactor Simulating Dynamic Physiological Conditions in the Human Ascending Colon: MimiCol3. Pharmaceutics 2022, 14, 1049. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, S.; Dillon, J.F. Microbial Biofilms in the Human Gastrointestinal Tract. J. Appl. Microbiol. 2007, 102, 1187–1196. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.L.; Wolin, M.J. Fermentation by the Human Large Intestine Microbial Community in an in Vitro Semicontinuous Culture System. Appl. Env. Microbiol. 1981, 42, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Verhoeckx, K.; Cotter, P.; López-Expósito, I.; Kleiveland, C.; Lea, T.; Mackie, A.; Requena, T.; Swiatecka, D.; Wichers, H. (Eds.) The Impact of Food Bioactives on Health; Springer International Publishing: Cham, Switzerland, 2015; ISBN 978-3-319-15791-7. [Google Scholar]
- Gibson, G.R.; Cummings, J.H.; Macfarlane, G.T. Use of a Three-Stage Continuous Culture System to Study the Effect of Mucin on Dissimilatory Sulfate Reduction and Methanogenesis by Mixed Populations of Human Gut Bacteria. Appl. Env. Microbiol. 1988, 54, 2750–2755. [Google Scholar] [CrossRef]
- Molly, K.; Vande Woestyne, M.; Verstraete, W. Development of a 5-Step Multi-Chamber Reactor as a Simulation of the Human Intestinal Microbial Ecosystem. Appl. Microbiol. Biotechnol. 1993, 39, 254–258. [Google Scholar] [CrossRef]
- Van de Wiele, T.; Van den Abbeele, P.; Ossieur, W.; Possemiers, S.; Marzorati, M. The Simulator of the human intestinal microbial ecosystem (SHIME®). In The Impact of Food Bioactives on Health; Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 305–317. ISBN 978-3-319-15791-7. [Google Scholar]
- Yoo, M.J.; Chen, X.D. GIT Physicochemical Modeling—A Critical Review. Int. J. Food Eng. 2006, 2. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Belzer, C.; Goossens, M.; Kleerebezem, M.; De Vos, W.M.; Thas, O.; De Weirdt, R.; Kerckhof, F.-M.; Van de Wiele, T. Butyrate-Producing Clostridium Cluster XIVa Species Specifically Colonize Mucins in an in Vitro Gut Model. ISME J. 2013, 7, 949–961. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Grootaert, C.; Marzorati, M.; Possemiers, S.; Verstraete, W.; Gérard, P.; Rabot, S.; Bruneau, A.; El Aidy, S.; Derrien, M.; et al. Microbial Community Development in a Dynamic Gut Model Is Reproducible, Colon Region Specific, and Selective for Bacteroidetes and Clostridium Cluster IX. Appl. Env. Microbiol. 2010, 76, 5237–5246. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Firrman, J.; Tanes, C.; Bittinger, K.; Thomas-Gahring, A.; Wu, G.D.; Van den Abbeele, P.; Tomasula, P.M. Establishing a Mucosal Gut Microbial Community in Vitro Using an Artificial Simulator. PLoS ONE 2018, 13, e0197692. [Google Scholar] [CrossRef]
- De Boever, P.; Wouters, R.; Vermeirssen, V.; Boon, N.; Verstraete, W. Development of a Six-Stage Culture System for Simulating the Gastrointestinal Microbiota of Weaned Infants. Microb. Ecol. Health Dis. 2001, 13, 111–123. [Google Scholar] [CrossRef]
- Minekus, M.; Smeets-Peeters, M.; Bernalier, A.; Marol-Bonnin, S.; Havenaar, R.; Marteau, P.; Alric, M.; Fonty, G.; Huis in’t Veld, J.H. A Computer-Controlled System to Simulate Conditions of the Large Intestine with Peristaltic Mixing, Water Absorption and Absorption of Fermentation Products. Appl. Microbiol. Biotechnol. 1999, 53, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Minekus, M.; Marteau, P.; Havenaar, R.; Veld, J.H.J.H. in’t A Multicompartmental Dynamic Computer-Controlled Model Simulating the Stomach and Small Intestine. Altern. Lab. Anim. 1995, 23, 197–209. [Google Scholar] [CrossRef]
- Minekus, M. The TNO Gastro-Intestinal Model (TIM). In The Impact of Food Bioactives on Health; Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 37–46. ISBN 978-3-319-15791-7. [Google Scholar]
- Venema, K.; van den Abbeele, P. Experimental Models of the Gut Microbiome. Best. Pract. Res. Clin. Gastroenterol. 2013, 27, 115–126. [Google Scholar] [CrossRef]
- Barroso, E.; Cueva, C.; Peláez, C.; Martínez-Cuesta, M.C.; Requena, T. Development of Human Colonic Microbiota in the Computer-Controlled Dynamic SIMulator of the GastroIntestinal Tract SIMGI. LWT Food Sci. Technol. 2015, 61, 283–289. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Macfarlane, S.; Gibson, G. Validation of a Three-Stage Compound Continuous Culture System for Investigating the Effect of Retention Time on the Ecology and Metabolism of Bacteria in the Human Colon. Microb. Ecol. 1998, 35, 180–187. [Google Scholar] [CrossRef]
- Ferreira-Lazarte, A.; Moreno, F.J.; Cueva, C.; Gil-Sánchez, I.; Villamiel, M. Behaviour of Citrus Pectin during Its Gastrointestinal Digestion and Fermentation in a Dynamic Simulator (Simgi®). Carbohydr. Polym. 2019, 207, 382–390. [Google Scholar] [CrossRef]
- Feria-Gervasio, D.; Tottey, W.; Gaci, N.; Alric, M.; Cardot, J.-M.; Peyret, P.; Martin, J.-F.; Pujos, E.; Sébédio, J.-L.; Brugère, J.-F. Three-Stage Continuous Culture System with a Self-Generated Anaerobia to Study the Regionalized Metabolism of the Human Gut Microbiota. J. Microbiol. Methods 2014, 96, 111–118. [Google Scholar] [CrossRef]
- Marzorati, M.; Vanhoecke, B.; De Ryck, T.; Sadaghian Sadabad, M.; Pinheiro, I.; Possemiers, S.; Van den Abbeele, P.; Derycke, L.; Bracke, M.; Pieters, J.; et al. The HMITM Module: A New Tool to Study the Host-Microbiota Interaction in the Human Gastrointestinal Tract in Vitro. BMC Microbiol. 2014, 14, 133. [Google Scholar] [CrossRef] [PubMed]
- Gościniak, A.; Eder, P.; Walkowiak, J.; Cielecka-Piontek, J. Artificial Gastrointestinal Models for Nutraceuticals Research—Achievements and Challenges: A Practical Review. Nutrients 2022, 14, 2560. [Google Scholar] [CrossRef]
- Zihler Berner, A.; Fuentes, S.; Dostal, A.; Payne, A.N.; Vazquez Gutierrez, P.; Chassard, C.; Grattepanche, F.; de Vos, W.M.; Lacroix, C. Novel Polyfermentor Intestinal Model (PolyFermS) for Controlled Ecological Studies: Validation and Effect of PH. PLoS ONE 2013, 8, e77772. [Google Scholar] [CrossRef]
- Beeck, R.; Glöckl, G.; Krause, J.; Schick, P.; Weitschies, W. Mimicking the Dynamic Colonic Microbiota in Vitro to Gain a Better Understanding on the in Vivo Metabolism of Xenobiotics: Degradation of Sulfasalazine. Int. J. Pharm. 2021, 603, 120704. [Google Scholar] [CrossRef] [PubMed]
- Seradj, D.-S.; Beeck, R.; Haase, A.; Krause, J.; Schick, P.; Weitschies, W. Influence of Different Diets on the Degradation of Sulfasalazine by Colon Bacteria Determined Using MimiCol3. Pharmaceuticals 2023, 16, 1128. [Google Scholar] [CrossRef]
- Parrish, J.; Lim, K.S.; Baer, K.; Hooper, G.J.; Woodfield, T.B.F. A 96-Well Microplate Bioreactor Platform Supporting Individual Dual Perfusion and High-Throughput Assessment of Simple or Biofabricated 3D Tissue Models. Lab. Chip 2018, 18, 2757–2775. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Abou-Samra, E.; Ning, Z.; Zhang, X.; Mayne, J.; Wang, J.; Cheng, K.; Walker, K.; Stintzi, A.; Figeys, D. An in Vitro Model Maintaining Taxon-Specific Functional Activities of the Gut Microbiome. Nat. Commun. 2019, 10, 4146. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.N.; Ingber, D.E. Microfluidic Organs-on-Chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef]
- Goyal, G.; Belgur, C.; Ingber, D.E. Human Organ Chips for Regenerative Pharmacology. Pharmacol. Res. Perspect. 2024, 12, e01159. [Google Scholar] [CrossRef]
- Esch, M.B.; Ueno, H.; Applegate, D.R.; Shuler, M.L. Modular, Pumpless Body-on-a-Chip Platform for the Co-Culture of GI Tract Epithelium and 3D Primary Liver Tissue. Lab. Chip 2016, 16, 2719–2729. [Google Scholar] [CrossRef]
- Chen, H.J.; Miller, P.; Shuler, M.L. A Pumpless Body-on-a-Chip Model Using a Primary Culture of Human Intestinal Cells and a 3D Culture of Liver Cells. Lab Chip 2018, 18, 2036–2046. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.; Kim, H.J. 3D in Vitro Morphogenesis of Human Intestinal Epithelium in a Gut-on-a-Chip or a Hybrid Chip with a Cell Culture Insert. Nat. Protoc. 2022, 17, 910–939. [Google Scholar] [CrossRef]
- Shah, P.; Fritz, J.V.; Glaab, E.; Desai, M.S.; Greenhalgh, K.; Frachet, A.; Niegowska, M.; Estes, M.; Jäger, C.; Seguin-Devaux, C.; et al. A Microfluidics-Based in Vitro Model of the Gastrointestinal Human-Microbe Interface. Nat. Commun. 2016, 7, 11535. [Google Scholar] [CrossRef] [PubMed]
- Valiei, A.; Aminian-Dehkordi, J.; Mofrad, M.R.K. Gut-on-a-Chip Models for Dissecting the Gut Microbiology and Physiology. APL Bioeng. 2023, 7, 011502. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Liu, T.; Liu, Q.; Lian, L.; Tang, G.; Mille, L.S.; García, F.R.; Engstrand, L.; Zhang, Y.S.; Du, J. A 3D Bioprinted Gut Anaerobic Model for Studying Bacteria–Host Interactions. Research 2023, 6, 0058. [Google Scholar] [CrossRef]
- Benam, K.H.; Dauth, S.; Hassell, B.; Herland, A.; Jain, A.; Jang, K.-J.; Karalis, K.; Kim, H.J.; MacQueen, L.; Mahmoodian, R.; et al. Engineered in Vitro Disease Models. Annu. Rev. Pathol. 2015, 10, 195–262. [Google Scholar] [CrossRef] [PubMed]
- Manfredonia, C.; Muraro, M.G.; Hirt, C.; Mele, V.; Governa, V.; Papadimitropoulos, A.; Däster, S.; Soysal, S.D.; Droeser, R.A.; Mechera, R.; et al. Maintenance of Primary Human Colorectal Cancer Microenvironment Using a Perfusion Bioreactor-Based 3D Culture System. Adv. Biosyst. 2019, 3, e1800300. [Google Scholar] [CrossRef] [PubMed]
- La Rocca, A.; De Gregorio, V.; Lagreca, E.; Vecchione, R.; Netti, P.A.; Imparato, G. Colorectal Cancer Bioengineered Microtissues as a Model to Replicate Tumor-ECM Crosstalk and Assess Drug Delivery Systems In Vitro. Int. J. Mol. Sci. 2023, 24, 5678. [Google Scholar] [CrossRef]
- Gouws, C.; Smit, T.; Willers, C.; Svitina, H.; Calitz, C.; Wrzesinski, K. Anticancer Potential of Sutherlandia frutescens and Xysmalobium undulatum in LS180 Colorectal Cancer Mini-Tumors. Molecules 2021, 26, 605. [Google Scholar] [CrossRef]
- Chen, Z.-Y.; Jiang, N.; Guo, S.; Li, B.-B.; Yang, J.-Q.; Chai, S.-B.; Yan, H.-F.; Sun, P.-M.; Zhang, T.; Sun, H.-W.; et al. Effect of Simulated Microgravity on Metabolism of HGC-27 Gastric Cancer Cells. Oncol. Lett. 2020, 19, 3439–3450. [Google Scholar] [CrossRef]
- Rembiałkowska, N.; Baczyńska, D.; Dubińska-Magiera, M.; Choromańska, A.; Bieżuńska-Kusiak, K.; Gajewska-Naryniecka, A.; Novickij, V.; Saczko, J.; Przystupski, D.; Kulbacka, J. RCCS Bioreactor-Based Modeled Microgravity Affects Gastric Cancer Cells and Improves the Chemotherapeutic Effect. Membranes 2022, 12, 448. [Google Scholar] [CrossRef] [PubMed]
- Roeder, F.; Meldolesi, E.; Gerum, S.; Valentini, V.; Rödel, C. Recent Advances in (Chemo-)Radiation Therapy for Rectal Cancer: A Comprehensive Review. Radiat. Oncol. 2020, 15, 262. [Google Scholar] [CrossRef] [PubMed]
- Hauer-Jensen, M.; Denham, J.W.; Andreyev, H.J.N. Radiation Enteropathy—Pathogenesis, Treatment, and Prevention. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 470–479. [Google Scholar] [CrossRef] [PubMed]
- Ichim, T.E.; Kesari, S.; Shafer, K. Protection from Chemotherapy- and Antibiotic-Mediated Dysbiosis of the Gut Microbiota by a Probiotic with Digestive Enzymes Supplement. Oncotarget 2018, 9, 30919–30935. [Google Scholar] [CrossRef] [PubMed]
- Verdier, C.; Denis, S.; Gasc, C.; Boucinha, L.; Uriot, O.; Delmas, D.; Dore, J.; Le Camus, C.; Schwintner, C.; Blanquet-Diot, S. An Oral FMT Capsule as Efficient as an Enema for Microbiota Reconstruction Following Disruption by Antibiotics, as Assessed in an In Vitro Human Gut Model. Microorganisms 2021, 9, 358. [Google Scholar] [CrossRef]
- Holma, R.; Korpela, R.; Sairanen, U.; Blom, M.; Rautio, M.; Poussa, T.; Saxelin, M.; Osterlund, P. Colonic Methane Production Modifies Gastrointestinal Toxicity Associated with Adjuvant 5-Fluorouracil Chemotherapy for Colorectal Cancer. J. Clin. Gastroenterol. 2013, 47, 45–51. [Google Scholar] [CrossRef]
- Blaustein, R.A.; Seed, P.C.; Hartmann, E.M. Biotransformation of Doxorubicin Promotes Resilience in Simplified Intestinal Microbial Communities. mSphere 2021, 6, e0006821. [Google Scholar] [CrossRef]
- Horvath, A.; Bausys, A.; Sabaliauskaite, R.; Stratilatovas, E.; Jarmalaite, S.; Schuetz, B.; Stiegler, P.; Bausys, R.; Stadlbauer, V.; Strupas, K. Distal Gastrectomy with Billroth II Reconstruction Is Associated with Oralization of Gut Microbiome and Intestinal Inflammation: A Proof-of-Concept Study. Ann. Surg. Oncol. 2021, 28, 1198–1208. [Google Scholar] [CrossRef]
- Horvath, A.; Rainer, F.; Bashir, M.; Leber, B.; Schmerboeck, B.; Klymiuk, I.; Groselj-Strele, A.; Durdevic, M.; Freedberg, D.E.; Abrams, J.A.; et al. Biomarkers for Oralization during Long-Term Proton Pump Inhibitor Therapy Predict Survival in Cirrhosis. Sci. Rep. 2019, 9, 12000. [Google Scholar] [CrossRef]
- Etienne-Mesmin, L.; Meslier, V.; Uriot, O.; Fournier, E.; Deschamps, C.; Denis, S.; David, A.; Jegou, S.; Morabito, C.; Quinquis, B.; et al. In Vitro Modelling of Oral Microbial Invasion in the Human Colon. Microbiol. Spectr. 2023, 11, e0434422. [Google Scholar] [CrossRef]
- Ravichandran, A.; Liu, Y.; Teoh, S.-H. Review: Bioreactor Design towards Generation of Relevant Engineered Tissues: Focus on Clinical Translation. J. Tissue Eng. Regen. Med. 2018, 12, e7–e22. [Google Scholar] [CrossRef]
- Yang, C.; Kong, L.; Zhang, Z. Bioreactor: Intelligent Platform for Drug Delivery. Nano Today 2022, 44, 101481. [Google Scholar] [CrossRef]
- Bisgin, A.; Mujde, C. Bioreactor-Based Tissue Models as an Alternative Approach in Cancer Research. In Handbook of Animal Models and its Uses in Cancer Research; Pathak, S., Banerjee, A., Bisgin, A., Eds.; Springer Nature: Singapore, 2022; pp. 1–16. ISBN 978-981-19128-2-5. [Google Scholar]
- McCarthy, M.; Brown, T.; Alarcon, A.; Williams, C.; Wu, X.; Abbott, R.D.; Gimble, J.; Frazier, T. Fat-On-A-Chip Models for Research and Discovery in Obesity and Its Metabolic Comorbidities. Tissue Eng. Part B Rev. 2020, 26, 586–595. [Google Scholar] [CrossRef]
- De Boever, P.; Deplancke, B.; Verstraete, W. Fermentation by Gut Microbiota Cultured in a Simulator of the Human Intestinal Microbial Ecosystem Is Improved by Supplementing a Soygerm Powder. J. Nutr. 2000, 130, 2599–2606. [Google Scholar] [CrossRef] [PubMed]
- Possemiers, S.; Verthé, K.; Uyttendaele, S.; Verstraete, W. PCR-DGGE-Based Quantification of Stability of the Microbial Community in a Simulator of the Human Intestinal Microbial Ecosystem. FEMS Microbiol. Ecol. 2004, 49, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Van den Abbeele, P.; Roos, S.; Eeckhaut, V.; MacKenzie, D.A.; Derde, M.; Verstraete, W.; Marzorati, M.; Possemiers, S.; Vanhoecke, B.; Van Immerseel, F.; et al. Incorporating a Mucosal Environment in a Dynamic Gut Model Results in a More Representative Colonization by Lactobacilli. Microb. Biotechnol. 2012, 5, 106–115. [Google Scholar] [CrossRef]
- Schönberger, M.; Hoffstetter, M. Emerging Trends in Medical Plastic Engineering and Manufacturing; Elsevier Inc.: Amsterdam, The Netherlands, 2016; ISBN 978-0-323-37023-3. [Google Scholar] [CrossRef]
- Ashammakhi, N.; Nasiri, R.; de Barros, N.R.; Tebon, P.; Thakor, J.; Goudie, M.; Shamloo, A.; Martin, M.G.; Khademhosseini, A. Gut-on-a-Chip: Current Progress and Future Opportunities. Biomaterials 2020, 255, 120196. [Google Scholar] [CrossRef]
- Marrero, D.; Pujol-Vila, F.; Vera, D.; Gabriel, G.; Illa, X.; Elizalde-Torrent, A.; Alvarez, M.; Villa, R. Gut-on-a-Chip: Mimicking and Monitoring the Human Intestine. Biosens. Bioelectron. 2021, 181, 113156. [Google Scholar] [CrossRef]
- Baquero, F.; Nombela, C. The Microbiome as a Human Organ. Clin. Microbiol. Infect. 2012, 18, 2–4. [Google Scholar] [CrossRef]
- Chen, L.; Wang, D.; Garmaeva, S.; Kurilshikov, A.; Vich Vila, A.; Gacesa, R.; Sinha, T.; Lifelines Cohort Study; Segal, E.; Weersma, R.K.; et al. The Long-Term Genetic Stability and Individual Specificity of the Human Gut Microbiome. Cell 2021, 184, 2302–2315.e12. [Google Scholar] [CrossRef]
- Johnson, K.V.-A. Gut Microbiome Composition and Diversity Are Related to Human Personality Traits. Hum. Microb. J. 2020, 15, 100069. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, N.; Kitano, S.; Puah, G.R.Y.; Kittelmann, S.; Hwang, I.Y.; Chang, M.W. Microbiome and Human Health: Current Understanding, Engineering, and Enabling Technologies. Chem. Rev. 2022, 123, 31–72. [Google Scholar] [CrossRef]
- Katsoulakis, E.; Wang, Q.; Wu, H.; Shahriyari, L.; Fletcher, R.; Liu, J.; Achenie, L.; Liu, H.; Jackson, P.; Xiao, Y.; et al. Digital Twins for Health: A Scoping Review. NPJ Digit. Med. 2024, 7, 77. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Matheyambath, A.C.; Polic, I.I.; LaPointe, G. Differential fermentation of raw and processed high-amylose and waxy maize starches in the Simulator of the Human Intestinal Microbial Ecosystem (SHIME®). J. Funct. Foods 2021, 86, 104735. [Google Scholar] [CrossRef]
- Roussel, C.; De Paepe, K.; Galia, W.; De Bodt, J.; Chalancon, S.; Leriche, F.; Ballet, N.; Denis, S.; Alric, M.; Van de Wiele, T.; et al. Spatial and temporal modulation of enterotoxigenic E. coli H10407 pathogenesis and interplay with microbiota in human gut models. BMC Biol. 2020, 18, 141. [Google Scholar] [CrossRef] [PubMed]
- Vanlancker, E.; Vanhoecke, B.; Stringer, A.; Van de Wiele, T. 5-Fluorouracil and irinotecan (SN-38) have limited impact on colon microbial functionality and composition in vitro. PeerJ. 2017, 5, e4017. [Google Scholar] [CrossRef]
- Arroyo, M.C.; Laurie, I.; Rotsaert, C.; Marzorati, M.; Risso, D.; Karnik, K. Age-Dependent Prebiotic Effects of Soluble Corn Fiber in M-SHIME® Gut Microbial Ecosystems. Plant Foods Hum. Nutr. 2023, 78, 213–220. [Google Scholar] [CrossRef]
- Nissen, L.; Casciano, F.; Gianotti, A. Intestinal fermentation in vitro models to study food-induced gut microbiota shift: An updated review. FEMS Microbiol. Lett. 2020, 367, fnaa097. [Google Scholar] [CrossRef]
- Hatanaka, M.; Nakamura, Y.; Maathuis, A.; Venema, K.; Murota, I.; Yamamoto, N. Influence of Bacillus subtilis C-3102 on microbiota in a dynamic in vitro model of the gastrointestinal tract simulating human conditions. Benef. Microbes 2012, 3, 229–236. [Google Scholar] [CrossRef]
- Dickinson, P.A.; Abu Rmaileh, R.; Ashworth, L.; Barker, R.A.; Burke, W.M.; Patterson, C.M.; Stainforth, N.; Yasin, M. An Investigation into the Utility of a Multi-compartmental, Dynamic, System of the Upper Gastrointestinal Tract to Support Formulation Development and Establish Bioequivalence of Poorly Soluble Drugs. AAPS J. 2012, 14, 196–205. [Google Scholar] [CrossRef]
- Venema, K.; Verhoeven, J.; Verbruggen, S.; Espinosa, L.; Courau, S. Probiotic survival during a multi-layered tablet development as tested in a dynamic, computer-controlled in vitro model of the stomach and small intestine (TIM-1). Lett. Appl. Microbiol. 2019, 69, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Maas, E.; Penders, J.; Venema, K. Studying Fungal-Bacterial Relationships in the Human Gut Using an In Vitro Model (TIM-2). J. Fungi 2023, 9, 174. [Google Scholar] [CrossRef]
- Bordonaro, M.; Venema, K.; Putri, A.K.; Lazarova, D. Approaches that ascertain the role of dietary compounds in colonic cancer cells. World J. Gastrointest. Oncol. 2014, 15, 1–10. [Google Scholar] [CrossRef]
- Gao, K.; Xu, A.; Krul, C.; Venema, K.; Liu, Y.; Niu, Y.; Lu, J.; Bensoussan, L.; Seeram, N.P.; Heber, D.; et al. Of the Major Phenolic Acids Formed during Human Microbial Fermentation of Tea, Citrus, and Soy Flavonoid Supplements, Only 3,4-Dihydroxyphenylacetic Acid Has Antiproliferative Activity. J. Nutr. 2006, 136, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Wang, M.; Venema, K.; Maathuis, A.; van der Heijden, R.; van der Greef, J.; Xu, G.; Hankemeier, T. Bioconversion of red ginseng saponins in the gastro-intestinal tract in vitro model studied by high-performance liquid chromatography–high resolution Fourier transform ion cyclotron resonance mass spectrometry. J. Chromatogr. A 2009, 1216, 2195–2203. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Heinsen, F.-A.; Koenen, M.E.; Venema, K.; Knecht, H.; Hellmig, S.; Schreiber, S.; Ott, S.J. Effects of probiotics and antibiotics on the intestinal homeostasis in a computer controlled model of the large intestine. BMC Microbiol. 2012, 12, 47. [Google Scholar] [CrossRef]
- Gil-Sánchez, I.; Cueva, C.; Tamargo, A.; Quintela, J.C.; de la Fuente, E.; Walker, A.W.; Moreno-Arribas, M.V.; Bartolomé, B. Application of the dynamic gastrointestinal simulator (simgi®) to assess the impact of probiotic supplementation in the metabolism of grape polyphenols. Food Res. Int. 2019, 129, 108790. [Google Scholar] [CrossRef]
- von Martels, J.Z.; Sadabad, M.S.; Bourgonje, A.R.; Blokzijl, T.; Dijkstra, G.; Faber, K.N.; Harmsen, H.J. The role of gut microbiota in health and disease: In vitro modeling of host-microbe interactions at the aerobe-anaerobe interphase of the human gut. Anaerobe 2017, 44, 3–12. [Google Scholar] [CrossRef]
- Makivuokko, H.; Nurmi, J.; Nurminen, P.; Stowell, J.; Rautonen, N. In Vitro Effects on Polydextrose by Colonic Bacteria and Caco-2 Cell Cyclooxygenase Gene Expression. Nutr. Cancer 2005, 52, 94–104. [Google Scholar] [CrossRef]
- Williams, C.; Walton, G.; Jiang, L.; Plummer, S.; Garaiova, I.; Gibson, G. Comparative Analysis of Intestinal Tract Models. Annu. Rev. Food Sci. Technol. 2015, 6, 329–350. [Google Scholar] [CrossRef]
- Bein, A.; Shin, W.; Jalili-Firoozinezhad, S.; Park, M.H.; Sontheimer-Phelps, A.; Tovaglieri, A.; Chalkiadaki, A.; Kim, H.J.; Ingber, D.E. Microfluidic Organ-on-a-Chip Models of Human Intestine. Cell. Mol. Gastroenterol. Hepatol. 2018, 5, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Sontheimer-Phelps, A.; Chou, D.B.; Tovaglieri, A.; Ferrante, T.C.; Duckworth, T.; Fadel, C.; Frismantas, V.; Sutherland, A.D.; Jalili-Firoozinezhad, S.; Kasendra, M.; et al. Human Colon-on-a-Chip Enables Continuous In Vitro Analysis of Colon Mucus Layer Accumulation and Physiology. Cell. Mol. Gastroenterol. Hepatol. 2020, 9, 507–526. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Žukauskaitė, K.; Li, M.; Horvath, A.; Jarmalaitė, S.; Stadlbauer, V. Cellular and Microbial In Vitro Modelling of Gastrointestinal Cancer. Cancers 2024, 16, 3113. https://doi.org/10.3390/cancers16173113
Žukauskaitė K, Li M, Horvath A, Jarmalaitė S, Stadlbauer V. Cellular and Microbial In Vitro Modelling of Gastrointestinal Cancer. Cancers. 2024; 16(17):3113. https://doi.org/10.3390/cancers16173113
Chicago/Turabian StyleŽukauskaitė, Kristina, Melissa Li, Angela Horvath, Sonata Jarmalaitė, and Vanessa Stadlbauer. 2024. "Cellular and Microbial In Vitro Modelling of Gastrointestinal Cancer" Cancers 16, no. 17: 3113. https://doi.org/10.3390/cancers16173113
APA StyleŽukauskaitė, K., Li, M., Horvath, A., Jarmalaitė, S., & Stadlbauer, V. (2024). Cellular and Microbial In Vitro Modelling of Gastrointestinal Cancer. Cancers, 16(17), 3113. https://doi.org/10.3390/cancers16173113





