The G-Protein-Coupled Estrogen Receptor Agonist G-1 Mediates Antitumor Effects by Activating Apoptosis Pathways and Regulating Migration and Invasion in Cervical Cancer Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. HaCaT Cells Transduced with E6 or E7 from HPV-16
2.3. Stimuli
2.4. RNA Extraction
2.5. RNA-Seq
2.6. Gene Set Enrichment Analysis
2.7. Immunofluorescence
2.8. Migration and Invasion Assays
2.9. Statistics
3. Results
3.1. G-1 Induces Transcription of Genes and Enrichment of Pathways Associated with Proliferation, Apoptosis, Metabolism, and Metastasis in the SiHa Cell Line
3.2. G-1 Increases Vimentin Expression without Altering α-SMA Levels
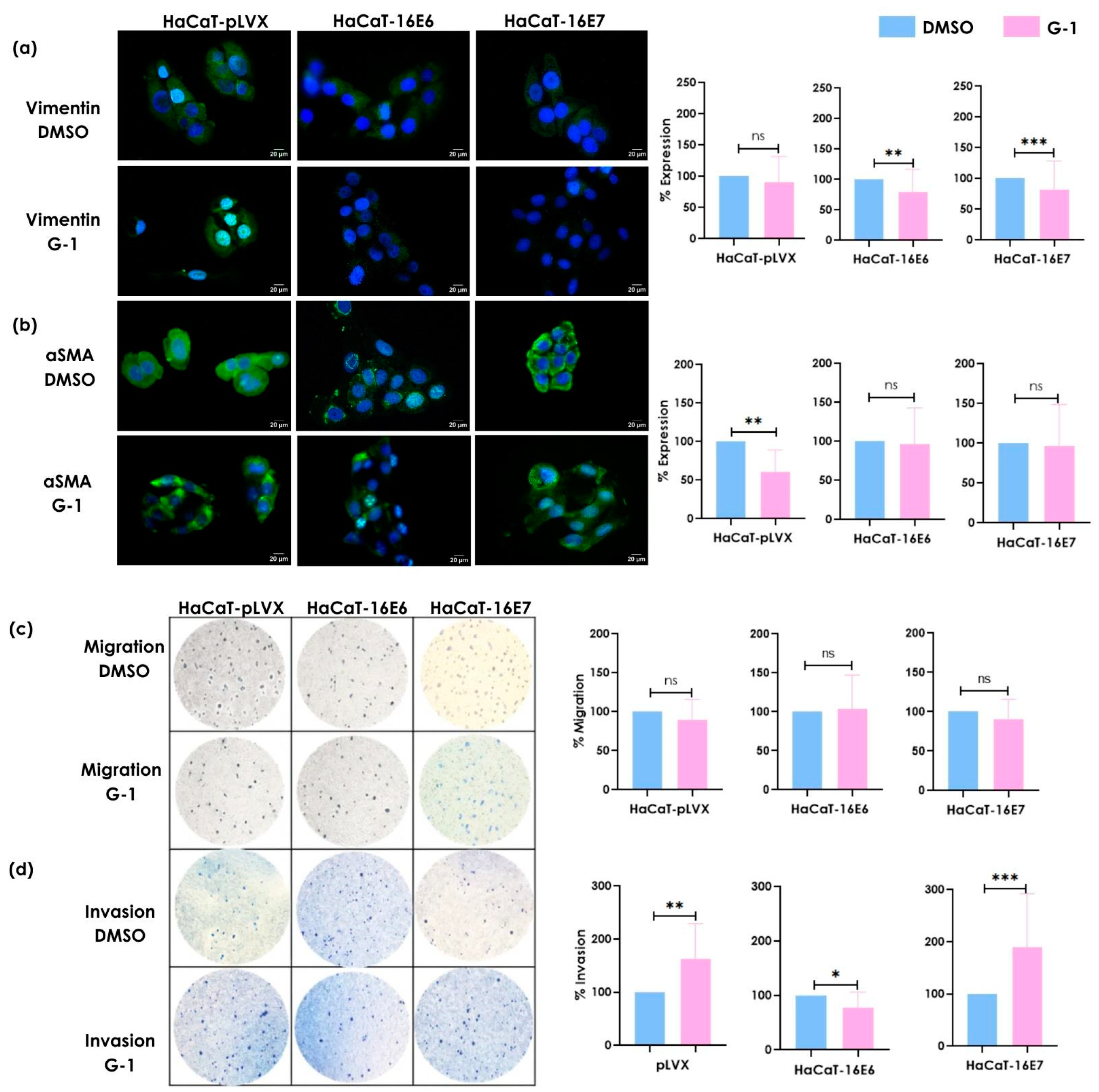
3.3. G-1 Modulates Vimentin and α-SMA Expression as Well as Invasion Processes in Keratinocytes
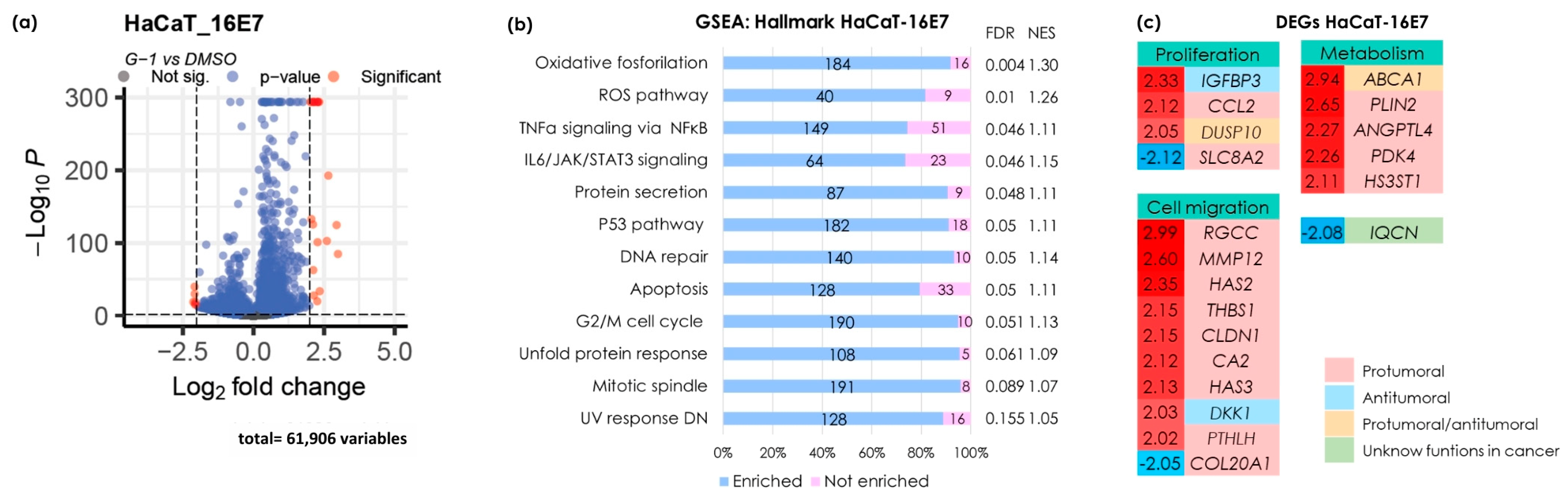
3.4. G-1 Triggers Gene Transcription and Activates Pathways Associated with Proliferation, Apoptosis, Metabolism, and Metastasis in the HaCaT-16E7 Cell Line
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Cancer Society. Cancer Facts & Figures 2024; American Cancer Society: Atlanta, GA, USA, 2024. [Google Scholar]
- Walboomers, J.M.M.; Jacobs, M.V.; Manos, M.M.; Bosch, F.X.; Kummer, J.A.; Shah, K.V.; Snijders, P.J.F.; Peto, J.; Meijer, C.J.L.M.; Muñoz, N. Human Papillomavirus Is a Necessary Cause of Invasive Cervical Cancer Worldwide. J. Pathol. 1999, 189, 12–19. [Google Scholar] [CrossRef]
- Smith, J.S.; Green, J.; De Gonzalez, A.B.; Appleby, P.; Peto, J.; Plummer, M.; Franceschi, S.; Beral, V. Cervical Cancer and Use of Hormonal Contraceptives: A Systematic Review. Lancet 2003, 361, 1159–1167. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-López, I.G.; Ramírez De Arellano, A.; Jave-Suárez, L.F.; Hernández-Silva, C.D.; García-Chagollan, M.; Hernández-Bello, J.; Lopez-Pulido, E.I.; Macias-Barragan, J.; Montoya-Buelna, M.; Muñoz-Valle, J.F.; et al. Interaction between 17β-Estradiol, Prolactin and Human Papillomavirus Induce E6/E7 Transcript and Modulate the Expression and Localization of Hormonal Receptors. Cancer Cell Int. 2019, 19, 227. [Google Scholar] [CrossRef] [PubMed]
- Spurgeon, M.E.; Den Boon, J.A.; Horswill, M.; Barthakur, S.; Forouzan, O.; Rader, J.S.; Beebe, D.J.; Roopra, A.; Ahlquist, P.; Lambert, P.F. Human Papillomavirus Oncogenes Reprogram the Cervical Cancer Microenvironment Independently of and Synergistically with Estrogen. Proc. Natl. Acad. Sci. USA 2017, 114, E9076–E9085. [Google Scholar] [CrossRef]
- Auborn, K.J.; Woodworth, C.; Dipaolo, J.A.; Bradlow, H.L. The Interaction between HPV Infection and Estrogen Metabolism in Cervical Carcinogenesis. Int. J. Cancer 1991, 49, 867–869. [Google Scholar] [CrossRef]
- Riera Leal, A.; Ortiz-Lazareno, P.C.; Jave-Suárez, L.F.; Ramírez De Arellano, A.; Aguilar-Lemarroy, A.; Ortiz-García, Y.M.; Barrón-Gallardo, C.A.; Solís-Martínez, R.; Luquin De Anda, S.; Muñoz-Valle, J.F.; et al. 17β-estradiol-induced Mitochondrial Dysfunction and Warburg Effect in Cervical Cancer Cells Allow Cell Survival under Metabolic Stress. Int. J. Oncol. 2019, 56, 33–46. [Google Scholar] [CrossRef]
- Rinaldi, S.; Plummer, M.; Biessy, C.; Castellsagué, X.; Overvad, K.; Krüger Kjær, S.; Tjønneland, A.; Clavel-Chapelon, F.; Chabbert-Buffet, N.; Mesrine, S.; et al. Endogenous Sex Steroids and Risk of Cervical Carcinoma: Results from the EPIC Study. Cancer Epidemiol. Biomark. Prev. 2011, 20, 2532–2540. [Google Scholar] [CrossRef] [PubMed]
- Brake, T.; Lambert, P.F. Estrogen Contributes to the Onset, Persistence, and Malignant Progression of Cervical Cancer in a Human Papillomavirus-Transgenic Mouse Model. Proc. Natl. Acad. Sci. USA 2005, 102, 2490–2495. [Google Scholar] [CrossRef]
- Chen, P.; Li, B.; Ou-Yang, L. Role of Estrogen Receptors in Health and Disease. Front. Endocrinol. 2022, 13, 839005. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.-H.; Wiedmeyer, K.; Shai, A.; Korach, K.S.; Lambert, P.F. Requirement for Estrogen Receptor α in a Mouse Model for Human Papillomavirus–Associated Cervical Cancer. Cancer Res. 2008, 68, 9928–9934. [Google Scholar] [CrossRef]
- Filardo, E.J.; Quinn, J.A.; Bland, K.I.; Frackelton, A.R. Estrogen-Induced Activation of Erk-1 and Erk-2 Requires the G Protein-Coupled Receptor Homolog, GPR30, and Occurs via Trans-Activation of the Epidermal Growth Factor Receptor through Release of HB-EGF. Mol. Endocrinol. 2000, 14, 1649–1660. [Google Scholar] [CrossRef] [PubMed]
- Pupo, M.; Vivacqua, A.; Perrotta, I.; Pisano, A.; Aquila, S.; Abonante, S.; Gasperi-Campani, A.; Pezzi, V.; Maggiolini, M. The Nuclear Localization Signal Is Required for Nuclear GPER Translocation and Function in Breast Cancer-Associated Fibroblasts (CAFs). Mol. Cell. Endocrinol. 2013, 376, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Pang, Y.; Filardo, E.J.; Dong, J. Identity of an Estrogen Membrane Receptor Coupled to a G Protein in Human Breast Cancer Cells. Endocrinology 2005, 146, 624–632. [Google Scholar] [CrossRef]
- Revankar, C.M.; Cimino, D.F.; Sklar, L.A.; Arterburn, J.B.; Prossnitz, E.R. A Transmembrane Intracellular Estrogen Receptor Mediates Rapid Cell Signaling. Science 2005, 307, 1625–1630. [Google Scholar] [CrossRef] [PubMed]
- Vo, D.-K.H.; Hartig, R.; Weinert, S.; Haybaeck, J.; Nass, N. G-Protein-Coupled Estrogen Receptor (GPER)-Specific Agonist G1 Induces ER Stress Leading to Cell Death in MCF-7 Cells. Biomolecules 2019, 9, 503. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Silva, C.D.H.; Leal, A.R.; Ortiz-Lazareno, P.C.; Suárez, L.F.J.; De Arellano, A.R.; Lopez-Pulido, E.I.; Barragan, J.G.M.; Buelna, M.M.; Rodríguez, J.R.D.; Chabay, P.; et al. GPER Overexpression in Cervical Cancer Versus Premalignant Lesions: Its Activation Induces Different Forms of Cell Death. Anticancer Agents Med. Chem. 2019, 19, 783–791. [Google Scholar] [CrossRef]
- Friese, K.; Kost, B.; Vattai, A.; Marmé, F.; Kuhn, C.; Mahner, S.; Dannecker, C.; Jeschke, U.; Heublein, S. The G Protein-Coupled Estrogen Receptor (GPER/GPR30) May Serve as a Prognostic Marker in Early-Stage Cervical Cancer. J. Cancer Res. Clin. Oncol. 2018, 144, 13–19. [Google Scholar] [CrossRef]
- Zhang, Q.; Wu, Y.; Zhang, Y.; Ji, X.; Hao, Q. Activation of G -protein Coupled Estrogen Receptor Inhibits the Proliferation of Cervical Cancer Cells via Sustained Activation of ERK1/2. Cell Biochem. Funct. 2015, 33, 134–142. [Google Scholar] [CrossRef]
- Hernández-Silva, C.D.; Villegas-Pineda, J.C.; Pereira-Suárez, A.L. Expression and Role of the G Protein-Coupled Estrogen Receptor (GPR30/GPER) in the Development and Immune Response in Female Reproductive Cancers. Front. Endocrinol. 2020, 11, 544. [Google Scholar] [CrossRef]
- Yager, J.D.; Davidson, N.E. Estrogen Carcinogenesis in Breast Cancer. N. Engl. J. Med. 2006, 354, 270–282. [Google Scholar] [CrossRef]
- Gaben, A.-M.; Saucier, C.; Bedin, M.; Redeuilh, G.; Mester, J. Mitogenic Activity of Estrogens in Human Breast Cancer Cells Does Not Rely on Direct Induction of Mitogen-Activated Protein Kinase/Extracellularly Regulated Kinase or Phosphatidylinositol 3-Kinase. Mol. Endocrinol. 2004, 18, 2700–2713. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, C.J.; Um, S.J.; Kim, T.Y.; Kim, E.J.; Park, T.C.; Kim, S.J.; Namkoong, S.E.; Park, J.S. Regulation of Cell Growth and HPV Genes by Exogenous Estrogen in Cervical Cancer Cells. Int. J. Gynecol. Cancer 2000, 10, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Di Fonte, R.; Strippoli, S.; Garofoli, M.; Cormio, G.; Serratì, S.; Loizzi, V.; Fasano, R.; Arezzo, F.; Volpicella, M.; Derakhshani, A.; et al. Cervical Cancer Benefits from Trabectedin Combination with the β-Blocker Propranolol: In Vitro and Ex Vivo Evaluations in Patient-Derived Organoids. Front. Cell Dev. Biol. 2023, 11, 1178316. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Delmas, D.; Vang, O.; Hsieh, T.; Lin, S.; Cheng, G.; Chiang, H.; Chen, C.E.; Tang, H.; Crawford, D.R.; et al. Mechanisms of Ceramide-induced COX-2-dependent Apoptosis in Human Ovarian Cancer OVCAR-3 Cells Partially Overlapped with Resveratrol. J. Cell. Biochem. 2013, 114, 1940–1954. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liang, L.; He, J.; He, Z.; Yue, C.; Jin, X.; Gao, M.; Xiao, S.; Zhou, Y. Fra-1 Inhibits Cell Growth and the Warburg Effect in Cervical Cancer Cells via STAT1 Regulation of the P53 Signaling Pathway. Front. Cell Dev. Biol. 2020, 8, 579629. [Google Scholar] [CrossRef]
- Song, X.; Zhou, L.; Yang, W.; Li, X.; Ma, J.; Qi, K.; Liang, R.; Li, M.; Xie, L.; Su, T.; et al. PHLDA1 Is a P53 Target Gene Involved in P53-Mediated Cell Apoptosis. Mol. Cell. Biochem. 2024, 479, 653–664. [Google Scholar] [CrossRef]
- Sano, E.; Kazaana, A.; Tadakuma, H.; Takei, T.; Yoshimura, S.; Hanashima, Y.; Ozawa, Y.; Yoshino, A.; Suzuki, Y.; Ueda, T. Interleukin-6 Sensitizes TNF-α and TRAIL/Apo2L Dependent Cell Death through Upregulation of Death Receptors in Human Cancer Cells. Biochim. Biophys. Acta BBA Mol. Cell Res. 2021, 1868, 119037. [Google Scholar] [CrossRef]
- Prossnitz, E.R.; Barton, M. The G Protein-Coupled Oestrogen Receptor GPER in Health and Disease: An Update. Nat. Rev. Endocrinol. 2023, 19, 407–424. [Google Scholar] [CrossRef]
- Ariazi, E.A.; Brailoiu, E.; Yerrum, S.; Shupp, H.A.; Slifker, M.J.; Cunliffe, H.E.; Black, M.A.; Donato, A.L.; Arterburn, J.B.; Oprea, T.I.; et al. The G Protein–Coupled Receptor GPR30 Inhibits Proliferation of Estrogen Receptor–Positive Breast Cancer Cells. Cancer Res. 2010, 70, 1184–1194. [Google Scholar] [CrossRef]
- Zhu, X.; Huang, L.; Gong, J.; Shi, C.; Wang, Z.; Ye, B.; Xuan, A.; He, X.; Long, D.; Zhu, X.; et al. NF-κB Pathway Link with ER Stress-Induced Autophagy and Apoptosis in Cervical Tumor Cells. Cell Death Discov. 2017, 3, 17059. [Google Scholar] [CrossRef]
- Lu, D.-N.; Zhang, W.-C.; Lin, Y.-Z.; Jiang, H.-Y.; He, R.; Li, S.-L.; Zhang, Y.-N.; Shao, C.-Y.; Zheng, C.-M.; Xu, J.-J.; et al. Single-Cell and Bulk RNA Sequencing Reveal Heterogeneity and Diagnostic Markers in Papillary Thyroid Carcinoma Lymph-Node Metastasis. J. Endocrinol. Investig. 2023, 47, 1513–1530. [Google Scholar] [CrossRef] [PubMed]
- Frensemeier, K.; Holzer, A.; Hoppe-Seyler, K.; Hoppe-Seyler, F. Dickkopf-1 Expression Is Repressed by Oncogenic Human Papillomaviruses (HPVS) and Regulates the Cisplatin Sensitivity of HPV-positive Cancer Cells in a JNK-dependent Manner. Int. J. Cancer 2022, 151, 2215–2228. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Li, Q.; Zhou, Q.; Zhang, S.; Chen, J.; Wang, Y.; Guo, J.; Gu, Y.; Gong, F.; Tan, Y.; et al. IQCN Disruption Causes Fertilization Failure and Male Infertility Due to Manchette Assembly Defect. EMBO Mol. Med. 2022, 14, e16501. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Hashimoto, K.; Kitano, S.; Yamashita, S.; Toda, A.; Nakamura, K.; Kinose, Y.; Kodama, M.; Sawada, K.; Kimura, T. Estrogen Induces Genomic Instability in High-Risk HPV-Infected Cervix and Promotes the Carcinogenesis of Cervical Adenocarcinoma. Biochem. Biophys. Res. Commun. 2023, 659, 80–90. [Google Scholar] [CrossRef]
- Lehman, T.A.; Modali, R.; Boukamp, P.; Stanek, J.; Bennett, W.P.; Welsh, J.A.; Metcalf, R.A.; Stampfer, M.R.; Fusenig, N.; Rogan, E.M.; et al. P53 Mutations in Human Immortalized Epithelial Cell Lines. Carcinogenesis 1993, 14, 833–839. [Google Scholar] [CrossRef]
- Holm, A.; Grände, P.-O.; Ludueña, R.F.; Olde, B.; Prasad, V.; Leeb-Lundberg, L.M.F.; Nilsson, B.-O. The G Protein-Coupled Oestrogen Receptor 1 Agonist G-1 Disrupts Endothelial Cell Microtubule Structure in a Receptor-Independent Manner. Mol. Cell. Biochem. 2012, 366, 239–249. [Google Scholar] [CrossRef]
- Wang, C.; Lv, X.; He, C.; Hua, G.; Tsai, M.-Y.; Davis, J.S. The G-Protein-Coupled Estrogen Receptor Agonist G-1 Suppresses Proliferation of Ovarian Cancer Cells by Blocking Tubulin Polymerization. Cell Death Dis. 2013, 4, e869. [Google Scholar] [CrossRef]
- Lv, X.; He, C.; Huang, C.; Hua, G.; Wang, Z.; Remmenga, S.W.; Rodabough, K.J.; Karpf, A.R.; Dong, J.; Davis, J.S.; et al. G-1 Inhibits Breast Cancer Cell Growth via Targeting Colchicine-Binding Site of Tubulin to Interfere with Microtubule Assembly. Mol. Cancer Ther. 2017, 16, 1080–1091. [Google Scholar] [CrossRef]
- Torres-López, L.; Olivas-Aguirre, M.; Villatoro-Gómez, K.; Dobrovinskaya, O. The G-Protein–Coupled Estrogen Receptor Agonist G-1 Inhibits Proliferation and Causes Apoptosis in Leukemia Cell Lines of T Lineage. Front. Cell Dev. Biol. 2022, 10, 811479. [Google Scholar] [CrossRef]
- Cardeal, L.B.D.S.; Boccardo, E.; Termini, L.; Rabachini, T.; Andreoli, M.A.; Di Loreto, C.; Filho, A.L.; Villa, L.L.; Maria-Engler, S.S. HPV16 Oncoproteins Induce MMPs/RECK-TIMP-2 Imbalance in Primary Keratinocytes: Possible Implications in Cervical Carcinogenesis. PLoS ONE 2012, 7, e33585. [Google Scholar] [CrossRef]
- Takasawa, K.; Takasawa, A.; Akimoto, T.; Magara, K.; Aoyama, T.; Kitajima, H.; Murakami, T.; Ono, Y.; Kyuno, D.; Suzuki, H.; et al. Regulatory Roles of Claudin-1 in Cell Adhesion and Microvilli Formation. Biochem. Biophys. Res. Commun. 2021, 565, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Akimoto, T.; Takasawa, A.; Takasawa, K.; Aoyama, T.; Murata, M.; Osanai, M.; Saito, T.; Sawada, N. Estrogen/GPR30 Signaling Contributes to the Malignant Potentials of ER-Negative Cervical Adenocarcinoma via Regulation of Claudin-1 Expression. Neoplasia 2018, 20, 1083–1093. [Google Scholar] [CrossRef] [PubMed]
- Ruckriegl, S.; Loris, J.; Wert, K.; Bauerschmitz, G.; Gallwas, J.; Gründker, C. Knockdown of G Protein-Coupled Estrogen Receptor 1 (GPER1) Enhances Tumor-Supportive Properties in Cervical Carcinoma Cells. Cancer Genom. Proteom. 2023, 20, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Toivola, D.M.; Strnad, P.; Habtezion, A.; Omary, M.B. Intermediate Filaments Take the Heat as Stress Proteins. Trends Cell Biol. 2010, 20, 79–91. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaxiola-Rubio, A.; Jave-Suárez, L.F.; Hernández-Silva, C.D.; Ramírez-de-Arellano, A.; Villegas-Pineda, J.C.; Lizárraga-Ledesma, M.d.J.; Ramos-Solano, M.; Diaz-Palomera, C.D.; Pereira-Suárez, A.L. The G-Protein-Coupled Estrogen Receptor Agonist G-1 Mediates Antitumor Effects by Activating Apoptosis Pathways and Regulating Migration and Invasion in Cervical Cancer Cells. Cancers 2024, 16, 3292. https://doi.org/10.3390/cancers16193292
Gaxiola-Rubio A, Jave-Suárez LF, Hernández-Silva CD, Ramírez-de-Arellano A, Villegas-Pineda JC, Lizárraga-Ledesma MdJ, Ramos-Solano M, Diaz-Palomera CD, Pereira-Suárez AL. The G-Protein-Coupled Estrogen Receptor Agonist G-1 Mediates Antitumor Effects by Activating Apoptosis Pathways and Regulating Migration and Invasion in Cervical Cancer Cells. Cancers. 2024; 16(19):3292. https://doi.org/10.3390/cancers16193292
Chicago/Turabian StyleGaxiola-Rubio, Abigail, Luis Felipe Jave-Suárez, Christian David Hernández-Silva, Adrián Ramírez-de-Arellano, Julio César Villegas-Pineda, Marisa de Jesús Lizárraga-Ledesma, Moisés Ramos-Solano, Carlos Daniel Diaz-Palomera, and Ana Laura Pereira-Suárez. 2024. "The G-Protein-Coupled Estrogen Receptor Agonist G-1 Mediates Antitumor Effects by Activating Apoptosis Pathways and Regulating Migration and Invasion in Cervical Cancer Cells" Cancers 16, no. 19: 3292. https://doi.org/10.3390/cancers16193292
APA StyleGaxiola-Rubio, A., Jave-Suárez, L. F., Hernández-Silva, C. D., Ramírez-de-Arellano, A., Villegas-Pineda, J. C., Lizárraga-Ledesma, M. d. J., Ramos-Solano, M., Diaz-Palomera, C. D., & Pereira-Suárez, A. L. (2024). The G-Protein-Coupled Estrogen Receptor Agonist G-1 Mediates Antitumor Effects by Activating Apoptosis Pathways and Regulating Migration and Invasion in Cervical Cancer Cells. Cancers, 16(19), 3292. https://doi.org/10.3390/cancers16193292






