A Comprehensive Review of the Antitumor Properties and Mechanistic Insights of Duocarmycin Analogs
Abstract
Simple Summary
Abstract
1. Introduction
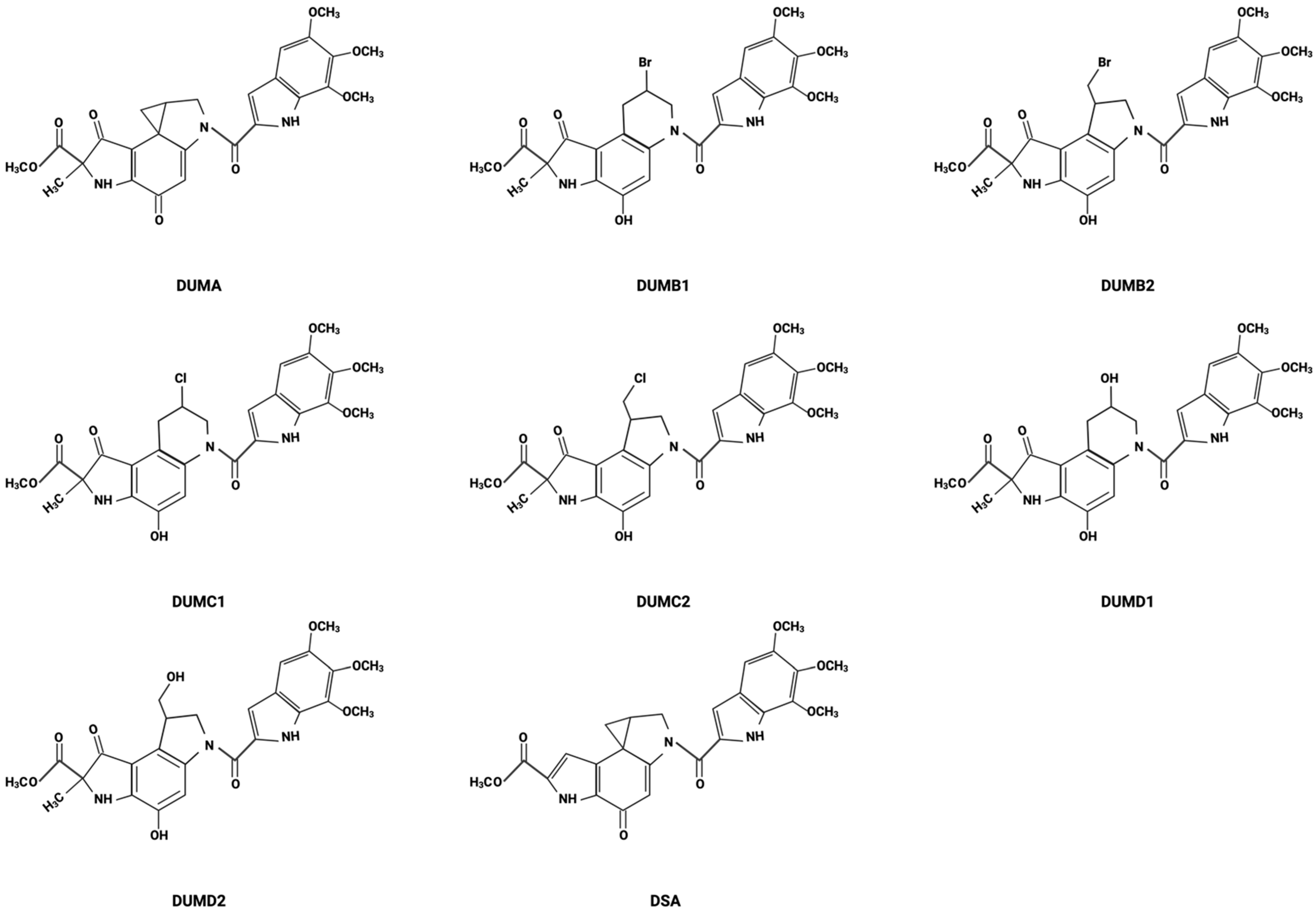
2. Discovery and Development
2.1. Historical Background of Duocarmycin Discovery
2.2. Isolation Details
2.3. Chemical Structure Elucidation
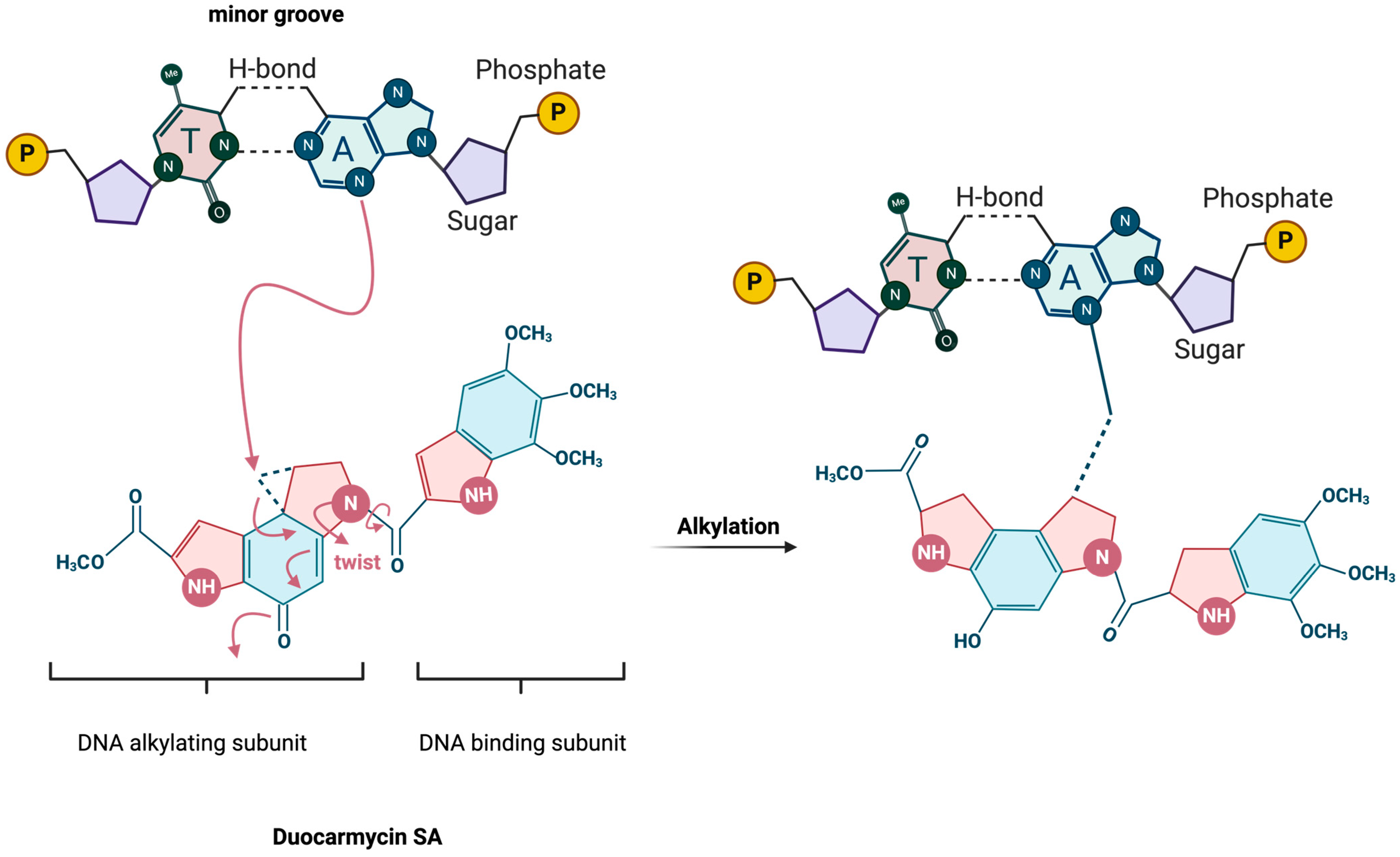
3. Mechanism of Action
3.1. DNA-Binding and Alkylation Process
3.2. Selectivity for Adenine Residues and AT-Rich Sequences
3.3. Biochemical Pathways Affected by Duocarmycin-Induced DNA Damage
4. Biological Activities
4.1. Cytotoxic Effects of DSA in Various Cancer Cell Lines
4.2. Exploring the Effects of DUMA on Human Lung Carcinoma
4.3. Comparison of Duocarmycin A with Another Duocarmycin Derivative
4.4. The Role of Reactive Oxygen Species in DUMA-Induced Apoptosis in Human Leukemia Cell Lines
4.5. Molecular and Cellular Effects of DSA on AML Cells
4.6. DSA and Proton Radiation Combination Therapy in GBM
4.7. Targeting Senescent Cancer Cells
5. Preclinical Studies
5.1. CC-1065
5.2. Adozelesin (U-73,975)
5.3. Carzelesin (U-80,244)
5.4. Bizelesin (U-77,779)
5.5. Pibrozelesin (KW-2189) DAerivative of Duocarmycin B2
5.6. Yatakemycin
6. Progress of Synthetic Analogs of Duocarmycin in Clinical Trials
7. Duocarmycin-Based Antibody–Drug Conjugates
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Felber, J.G.; Thorn-Seshold, O. 40 Years of Duocarmycins: A Graphical Structure/Function Review of Their Chemical Evolution, from SAR to Prodrugs and ADCs. JACS Au 2022, 2, 2636–2644. [Google Scholar] [CrossRef] [PubMed]
- Nani, R.R.; Gorka, A.P.; Nagaya, T.; Yamamoto, T.; Ivanic, J.; Kobayashi, H.; Schnermann, M.J. In Vivo Activation of Duocarmycin-Antibody Conjugates by Near-Infrared Light. ACS Cent. Sci. 2017, 3, 329–337. [Google Scholar] [CrossRef] [PubMed]
- MacMillan, K.S.; Nguyen, T.; Hwang, I.; Boger, D.L. Total synthesis and evaluation of iso-duocarmycin SA and iso-yatakemycin. J. Am. Chem. Soc. 2009, 131, 1187–1194. [Google Scholar] [CrossRef]
- Yao, H.P.; Zhao, H.; Hudson, R.; Tong, X.M.; Wang, M.H. Duocarmycin-based antibody-drug conjugates as an emerging biotherapeutic entity for targeted cancer therapy: Pharmaceutical strategy and clinical progress. Drug Discov. Today 2021, 26, 1857–1874. [Google Scholar] [CrossRef]
- Boyle, K.E.; Boger, D.L.; Wroe, A.; Vazquez, M. Duocarmycin SA, a potent antitumor antibiotic, sensitizes glioblastoma cells to proton radiation. Bioorganic Med. Chem. Lett. 2018, 28, 2688–2692. [Google Scholar] [CrossRef] [PubMed]
- Pourquier, P.; Waltman, J.L.; Urasaki, Y.; Loktionova, N.A.; Pegg, A.E.; Nitiss, J.L.; Pommier, Y. Topoisomerase I-mediated cytotoxicity of N-methyl-N’-nitro-N-nitrosoguanidine: Trapping of topoisomerase I by the O6-methylguanine. Cancer Res. 2001, 61, 53–58. [Google Scholar]
- Deans, A.J.; West, S.C. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer 2011, 11, 467–480. [Google Scholar] [CrossRef]
- Sauter, B.; Gillingham, D. DNA Damaging Agents in Chemical Biology and Cancer. Chimia 2020, 74, 693–698. [Google Scholar] [CrossRef]
- Lee, S.Y. Temozolomide resistance in glioblastoma multiforme. Genes Dis. 2016, 3, 198–210. [Google Scholar] [CrossRef]
- Kitange, G.J.; Carlson, B.L.; Schroeder, M.A.; Grogan, P.T.; Lamont, J.D.; Decker, P.A.; Wu, W.; James, C.D.; Sarkaria, J.N. Induction of MGMT expression is associated with temozolomide resistance in glioblastoma xenografts. Neuro Oncol. 2009, 11, 281–291. [Google Scholar] [CrossRef]
- Harrabi, S.; Combs, S.E.; Brons, S.; Haberer, T.; Debus, J.; Weber, K.J. Temozolomide in combination with carbon ion or photon irradiation in glioblastoma multiforme cell lines—Does scheduling matter? Int. J. Radiat. Biol. 2013, 89, 692–697. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.C.; Cho, H.Y.; Wang, W.; Nguyen, J.; Jhaveri, N.; Rosenstein-Sisson, R.; Hofman, F.M.; Schönthal, A.H. A novel temozolomide analog, NEO212, with enhanced activity against MGMT-positive melanoma in vitro and in vivo. Cancer Lett. 2015, 358, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Janku, F. Tumor heterogeneity in the clinic: Is it a real problem? Ther. Adv. Med. Oncol. 2014, 6, 43–51. [Google Scholar] [CrossRef]
- Parker, N.R.; Khong, P.; Parkinson, J.F.; Howell, V.M.; Wheeler, H.R. Molecular heterogeneity in glioblastoma: Potential clinical implications. Front. Oncol. 2015, 5, 55. [Google Scholar] [CrossRef]
- Pawlik, T.M.; Keyomarsi, K. Role of cell cycle in mediating sensitivity to radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2004, 59, 928–942. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, Y.; Nakatani, K.; Ito, Y.; Terashima, S. First total synthesis of dl-duocarmycin A. Tetrahedron Lett. 1990, 31, 6699–6702. [Google Scholar] [CrossRef]
- Boger, D.L.; McKie, J.A.; Nishi, T.; Ogiku, T. Enantioselective Total Synthesis of (+)-Duocarmycin A, epi-(+)-Duocarmycin A, and Their Unnatural Enantiomers. J. Am. Chem. Soc. 1996, 118, 2301–2302. [Google Scholar] [CrossRef]
- Dale, L.; Boger, K.M. Total synthesis of (+)-duocarmycin SA. J. Am. Chem. Soc. 1992, 114, 10056–10058. [Google Scholar]
- Ichimura, M.; Ogawa, T.; Katsumata, S.; Takahashi, K.; Takahashi, I.; Nakano, H. Duocarmycins, new antitumor antibiotics produced by Streptomyces; producing organisms and improved production. J. Antibiot. 1991, 44, 1045–1053. [Google Scholar] [CrossRef]
- Boger, D.L.; Boyce, C.W.; Garbaccio, R.M.; Goldberg, J.A. CC-1065 and the Duocarmycins: Synthetic Studies. Chem. Rev. 1997, 97, 787–828. [Google Scholar] [CrossRef]
- Ichimura, M.; Ogawa, T.; Takahashi, K.; Kobayashi, E.; Kawamoto, I.; Yasuzawa, T.; Takahashi, I.; Nakano, H. Duocarmycin SA, a new antitumor antibiotic from Streptomyces sp. J. Antibiot. 1990, 43, 1037–1038. [Google Scholar] [CrossRef] [PubMed]
- Sekurova, O.N.; Schneider, O.; Zotchev, S.B. Novel bioactive natural products from bacteria via bioprospecting, genome mining and metabolic engineering. Microb. Biotechnol. 2019, 12, 828–844. [Google Scholar] [CrossRef] [PubMed]
- Boger, D.L.; Johnson, D.S. CC-1065 and the duocarmycins: Unraveling the keys to a new class of naturally derived DNA alkylating agents. Proc. Natl. Acad. Sci. USA 1995, 92, 3642–3649. [Google Scholar] [CrossRef] [PubMed]
- Boger, D.L.; Santillan, A.; Searcey, M.; Jin, Q. Synthesis and Evaluation of Duocarmycin and CC-1065 Analogues Containing Modifications in the Subunit Linking Amide. J. Org. Chem. 1999, 64, 5241–5244. [Google Scholar] [CrossRef]
- Patil, P.C.; Satam, V.; Lee, M. A Short Review on the Synthetic Strategies of Duocarmycin Analogs that are Powerful DNA Alkylating Agents. Anticancer Agents Med. Chem. 2015, 15, 616–630. [Google Scholar] [CrossRef]
- Takahashi, I.; Takahashi, K.; Ichimura, M.; Morimoto, M.; Asano, K.; Kawamoto, I.; Tomita, F.; Nakano, H. Duocarmycin A, a new antitumor antibiotic from Streptomyces. J. Antibiot. 1988, 41, 1915–1917. [Google Scholar] [CrossRef]
- Sugiyama, H.; Ohmori, K.; Chan, K.L.; Hosoda, M.; Asai, A.; Saito, H.; Saito, I. A novel guanine N3 alkylation by antitumor antibiotic duocarmycin A. Tetrahedron Lett. 1993, 34, 2179–2182. [Google Scholar] [CrossRef]
- Gomi, K.; Kobayashi, E.; Miyoshi, K.; Ashizawa, T.; Okamoto, A.; Ogawa, T.; Katsumata, S.; Mihara, A.; Okabe, M.; Hirata, T. Anticellular and antitumor activity of duocarmycins, novel antitumor antibiotics. Jpn J. Cancer Res. 1992, 83, 113–120. [Google Scholar] [CrossRef]
- Shrestha, B.; Nath, D.K.; Maharjan, A.; Poudel, A.; Pradhan, R.N.; Aryal, S. Isolation and Characterization of Potential Antibiotic-Producing Actinomycetes from Water and Soil Sediments of Different Regions of Nepal. Int. J. Microbiol. 2021, 2021, 5586165. [Google Scholar] [CrossRef]
- Amishiro, N.; Okamoto, A.; Murakata, C.; Tamaoki, T.; Okabe, M.; Saito, H. Synthesis and antitumor activity of duocarmycin derivatives: Modification of segment-A of A-ring pyrrole compounds. J. Med. Chem. 1999, 42, 2946–2960. [Google Scholar] [CrossRef]
- Eis, P.S.; Smith, J.A.; Rydzewski, J.M.; Case, D.A.; Boger, D.L.; Chazin, W.J. High resolution solution structure of a DNA duplex alkylated by the antitumor agent duocarmycin SA. J. Mol. Biol. 1997, 272, 237–252. [Google Scholar] [CrossRef] [PubMed]
- Schuster, H.J.; Krewer, B.; von Hof, J.M.; Schmuck, K.; Schuberth, I.; Alves, F.; Tietze, L.F. Synthesis of the first spacer containing prodrug of a duocarmycin analogue and determination of its biological activity. Org. Biomol. Chem. 2010, 8, 1833–1842. [Google Scholar] [CrossRef] [PubMed]
- Sakata, J.; Tokuyama, H. Recent Progress on the Total Synthesis of Duocarmycins A and SA, Yatakemycin, and PDE-I and PDE-II. In Cutting-Edge Organic Synthesis and Chemical Biology of Bioactive Molecules: The Shape of Organic Synthesis to Come; Kobayashi, Y., Ed.; Springer: Singapore, 2019; pp. 101–124. [Google Scholar]
- Ichimura, M.; Ogawa, T.; Takahashi, K.; Mihara, A.; Takahashi, I.; Nakano, H. Interconversion and stability of duocarmycins, a new family of antitumor antibiotics: Correlation to their cytotoxic and antimicrobial activities in vitro. Oncol Res. 1993, 5, 165–171. [Google Scholar] [PubMed]
- Puyo, S.; Montaudon, D.; Pourquier, P. From old alkylating agents to new minor groove binders. Crit. Rev. Oncol. Hematol. 2014, 89, 43–61. [Google Scholar] [CrossRef]
- Boger, D.L.; Garbaccio, R.M. Shape-Dependent Catalysis: Insights into the Source of Catalysis for the CC-1065 and Duocarmycin DNA Alkylation Reaction. Acc. Chem. Res. 1999, 32, 1043–1052. [Google Scholar] [CrossRef]
- Bhaduri, S.; Ranjan, N.; Arya, D.P. An overview of recent advances in duplex DNA recognition by small molecules. Beilstein J. Org. Chem. 2018, 14, 1051–1086. [Google Scholar] [CrossRef] [PubMed]
- Tichenor, M.S.; Trzupek, J.D.; Kastrinsky, D.B.; Shiga, F.; Hwang, I.; Boger, D.L. Asymmetric total synthesis of (+)- and ent-(-)-yatakemycin and duocarmycin SA: Evaluation of yatakemycin key partial structures and its unnatural enantiomer. J. Am. Chem. Soc. 2006, 128, 15683–15696. [Google Scholar] [CrossRef]
- Wrasidlo, W.; Johnson, D.; Boger, D. Induction of Endonucleolytic DNA Fragmentation and Apoptosis by the Duocarmycins. Bioorganic Med. Chem. Lett. 1994, 4, 631–636. [Google Scholar] [CrossRef]
- Wang, G.; Vasquez, K.M. Effects of Replication and Transcription on DNA Structure-Related Genetic Instability. Genes 2017, 8, 17. [Google Scholar] [CrossRef]
- Peng, Y.; Pei, H. DNA alkylation lesion repair: Outcomes and implications in cancer chemotherapy. J. Zhejiang Univ. Sci. B 2021, 22, 47–62. [Google Scholar] [CrossRef]
- Asai, A.; Yano, K.; Mizukami, T.; Nakano, H. Characterization of a duocarmycin-DNA adduct-recognizing protein in cancer cells. Cancer Res. 1999, 59, 5417–5420. [Google Scholar] [PubMed]
- Ghosh, N.; Sheldrake, H.M.; Searcey, M.; Pors, K. Chemical and biological explorations of the family of CC-1065 and the duocarmycin natural products. Curr. Top. Med. Chem. 2009, 9, 1494–1524. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Robertson, W.M.; Kastrinsky, D.B.; Hwang, I.; Boger, D.L. Synthesis and evaluation of a series of C5’-substituted duocarmycin SA analogs. Bioorganic Med. Chem. Lett. 2010, 20, 2722–2725. [Google Scholar] [CrossRef] [PubMed]
- Boger, D.; Johnson, D. CC-1065 and the Duocarmycins: Understanding their Biological Function through Mechanistic Studies. Angew. Chem. Int. Ed. Engl. 1996, 92, 3642–3649. [Google Scholar] [CrossRef]
- Mullins, E.A.; Shi, R.; Eichman, B.F. Toxicity and repair of DNA adducts produced by the natural product yatakemycin. Nat. Chem. Biol. 2017, 13, 1002–1008. [Google Scholar] [CrossRef] [PubMed]
- Torgovnick, A.; Schumacher, B. DNA repair mechanisms in cancer development and therapy. Front. Genet. 2015, 6, 157. [Google Scholar] [CrossRef]
- Nagamura, S.; Asai, A.; Kobayashi, E.; Gomi, K.; Saito, H. Studies on duocarmycin SA and its derivatives. Bioorganic Med. Chem. 1997, 5, 623–630. [Google Scholar] [CrossRef]
- Okamoto, A.; Asai, A.; Saito, H.; Okabe, M.; Gomi, K. Differential effect of duocarmycin A and its novel derivative DU-86 on DNA strand breaks in HeLa S3 cells. Jpn J. Cancer Res. 1994, 85, 1304–1311. [Google Scholar] [CrossRef]
- Chen, W.A.; Williams, T.G.; So, L.; Drew, N.; Fang, J.; Ochoa, P.; Nguyen, N.; Jawhar, Y.; Otiji, J.; Duerksen-Hughes, P.J.; et al. Duocarmycin SA Reduces Proliferation and Increases Apoptosis in Acute Myeloid Leukemia Cells In Vitro. Int. J. Mol. Sci. 2024, 25, 4342. [Google Scholar] [CrossRef]
- Hirota, M.; Fujiwara, T.; Mineshita, S.; Sugiyama, H.; Teraoka, H. Distamycin A enhances the cytotoxicity of duocarmycin A and suppresses duocarmycin A-induced apoptosis in human lung carcinoma cells. Int. J. Biochem. Cell Biol. 2007, 39, 988–996. [Google Scholar] [CrossRef]
- Tada-Oikawa, S.; Oikawa, S.; Kawanishi, M.; Yamada, M.; Kawanishi, S. Generation of hydrogen peroxide precedes loss of mitochondrial membrane potential during DNA alkylation-induced apoptosis. FEBS Lett. 1999, 442, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Kohli, J.; Demaria, M. Senescent Cells in Cancer Therapy: Friends or Foes? Trends Cancer 2020, 6, 838–857. [Google Scholar] [CrossRef]
- Poblocka, M.; Bassey, A.L.; Smith, V.M.; Falcicchio, M.; Manso, A.S.; Althubiti, M.; Sheng, X.; Kyle, A.; Barber, R.; Frigerio, M.; et al. Targeted clearance of senescent cells using an antibody-drug conjugate against a specific membrane marker. Sci. Rep. 2021, 11, 20358. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, A.; Guiho, R.; Herranz, N.; Uren, A.; Withers, D.J.; Martínez-Barbera, J.P.; Tietze, L.F.; Gil, J. Galactose-modified duocarmycin prodrugs as senolytics. Aging Cell 2020, 19, e13133. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Gao, F.; Gnawali, G.; Xu, H.; Dong, Y.; Meng, X.; Li, W.; Wang, Z.; Lopez, B.; Carew, J.S.; et al. Selective Elimination of Senescent Cancer Cells by Galacto-Modified PROTACs. J. Med. Chem. 2024, 67, 7301–7311. [Google Scholar] [CrossRef] [PubMed]
- Hanka, L.J.; Dietz, A.; Gerpheide, S.A.; Kuentzel, S.L.; Martin, D.G. CC-1065 (NSC-298223), a new antitumor antibiotic. Production, in vitro biological activity, microbiological assays and taxonomy of the producing microorganism. J. Antibiot. 1978, 31, 1211–1217. [Google Scholar] [CrossRef]
- Li, L.H.; Swenson, D.H.; Schpok, S.L.; Kuentzel, S.L.; Dayton, B.D.; Krueger, W.C. CC-1065 (NSC 298223), a novel antitumor agent that interacts strongly with double-stranded DNA. Cancer Res. 1982, 42, 999–1004. [Google Scholar] [PubMed]
- Swenson, D.H.; Li, L.H.; Hurley, L.H.; Rokem, J.S.; Petzold, G.L.; Dayton, B.D.; Wallace, T.; Lin, A.; Krueger, W. Mechanism of interaction of CC-1065 (NSC 298223) with DNA. Cancer Res. 1982, 42, 2821–2828. [Google Scholar]
- Hurley, L.H.; Reynolds, V.L.; Swenson, D.H.; Petzold, G.L.; Scahill, T.A. Reaction of the antitumor antibiotic CC-1065 with DNA: Structure of a DNA adduct with DNA sequence specificity. Science 1984, 226, 843–844. [Google Scholar] [CrossRef]
- Bhuyan, B.K.; Newell, K.A.; Crampton, S.L.; Von Hoff, D.D. CC-1065 (NSC 298223), a most potent antitumor agent: Kinetics of inhibition of growth, DNA synthesis, and cell survival. Cancer Res. 1982, 42, 3532–3537. [Google Scholar]
- Harbach, P.R.; Trzos, R.J.; Mazurek, J.H.; Zimmer, D.M.; Petzold, G.L.; Bhuyan, B.K. Genotoxicity of the antitumor antibiotic CC-1065. Mutagenesis 1986, 1, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.G.; Biles, C.; Gerpheide, S.A.; Hanka, L.J.; Krueger, W.C.; McGovren, J.P.; Mizsak, S.A.; Neil, G.L.; Stewart, J.C.; Visser, J. CC-1065 (NSC 298223), a potent new antitumor agent improved production and isolation, characterization and antitumor activity. J. Antibiot. 1981, 34, 1119–1125. [Google Scholar] [CrossRef] [PubMed]
- McGovren, J.P.; Clarke, G.L.; Pratt, E.A.; DeKoning, T.F. Preliminary toxicity studies with the DNA-binding antibiotic, CC-1065. J. Antibiot. 1984, 37, 63–70. [Google Scholar] [CrossRef]
- Li, L.H.; Kelly, R.C.; Warpehoski, M.A.; McGovren, J.P.; Gebhard, I.; DeKoning, T.F. Adozelesin, a selected lead among cyclopropylpyrroloindole analogs of the DNA-binding antibiotic, CC-1065. Investig. New Drugs 1991, 9, 137–148. [Google Scholar] [CrossRef]
- Ayash, L.; Korbut, T.; Herman, T.S.; Teicher, B.A. Combination of the minor groove-binder U73-975 or the intercalator mitoxantrone with antitumor alkylating agents in MCF-7 or MCF-7/CP cells. Cancer Lett. 1991, 61, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.N.; Sevin, B.U.; Averette, H.; Perras, J.; Ramos, R.; Donato, D. Spectrum of cell-cycle kinetics of alkylating agent adolezesin in gynecological cancer cell lines: Correlation with drug-induced cytotoxicity. J. Cancer Res. Clin. Oncol. 1992, 118, 515–522. [Google Scholar] [CrossRef]
- Wang, Y.; Beerman, T.A.; Kowalski, D. Antitumor drug adozelesin differentially affects active and silent origins of DNA replication in yeast checkpoint kinase mutants. Cancer Res. 2001, 61, 3787–3794. [Google Scholar]
- Liu, J.S.; Kuo, S.R.; Beerman, T.A.; Melendy, T. Induction of DNA damage responses by adozelesin is S phase-specific and dependent on active replication forks. Mol. Cancer Ther. 2003, 2, 41–47. [Google Scholar]
- Li, L.H.; DeKoning, T.F.; Kelly, R.C.; Krueger, W.C.; McGovren, J.P.; Padbury, G.E.; Petzold, G.L.; Wallace, T.L.; Ouding, R.J.; Prairie, M.D. Cytotoxicity and antitumor activity of carzelesin, a prodrug cyclopropylpyrroloindole analogue. Cancer Res. 1992, 52, 4904–4913. [Google Scholar]
- Hightower, R.D.; Sevin, B.U.; Perras, J.; Nguyen, H.; Angioli, R.; Untch, M.; Averette, H. In vitro evaluation of the novel chemotherapeutic agents U-73,975, U-77,779, and U-80,244 in gynecologic cancer cell lines. Cancer Investig. 1993, 11, 276–282. [Google Scholar] [CrossRef]
- Houghton, P.J.; Cheshire, P.J.; Hallman, J.D., 2nd; Houghton, J.A. Therapeutic efficacy of the cyclopropylpyrroloindole, carzelesin, against xenografts derived from adult and childhood solid tumors. Cancer Chemother Pharmacol. 1995, 36, 45–52. [Google Scholar] [CrossRef]
- Woynarowski, J.M.; Trevino, A.V.; Rodriguez, K.A.; Hardies, S.C.; Benham, C.J. AT-rich islands in genomic DNA as a novel target for AT-specific DNA-reactive antitumor drugs. J. Biol. Chem. 2001, 276, 40555–40566. [Google Scholar] [CrossRef] [PubMed]
- Carter, C.A.; Waud, W.R.; Li, L.H.; DeKoning, T.F.; McGovren, J.P.; Plowman, J. Preclinical antitumor activity of bizelesin in mice. Clin. Cancer Res. 1996, 2, 1143–1149. [Google Scholar]
- Butryn, R.K.; Smith, K.S.; Adams, E.G.; Abraham, I.; Stackpole, J.; Sampson, K.E.; Bhuyan, B.K. V79 Chinese hamster lung cells resistant to the bis-alkylator bizelesin are multidrug-resistant. Cancer Chemother Pharmacol. 1994, 34, 44–50. [Google Scholar] [CrossRef]
- Lee, C.S.; Gibson, N.W. DNA damage and differential cytotoxicity produced in human carcinoma cells by CC-1065 analogues, U-73,975 and U-77,779. Cancer Res. 1991, 51, 6586–6591. [Google Scholar] [PubMed]
- Lee, C.S.; Gibson, N.W. DNA interstrand cross-links induced by the cyclopropylpyrroloindole antitumor agent bizelesin are reversible upon exposure to alkali. Biochemistry 1993, 32, 9108–9114. [Google Scholar] [CrossRef]
- Cao, P.R.; McHugh, M.M.; Melendy, T.; Beerman, T. The DNA minor groove-alkylating cyclopropylpyrroloindole drugs adozelesin and bizelesin induce different DNA damage response pathways in human colon carcinoma HCT116 cells. Mol. Cancer Ther. 2003, 2, 651–659. [Google Scholar]
- Ogasawara, H.; Nishio, K.; Takeda, Y.; Ohmori, T.; Kubota, N.; Funayama, Y.; Ohira, T.; Kuraishi, Y.; Isogai, Y.; Saijo, N. A novel antitumor antibiotic, KW-2189 is activated by carboxyl esterase and induces DNA strand breaks in human small cell lung cancer cells. Jpn J. Cancer Res. 1994, 85, 418–425. [Google Scholar] [CrossRef]
- Kobayashi, E.; Okamoto, A.; Asada, M.; Okabe, M.; Nagamura, S.; Asai, A.; Saito, H.; Gomi, K.; Hirata, T. Characteristics of antitumor activity of KW-2189, a novel water-soluble derivative of duocarmycin, against murine and human tumors. Cancer Res. 1994, 54, 2404–2410. [Google Scholar]
- Ogasawara, H.; Nishio, K.; Kanzawa, F.; Lee, Y.S.; Funayama, Y.; Ohira, T.; Kuraishi, Y.; Isogai, Y.; Saijo, N. Intracellular carboxyl esterase activity is a determinant of cellular sensitivity to the antineoplastic agent KW-2189 in cell lines resistant to cisplatin and CPT-11. Jpn J. Cancer Res. 1995, 86, 124–129. [Google Scholar] [CrossRef]
- Igarashi, Y.; Futamata, K.; Fujita, T.; Sekine, A.; Senda, H.; Naoki, H.; Furumai, T. Yatakemycin, a novel antifungal antibiotic produced by Streptomyces sp. TP-A0356. J. Antibiot. 2003, 56, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Fleming, G.F.; Ratain, M.J.; O’Brien, S.M.; Schilsky, R.L.; Hoffman, P.C.; Richards, J.M.; Vogelzang, N.J.; Kasunic, D.A.; Earhart, R.H. Phase I study of adozelesin administered by 24-hour continuous intravenous infusion. J. Natl. Cancer Inst. 1994, 86, 368–372. [Google Scholar] [CrossRef] [PubMed]
- Shamdas, G.J.; Alberts, D.S.; Modiano, M.; Wiggins, C.; Power, J.; Kasunic, D.A.; Elfring, G.L.; Earhart, R.H. Phase I study of adozelesin (U-73,975) in patients with solid tumors. Anticancer Drugs 1994, 5, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Foster, B.J.; LoRusso, P.M.; Poplin, E.; Zalupski, M.; Valdivieso, M.; Wozniak, A.; Flaherty, L.; Kasunic, D.A.; Earhart, R.H.; Baker, L.H. Phase I trial of Adozelesin using the treatment schedule of daily x5 every 3 weeks. Investig. New Drugs 1996, 13, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Burris, H.A.; Dieras, V.C.; Tunca, M.; Earhart, R.H.; Eckardt, J.R.; Rodriguez, G.I.; Shaffer, D.S.; Fields, S.M.; Campbell, E.; Schaaf, L.; et al. Phase I study with the DNA sequence-specific agent adozelesin. Anticancer Drugs 1997, 8, 588–596. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Bryan, W.J.; Miller, L.L.; Chang, A.Y.; Gradishar, W.J.; Kufe, D.W.; Hortobagyi, G.N. Phase II study of adozelesin in untreated metastatic breast cancer. Anticancer Drugs 1998, 9, 779–782. [Google Scholar] [CrossRef]
- Wolff, I.; Bench, K.; Beijnen, J.H.; Bruntsch, U.; Cavalli, F.; de Jong, J.; Groot, Y.; Van Tellingen, O.; Wanders, J.; Sessa, C. Phase I clinical and pharmacokinetic study of carzelesin (U-80244) given daily for five consecutive days. Clin. Cancer Res. 1996, 2, 1717–1723. [Google Scholar]
- van Tellingen, O.; Punt, C.J.; Awada, A.; Wagener, D.J.; Piccart, M.J.; Schaaf, L.J.; Nooijen, W.J.; Beijnen, J.H. A clinical pharmacokinetics study of carzelesin given by short-term intravenous infusion in a phase I study. Cancer Chemother Pharmacol. 1998, 41, 377–384. [Google Scholar] [CrossRef]
- Awada, A.; Punt, C.J.; Piccart, M.J.; Van Tellingen, O.; Van Manen, L.; Kerger, J.; Groot, Y.; Wanders, J.; Verweij, J.; Wagener, D.J.T. Phase I study of Carzelesin (U-80,244) given (4-weekly) by intravenous bolus schedule. Br. J. Cancer 1999, 79, 1454–1461. [Google Scholar] [CrossRef][Green Version]
- Pavlidis, N.; Aamdal, S.; Awada, A.; Calvert, H.; Fumoleau, P.; Sorio, R.; Punt, C.; Verweij, J.; Van Oosterom, A.; Morant, R.; et al. Carzelesin phase II study in advanced breast, ovarian, colorectal, gastric, head and neck cancer, non-Hodgkin’s lymphoma and malignant melanoma: A study of the EORTC early clinical studies group (ECSG). Cancer Chemother Pharmacol. 2000, 46, 167–171. [Google Scholar] [CrossRef]
- Pitot, H.C.; Reid, J.M.; Sloan, J.A.; Ames, M.M.; Adjei, A.A.; Rubin, J.; Bagniewski, P.G.; Atherton, P.; Rayson, D.; Goldberg, R.M.; et al. A Phase I study of bizelesin (NSC 615291) in patients with advanced solid tumors. Clin. Cancer Res. 2002, 8, 712–717. [Google Scholar]
- Schwartz, G.H.; Patnaik, A.; Hammond, L.A.; Rizzo, J.; Berg, K.; Von Hoff, D.D.; Rowinsky, E.K. A phase I study of bizelesin, a highly potent and selective DNA-interactive agent, in patients with advanced solid malignancies. Ann. Oncol. 2003, 14, 775–782. [Google Scholar] [CrossRef]
- Alberts, S.R.; Erlichman, C.; Reid, J.M.; Sloan, J.A.; Ames, M.M.; Richardson, R.L.; Goldberg, R.M. Phase I study of the duocarmycin semisynthetic derivative KW-2189 given daily for five days every six weeks. Clin. Cancer Res. 1998, 4, 2111–2117. [Google Scholar]
- Small, E.J.; Figlin, R.; Petrylak, D.; Vaughn, D.J.; Sartor, O.; Horak, I.; Pincus, R.; Kremer, A.; Bowden, C. A phase II pilot study of KW-2189 in patients with advanced renal cell carcinoma. Investig. New Drugs 2000, 18, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Markovic, S.N.; Suman, V.J.; Vukov, A.M.; Fitch, T.R.; Hillman, D.W.; Adjei, A.A.; Alberts, S.R.; Kaur, J.S.; Braich, T.A.; Leitch, J.M.; et al. Phase II trial of KW2189 in patients with advanced malignant melanoma. Am. J. Clin. Oncol. 2002, 25, 308–312. [Google Scholar] [CrossRef]
- Alberts, S.R.; Suman, V.J.; Pitot, H.C.; Camoriano, J.K.; Rubin, J. Use of KW-2189, a DNA minor groove-binding agent, in patients with hepatocellular carcinoma: A north central cancer treatment group (NCCTG) phase II clinical trial. J. Gastrointest. Cancer 2007, 38, 10–14. [Google Scholar] [CrossRef]
- Khongorzul, P.; Ling, C.J.; Khan, F.U.; Ihsan, A.U.; Zhang, J. Antibody-Drug Conjugates: A Comprehensive Review. Mol. Cancer Res. 2020, 18, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Yaghoubi, S.; Karimi, M.H.; Lotfinia, M.; Gharibi, T.; Mahi-Birjand, M.; Kavi, E.; Hosseini, F.; Sepehr, K.S.; Khatami, M.; Bagheri, N.; et al. Potential drugs used in the antibody-drug conjugate (ADC) architecture for cancer therapy. J. Cell. Physiol. 2020, 235, 31–64. [Google Scholar] [CrossRef]
- Banerji, U.; van Herpen, C.M.L.; Saura, C.; Thistlethwaite, F.; Lord, S.; Moreno, V.; Macpherson, I.R.; Boni, V.; Rolfo, C.; de Vries, E.G.E.; et al. Trastuzumab duocarmazine in locally advanced and metastatic solid tumours and HER2-expressing breast cancer: A phase 1 dose-escalation and dose-expansion study. Lancet Oncol. 2019, 20, 1124–1135. [Google Scholar] [CrossRef]
- Owonikoko, T.K.; Hussain, A.; Stadler, W.M.; Smith, D.C.; Kluger, H.; Molina, A.M.; Gulati, P.; Shah, A.; Ahlers, C.M.; Cardarelli, P.M.; et al. First-in-human multicenter phase I study of BMS-936561 (MDX-1203), an antibody-drug conjugate targeting CD70. Cancer Chemother Pharmacol. 2016, 77, 155–162. [Google Scholar] [CrossRef]
- Scribner, J.A.; Brown, J.G.; Son, T.; Chiechi, M.; Li, P.; Sharma, S.; Li, H.; De Costa, A.; Li, Y.; Chen, Y.; et al. Preclinical Development of MGC018, a Duocarmycin-based Antibody-drug Conjugate Targeting B7-H3 for Solid Cancer. Mol. Cancer Ther. 2020, 19, 2235–2244. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Lu, Y.; Yao, Y.; Liu, Y.; Wang, Y.; Lai, Q.; Zhang, R.; Li, W.; Wang, R.; Fu, Y.; et al. Promiximab-duocarmycin, a new CD56 antibody-drug conjugates, is highly efficacious in small cell lung cancer xenograft models. Oncotarget 2018, 9, 5197–5207. [Google Scholar] [CrossRef] [PubMed]
- Lutje, S.; Gerrits, D.; Molkenboer-Kuenen, J.D.; Herrmann, K.; Fracasso, G.; Colombatti, M.; Boerman, O.C.; Heskamp, S. Characterization of Site-Specifically Conjugated Monomethyl Auristatin E- and Duocarmycin-Based Anti-PSMA Antibody-Drug Conjugates for Treatment of PSMA-Expressing Tumors. J. Nucl. Med. 2018, 59, 494–501. [Google Scholar] [CrossRef]
- Nagaya, T.; Gorka, A.P.; Nani, R.R.; Okuyama, S.; Ogata, F.; Maruoka, Y.; Choyke, P.L.; Schnermann, M.J.; Kobayashi, H. Molecularly Targeted Cancer Combination Therapy with Near-Infrared Photoimmunotherapy and Near-Infrared Photorelease with Duocarmycin-Antibody Conjugate. Mol. Cancer Ther. 2018, 17, 661–670. [Google Scholar] [CrossRef]
- Powderly, J.D.; Jang, S.; Lohr, J.; Spira, A.I.; Bohac, G.C.; Sharma, M. Preliminary dose escalation results from a phase I/II, first-in-human study of MGC018 (anti-B7-H3 antibody-drug conjugate) in patients with advanced solid tumors. Clin. Oncol. 2020, 38, 3071. [Google Scholar] [CrossRef]
- Zhao, R.Y.; Erickson, H.K.; Leece, B.A.; Reid, E.E.; Goldmacher, V.S.; Lambert, J.M.; Chari, R.V.J. Synthesis and biological evaluation of antibody conjugates of phosphate prodrugs of cytotoxic DNA alkylators for the targeted treatment of cancer. J. Med. Chem. 2012, 55, 766–782. [Google Scholar] [CrossRef]
- Jin, J.; Park, G.; Park, J.B.; Kim, S.; Kim, H.; Chung, J. An anti-EGFR x cotinine bispecific antibody complexed with cotinine-conjugated duocarmycin inhibits growth of EGFR-positive cancer cells with KRAS mutations. Exp. Mol. Med. 2018, 50, 1–14. [Google Scholar] [CrossRef]
- Pillow, T.H.; Tercel, M. Duocarmycin–PBD Dimers as Antibody–Drug Conjugate (ADC) Payloads. In Cytotoxic Payloads for Antibody—Drug Conjugates; Thurston, D.E., Jackson, P.J.M., Eds.; The Royal Society of Chemistry: London, UK, 2019; pp. 241–258. [Google Scholar]

| Type of Cancer | Cell Line | Duocarmycin Analogs | Incubation Time | Assay/Endpoint | IC50 | Reference |
|---|---|---|---|---|---|---|
| Human uterine cervix carcinoma | HeLa S3 | DUMA | 1 h | Growth Inhibition | 0.12 nM | [49] |
| 0.0058 nM | [48] | |||||
| 0.006 nM | [28] | |||||
| DUMB1 | 1 h | Growth Inhibition | 0.035 nM | [28] | ||
| DUMB2 | 1 h | Growth Inhibition | 0.1 nM | [28] | ||
| DUMC1 | 1 h | Growth Inhibition | 8.5 nM | [28] | ||
| DUMB2 | 1 h | Growth Inhibition | 0.57 nM | [28] | ||
| DU-86 | 1 h | Growth Inhibition | 0.23 nM | [49] | ||
| 0.0052 nM | [48] | |||||
| DSA | 1 h | Growth Inhibition | 0.00069 nM | [48] | ||
| Human glioblastoma | U-138 | DSA | 14 days | Colony Formation | 0.0018 nM | [5] |
| 72 h | Growth Inhibition | 0.4 nM | [5] | |||
| U-251 | DUMC2 | N/A | Cytotoxicity Profiles | 0.794 nM | [39] | |
| DSA | N/A | Cytotoxicity Profiles | ~0.63 nM | [39] | ||
| Human acute myeloid leukemia | Molm-14 | DSA | 72 h | MTT | 0.01112 nM | [50] |
| 48 h | Annexin-V Staining | ~0.1 nM | [50] | |||
| 7 days | Colony Formation | 0.02 nM | [50] | |||
| N/A | Cytotoxicity Profiles | ~0.001 nM | [39] | |||
| DUMC2 | N/A | Cytotoxicity Profiles | ~1.58 nM | [39] | ||
| HL-60 | DSA | 72 h | MTT | 0.1227 nM | [50] | |
| 48 h | Annexin-V Staining | ~0.25 nM | [50] | |||
| 7 days | Colony Formation | 0.05 nM | [50] | |||
| N/A | Cytotoxicity Profiles | ~0.0158 nM | [39] | |||
| Molt-4 | DUMC2 | N/A | Cytotoxicity Profiles | ~0.001 nM | [39] | |
| DSA | N/A | Cytotoxicity Profiles | ~0.001 nM | [39] | ||
| Mouse lymphocytic leukemia | L1210 | DSA | N/A | Cytotoxicity Profiles | ~0.001 nM | [39] |
| Human renal adenocarcinoma | 786-0 | DUMC2 | N/A | Cytotoxicity Profiles | ~63.1 nM | [39] |
| DSA | N/A | Cytotoxicity Profiles | ~0.001 nM | [39] | ||
| Human prostatic adenocarcinoma | PC-3 | DUMC2 | N/A | Cytotoxicity Profiles | ~3.16 nM | [39] |
| DSA | N/A | Cytotoxicity Profiles | ~0.00126 nM | [39] | ||
| Human pancreatic adenocarcinoma | Capan-1 | DUMC2 | N/A | Cytotoxicity Profiles | ~0.0631 nM | [39] |
| DSA | N/A | Cytotoxicity Profiles | ~0.001 nM | [39] | ||
| Human colorectal adenocarcinoma | HT-29 | DUMC2 | N/A | Cytotoxicity Profiles | ~31.6 nM | [39] |
| DSA | N/A | Cytotoxicity Profiles | ~5 nM | [39] | ||
| Human breast adenocarcinoma | MCF-7 | DUMC2 | N/A | Cytotoxicity Profiles | ~79.4 nM | [39] |
| DSA | N/A | Cytotoxicity Profiles | ~0.79 nM | [39] | ||
| Human ovarian adenocarcinoma | OVCAR-3 | DUMC2 | N/A | Cytotoxicity Profiles | ~1 nM | [39] |
| DSA | N/A | Cytotoxicity Profiles | ~5 nM | [39] | ||
| Human lung adenocarcinoma | H322 | DUMC2 | N/A | Cytotoxicity Profiles | ~3.98 nM | [39] |
| DSA | N/A | Cytotoxicity Profiles | ~2.51 nM | [39] | ||
| Human lung squamous cell carcinoma | UCLA-P3 | DUMC2 | N/A | Cytotoxicity Profiles | ~25.1 nM | [39] |
| DSA | N/A | Cytotoxicity Profiles | ~2 nM | [39] | ||
| Human cervical squamous cell carcinoma | SiHA | DUMC2 | N/A | Cytotoxicity Profiles | ~31.6 nM | [39] |
| DSA | N/A | Cytotoxicity Profiles | ~0.4 nM | [39] | ||
| Human breast ductal carcinoma | BT-549 | DUMC2 | N/A | Cytotoxicity Profiles | ~0.891 nM | [39] |
| DSA | N/A | Cytotoxicity Profiles | ~0.063 nM | [39] | ||
| Human melanoma | SK-MEL-28 | DUMC2 | N/A | Cytotoxicity Profiles | ~7.94 nM | [39] |
| DSA | N/A | Cytotoxicity Profiles | ~1 nM | [39] | ||
| Human amelanotic melanoma | M24-MET | DUMC2 | N/A | Cytotoxicity Profiles | ~8.91 nM | [39] |
| DSA | N/A | Cytotoxicity Profiles | ~1.26 nM | [39] | ||
| Human mammary epithelial cells | HMEC | DUMC2 | N/A | Cytotoxicity Profiles | ~2.51 nM | [39] |
| DSA | N/A | Cytotoxicity Profiles | ~0.5 nM | [39] | ||
| Human dermal fibroblasts | NHDF | DUMC2 | N/A | Cytotoxicity Profiles | ~1000 nM | [39] |
| DSA | N/A | Cytotoxicity Profiles | ~0.001 nM | [39] | ||
| Human B lymphoblast | RPMI 7666 | DUMC2 | N/A | Cytotoxicity Profiles | ~1000 nM | [39] |
| DSA | N/A | Cytotoxicity Profiles | ~316 nM | [39] | ||
| Hamster epithelial-like ovarian cells | CHO | DUMC2 | N/A | Cytotoxicity Profiles | ~50.1 nM | [39] |
| DSA | N/A | Cytotoxicity profiles | ~15.8 nM | [39] |
| Drug Name | Clinical Trial | Results | Reference |
|---|---|---|---|
| Adozelesin | Phase I |
| [83] |
| Adozelesin | Phase I |
| [84] |
| Adozelesin | Phase I |
| [85] |
| Adozelesin | Phase I |
| [86] |
| Adozelesin | Phase II |
| [87] |
| Carzelesin | Phase I |
| [88] |
| Carzelesin | Phase I |
| [89] |
| Carzelesin | Phase I |
| [90] |
| Carzelesin | Phase II |
| [91] |
| Bizelesin | Phase I |
| [92] |
| Bizelesin | Phase I |
| [93] |
| Pibrozelesin (KW-2189) | Phase I |
| [94] |
| Pibrozelesin (KW-2189) | Phase II |
| [95] |
| Pibrozelesin (KW-2189) | Phase II |
| [96] |
| Pibrozelesin (KW-2189) | Phase II |
| [97] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morcos, A.; Jung, Y.; Galvan Bustillos, J.; Fuller, R.N.; Caba Molina, D.; Bertucci, A.; Boyle, K.E.; Vazquez, M.E.; Wall, N.R. A Comprehensive Review of the Antitumor Properties and Mechanistic Insights of Duocarmycin Analogs. Cancers 2024, 16, 3293. https://doi.org/10.3390/cancers16193293
Morcos A, Jung Y, Galvan Bustillos J, Fuller RN, Caba Molina D, Bertucci A, Boyle KE, Vazquez ME, Wall NR. A Comprehensive Review of the Antitumor Properties and Mechanistic Insights of Duocarmycin Analogs. Cancers. 2024; 16(19):3293. https://doi.org/10.3390/cancers16193293
Chicago/Turabian StyleMorcos, Ann, Yeonkyu Jung, Joab Galvan Bustillos, Ryan N. Fuller, David Caba Molina, Antonella Bertucci, Kristopher E. Boyle, Marcelo E. Vazquez, and Nathan R. Wall. 2024. "A Comprehensive Review of the Antitumor Properties and Mechanistic Insights of Duocarmycin Analogs" Cancers 16, no. 19: 3293. https://doi.org/10.3390/cancers16193293
APA StyleMorcos, A., Jung, Y., Galvan Bustillos, J., Fuller, R. N., Caba Molina, D., Bertucci, A., Boyle, K. E., Vazquez, M. E., & Wall, N. R. (2024). A Comprehensive Review of the Antitumor Properties and Mechanistic Insights of Duocarmycin Analogs. Cancers, 16(19), 3293. https://doi.org/10.3390/cancers16193293







