ANKRD1 Promotes Breast Cancer Metastasis by Activating NF-κB-MAGE-A6 Pathway
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cancer Cells and Cell Culture
2.2. RNA Extraction and Real-Time PCR
2.3. Protein Extraction and Western Blot Analysis
2.4. Plasmids, shRNA Lentivirus and siRNA Transfection
2.5. Cell Proliferation Assay
2.6. In Vitro Wound Healing, Transwell Migration and Invasion Assay
2.7. In Vivo Imaging and Mouse Pulmonary Metastasis Model
2.8. Immunohistochemistry (IHC)
2.9. Hematoxylin and Eosin (H&E) Staining
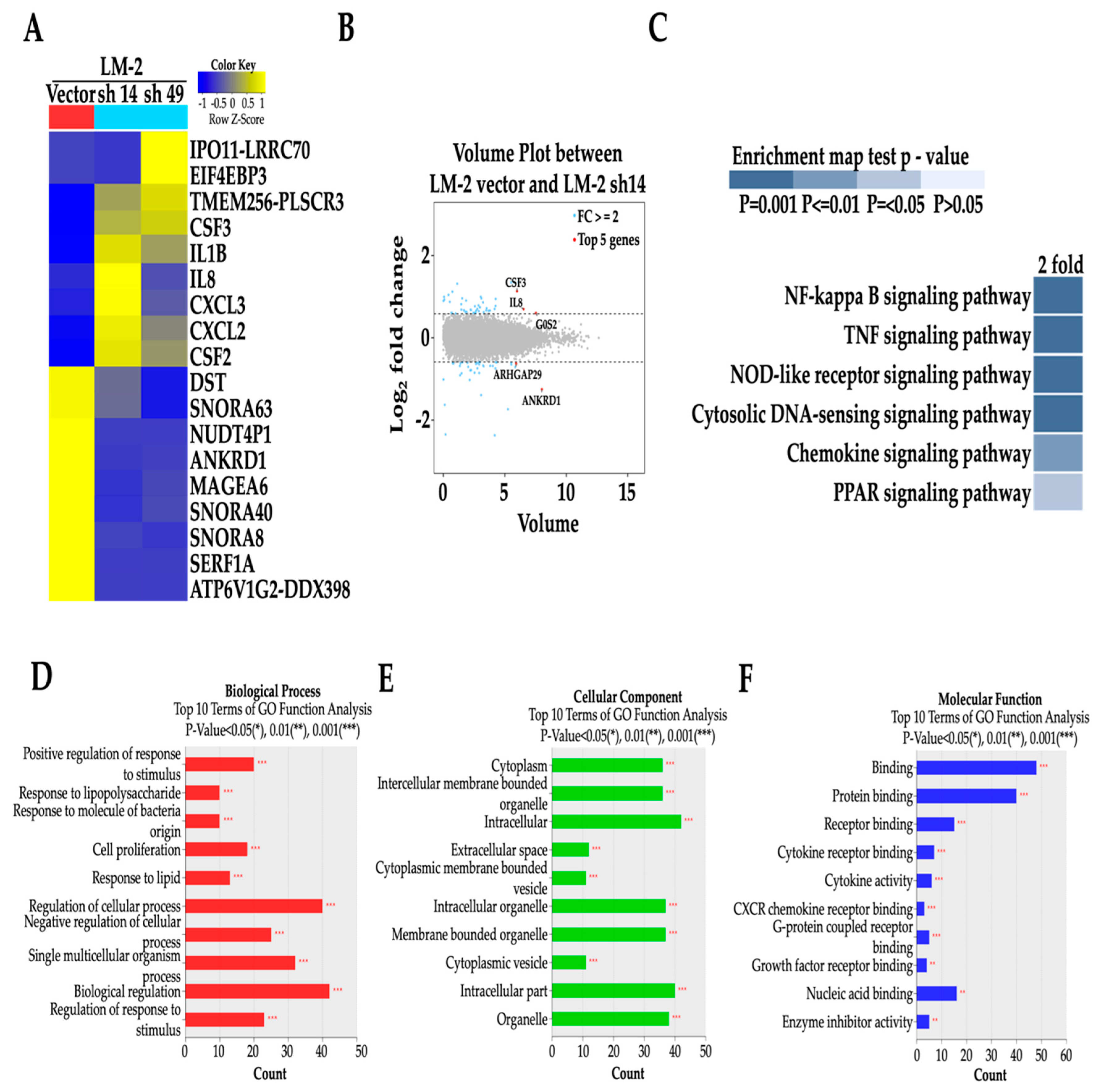
2.10. RNA-SEQ
2.11. Statistical Analysis
3. Results
3.1. ANKRD1 Is Upregulated in Highly Metastatic Breast Cancer Cell Lines
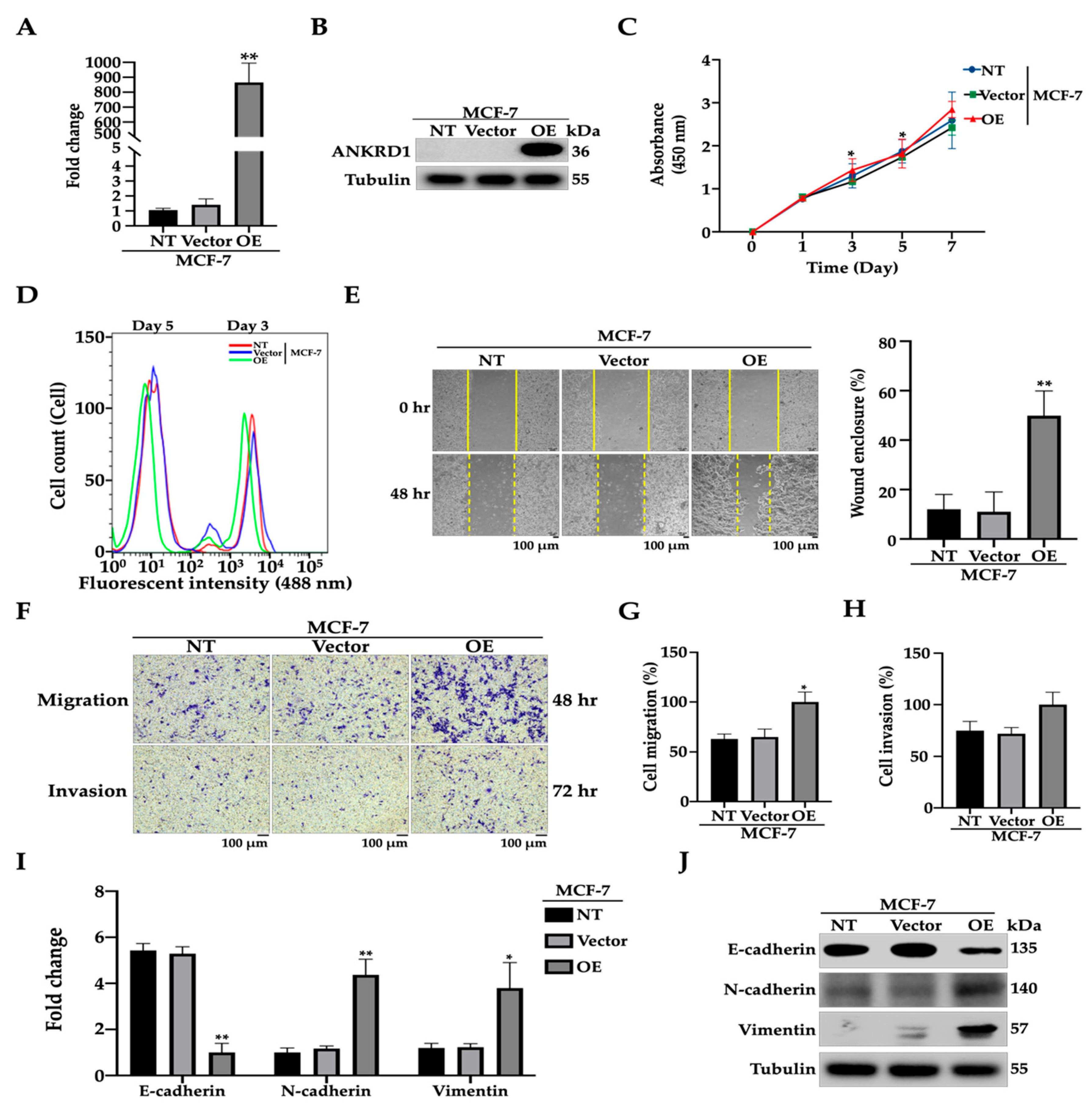
3.2. ANKRD1 Induces Cell Migration in Weakly Metastatic Breast Cancer Cells
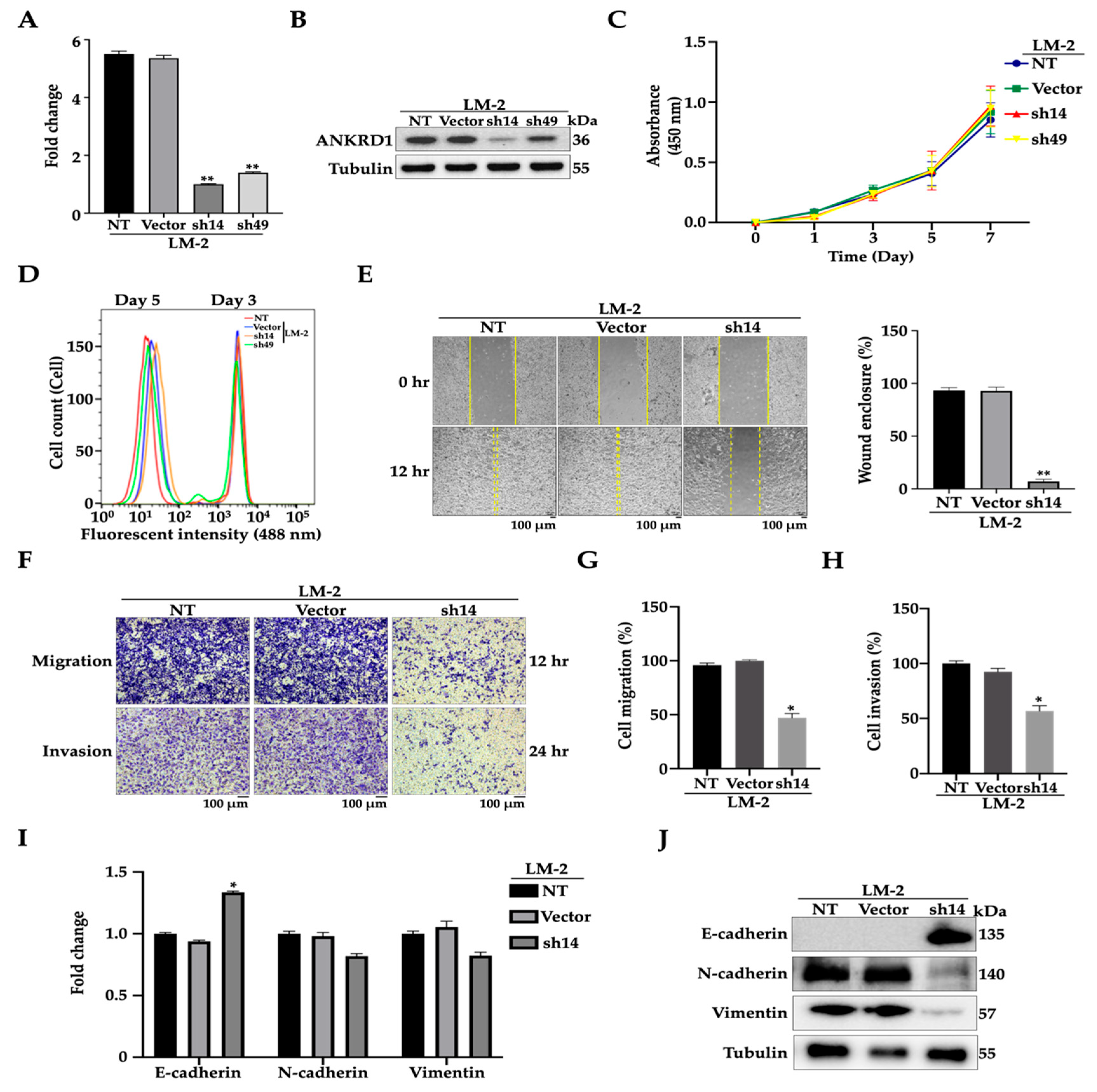
3.3. Knockdown of ANKRD1 Suppresses Migration and Invasion of Highly Metastatic LM-2 Cells
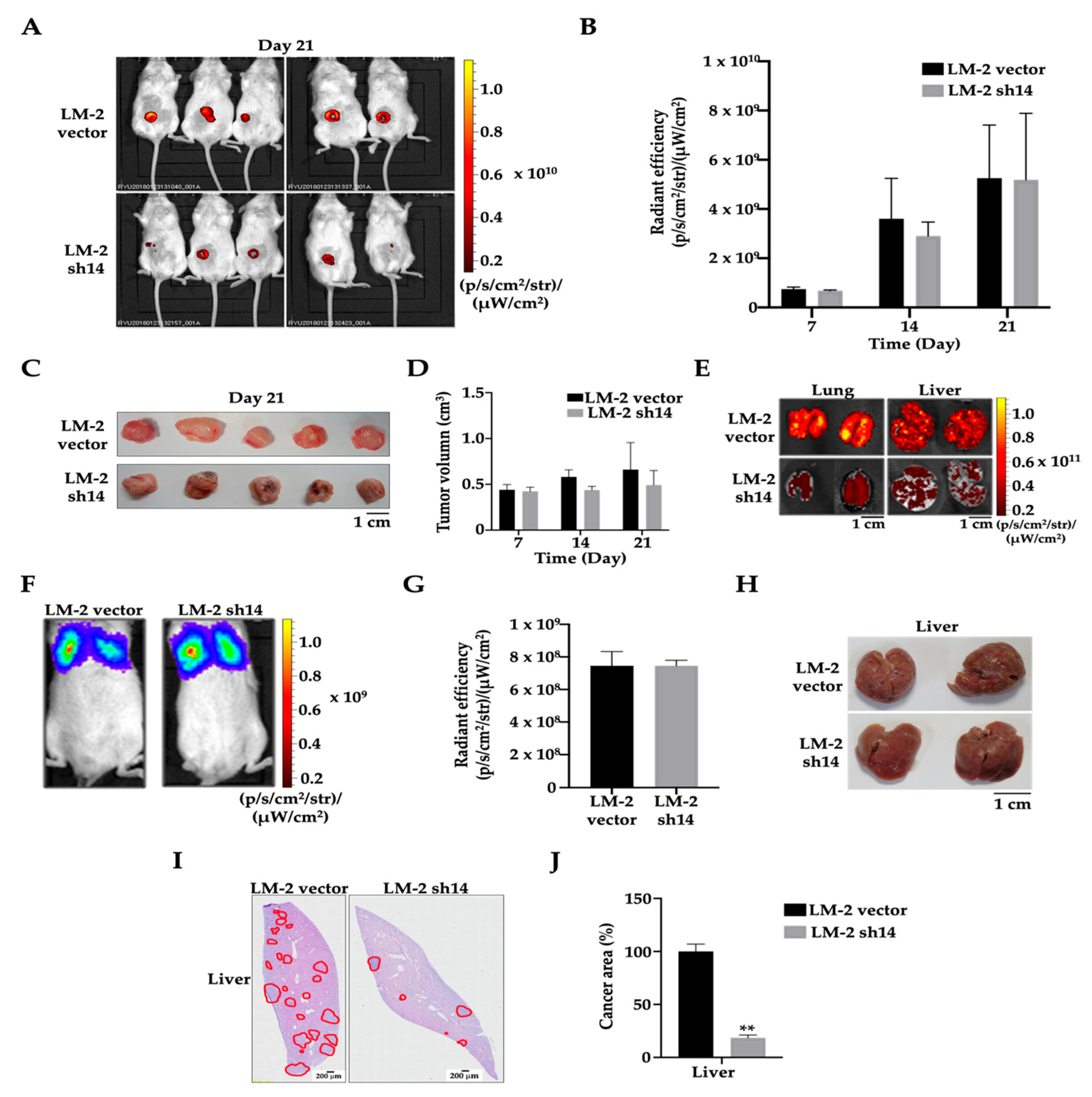
3.4. ANKRD1 Knockdown Suppresses Tumorigenesis and Metastasis
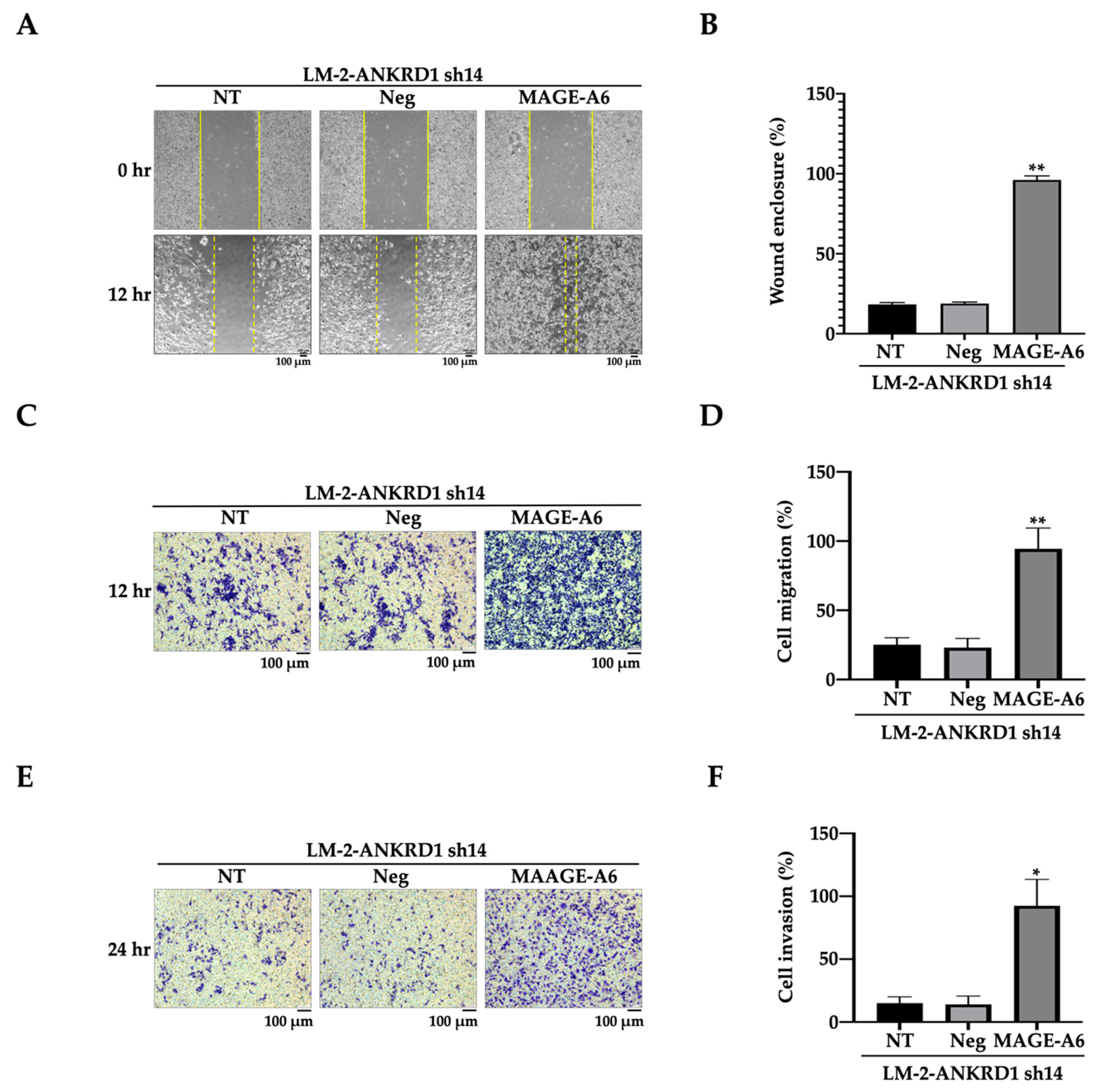
3.5. MAGE-A6 Acts Downstream of ANKRD1
3.6. MAGE-A6 Knockdown Reduces Cell Migration–Invasion
3.7. Recombinant MAGE-A6 Induces Cell Migration–Invasion
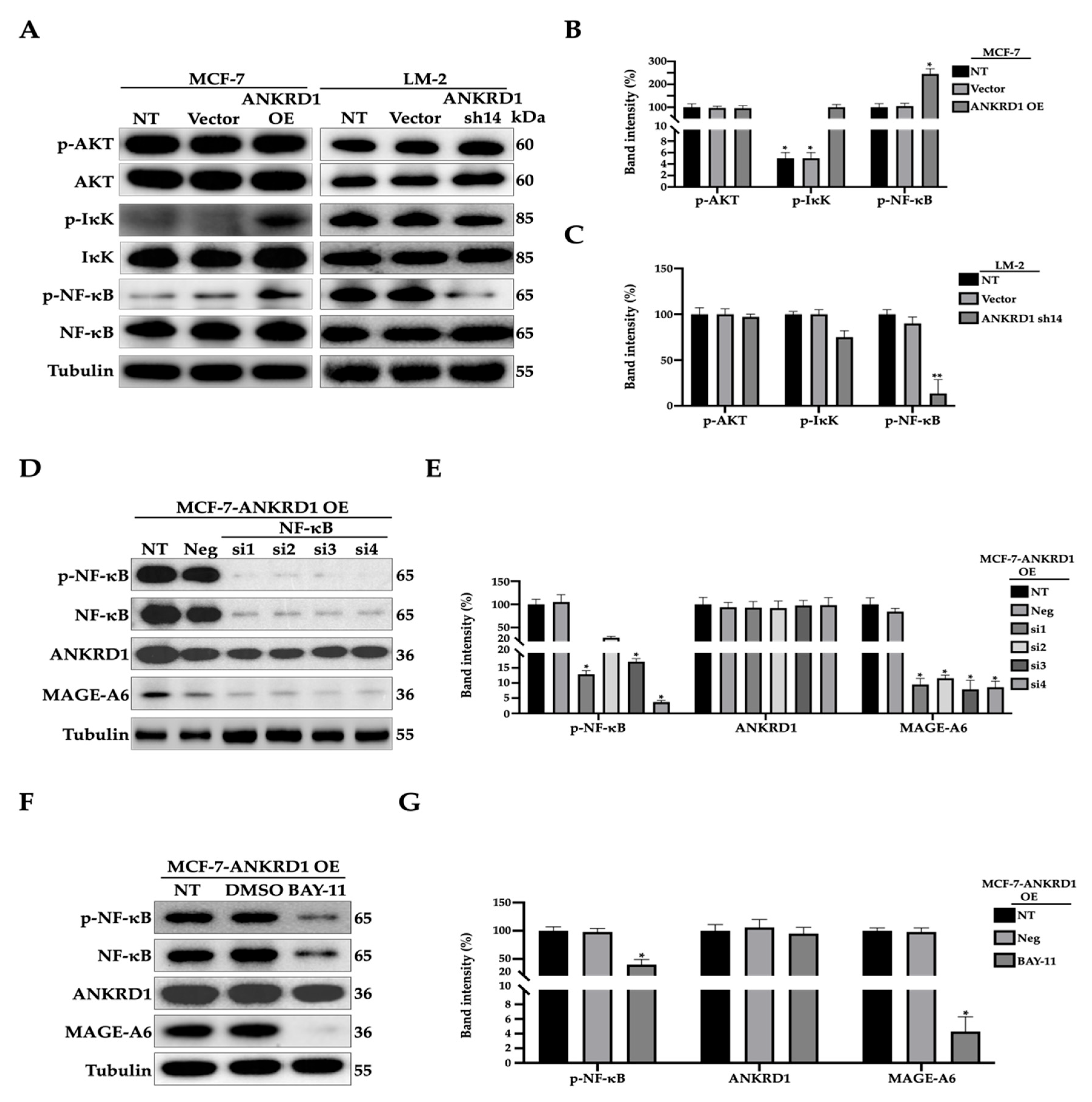
3.8. MAGE-A6 Acts as Downstream of NF-κB Pathway
3.9. MAGE-A6 Leads to Poor Prognosis in Breast Cancer
4. Discussion
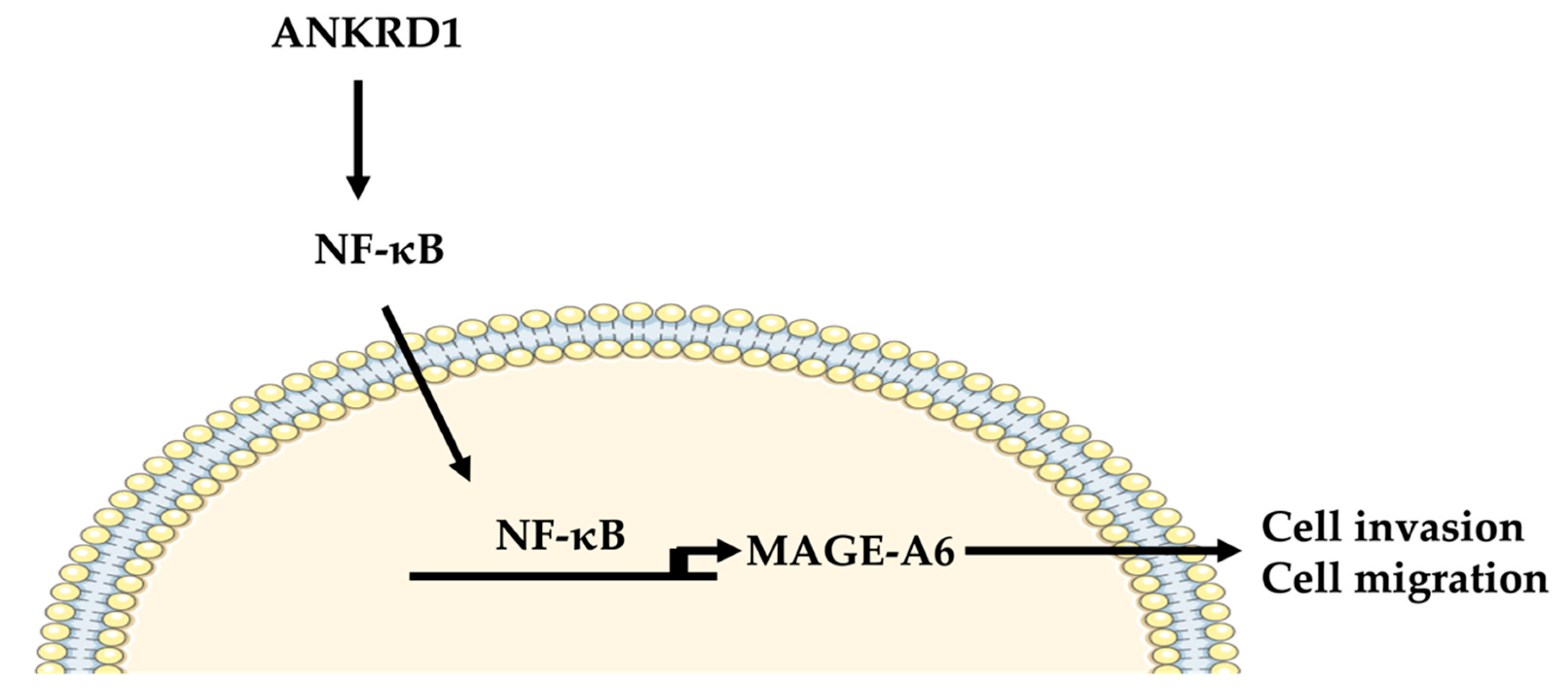
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lei, S.; Zheng, R.; Zhang, S.; Wang, S.; Chen, R.; Sun, K.; Zeng, H.; Zhou, J.; Wei, W. Global Patterns of Breast Cancer Incidence and Mortality: A Population-Based Cancer Registry Data Analysis from 2000 to 2020. Cancer Commun. 2021, 41, 1183–1194. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Goncalves, H., Jr.; Guerra, M.R.; Duarte Cintra, J.R.; Fayer, V.A.; Brum, I.V.; Bustamante Teixeira, M.T. Survival Study of Triple-Negative and Non-Triple-Negative Breast Cancer in a Brazilian Cohort. Clin. Med. Insights Oncol. 2018, 12, 1179554918790563. [Google Scholar] [CrossRef]
- Lebert, J.M.; Lester, R.; Powell, E.; Seal, M.; McCarthy, J. Advances in the Systemic Treatment of Triple-Negative Breast Cancer. Curr. Oncol. 2018, 25, S142–S150. [Google Scholar] [CrossRef] [PubMed]
- Anders, C.; Carey, L.A. Understanding and Treating Triple-Negative Breast Cancer. Oncology 2008, 22, 1233–1243; discussion 1239–1240. [Google Scholar] [PubMed]
- Anders, C.K.; Abramson, V.; Tan, T.; Dent, R. The Evolution of Triple-Negative Breast Cancer: From Biology to Novel Therapeutics. Am. Soc. Clin. Oncol. Educ. Book 2016, 35, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Diana, A.; Franzese, E.; Centonze, S.; Carlino, F.; della Corte, C.M.; Ventriglia, J.; Petrillo, A.; de Vita, F.; Alfano, R.; Ciardiello, F.; et al. Triple-Negative Breast Cancers: Systematic Review of the Literature on Molecular and Clinical Features with a Focus on Treatment with Innovative Drugs. Curr. Oncol. Rep. 2018, 20, 76. [Google Scholar] [CrossRef]
- Plasilova, M.L.; Hayse, B.; Killelea, B.K.; Horowitz, N.R.; Chagpar, A.B.; Lannin, D.R. Features of Triple-Negative Breast Cancer: Analysis of 38,813 Cases from the National Cancer Database. Medicine 2016, 95, e4614. [Google Scholar] [CrossRef]
- Yao, H.; He, G.; Yan, S.; Chen, C.; Song, L.; Rosol, T.J.; Deng, X. Triple-Negative Breast Cancer: Is There a Treatment on the Horizon? Oncotarget 2017, 8, 1913–1924. [Google Scholar] [CrossRef]
- O’Toole, S.A.; Beith, J.M.; Millar, E.K.; West, R.; McLean, A.; Cazet, A.; Swarbrick, A.; Oakes, S.R. Therapeutic Targets in Triple Negative Breast Cancer. J. Clin. Pathol. 2013, 66, 530–542. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M. Molecular Stratification of Triple-Negative Breast Cancers. Oncologist 2011, 16 (Suppl. S1), 61–70. [Google Scholar] [CrossRef] [PubMed]
- Mouh, F.Z.; Mzibri, M.E.; Slaoui, M.; Amrani, M. Recent Progress in Triple Negative Breast Cancer Research. Asian Pac. J. Cancer Prev. 2016, 17, 1595–1608. [Google Scholar] [CrossRef]
- Klein, C.A. Parallel Progression of Primary Tumours and Metastases. Nat. Rev. Cancer 2009, 9, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Xue, X.; Hu, C.; Xu, H.; Kou, D.; Li, R.; Li, M. Comparison of Clinicopathological Features and Prognosis in Triple-Negative and Non-Triple Negative Breast Cancer. J. Cancer 2016, 7, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Davion, S.M.; Siziopikou, K.P.; Sullivan, M.E. Cytokeratin 7: A Re-Evaluation of the “tried and True” in Triple-Negative Breast Cancers. Histopathology 2012, 61, 660–666. [Google Scholar] [CrossRef] [PubMed]
- Valastyan, S.; Weinberg, R.A. Tumor Metastasis: Molecular Insights and Evolving Paradigms. Cell 2011, 147, 275–292. [Google Scholar] [CrossRef]
- Talmadge, J.E.; Fidler, I.J. AACR Centennial Series: The Biology of Cancer Metastasis: Historical Perspective. Cancer Res. 2010, 70, 5649–5669. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular Principles of Metastasis: A Hallmark of Cancer Revisited. Signal Transduct. Target. Ther. 2020, 5, 28. [Google Scholar] [CrossRef]
- De Craene, B.; Berx, G. Regulatory Networks Defining EMT during Cancer Initiation and Progression. Nat. Rev. Cancer 2013, 13, 97–110. [Google Scholar] [CrossRef]
- Kikuchi, M.; Yamashita, K.; Waraya, M.; Minatani, N.; Ushiku, H.; Kojo, K.; Ema, A.; Kosaka, Y.; Katoh, H.; Sengoku, N.; et al. Epigenetic Regulation of ZEB1-RAB25/ESRP1 Axis Plays a Critical Role in Phenylbutyrate Treatment-Resistant Breast Cancer. Oncotarget 2016, 7, 1741–1753. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Seike, M.; Chiba, M.; Takahashi, S.; Nakamichi, S.; Matsumoto, M.; Takeuchi, S.; Minegishi, Y.; Noro, R.; Kunugi, S.; et al. Ankyrin Repeat Domain 1 Overexpression Is Associated with Common Resistance to Afatinib and Osimertinib in EGFR-Mutant Lung Cancer. Sci. Rep. 2018, 8, 14896. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.K.; Bang, M.-L.; Witt, C.C.; Labeit, D.; Trombitas, C.; Watanabe, K.; Granzier, H.; McElhinny, A.S.; Gregorio, C.C.; Labeit, S. The Muscle Ankyrin Repeat Proteins: CARP, Ankrd2/Arpp and DARP as a Family of Titin Filament-Based Stress Response Molecules. J. Mol. Biol. 2003, 333, 951–964. [Google Scholar] [CrossRef] [PubMed]
- Bang, M.L.; Mudry, R.E.; McElhinny, A.S.; Trombitás, K.; Geach, A.J.; Yamasaki, R.; Sorimachi, H.; Granzier, H.; Gregorio, C.C.; Labeit, S. Myopalladin, a Novel 145-Kilodalton Sarcomeric Protein with Multiple Roles in Z-Disc and I-Band Protein Assemblies. J. Cell Biol. 2001, 153, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Gnimassou, O.; Francaux, M.; Deldicque, L. Hippo Pathway and Skeletal Muscle Mass Regulation in Mammals: A Controversial Relationship. Front. Physiol. 2017, 8, 190. [Google Scholar] [CrossRef]
- Maugeri-Sacca, M.; de Maria, R. The Hippo Pathway in Normal Development and Cancer. Pharmacol. Ther. 2018, 186, 60–72. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, H.; Meyer, C.; Li, J.; Nadalin, S.; Königsrainer, A.; Weng, H.; Dooley, S.; Ten Dijke, P. Transforming Growth Factor-β (TGF-β)-Mediated Connective Tissue Growth Factor (CTGF) Expression in Hepatic Stellate Cells Requires Stat3 Signaling Activation. J. Biol. Chem. 2013, 288, 30708–30719. [Google Scholar] [CrossRef]
- Gressner, O.A.; Lahme, B.; Demirci, I.; Gressner, A.M.; Weiskirchen, R. Differential Effects of TGF-Beta on Connective Tissue Growth Factor (CTGF/CCN2) Expression in Hepatic Stellate Cells and Hepatocytes. J. Hepatol. 2007, 47, 699–710. [Google Scholar] [CrossRef]
- Zou, Y.; Evans, S.; Chen, J.; Kuo, H.C.; Harvey, R.P.; Chien, K.R. CARP, a Cardiac Ankyrin Repeat Protein, Is Downstream in the Nkx2-5 Homeobox Gene Pathway. Development 1997, 124, 793–804. [Google Scholar] [CrossRef]
- Kojic, S.; Nestorovic, A.; Rakicevic, L.; Belgrano, A.; Stankovic, M.; Divac, A.; Faulkner, G. A Novel Role for Cardiac Ankyrin Repeat Protein Ankrd1/CARP as a Co-Activator of the P53 Tumor Suppressor Protein. Arch. Biochem. Biophys. 2010, 502, 60–67. [Google Scholar] [CrossRef]
- Zhang, N.; Ye, F.; Zhu, W.; Hu, D.; Xiao, C.; Nan, J.; Su, S.; Wang, Y.; Liu, M.; Gao, K.; et al. Cardiac Ankyrin Repeat Protein Attenuates Cardiomyocyte Apoptosis by Upregulation of Bcl-2 Expression. Biochim. Biophys. Acta 2016, 1863, 3040–3049. [Google Scholar] [CrossRef]
- Ling, S.S.M.; Chen, Y.-T.; Wang, J.; Richards, A.M.; Liew, O.W. Ankyrin Repeat Domain 1 Protein: A Functionally Pleiotropic Protein with Cardiac Biomarker Potential. Int. J. Mol. Sci. 2017, 18, 1362. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-H.; Bauman, W.A.; Cardozo, C. ANKRD1 Modulates Inflammatory Responses in C2C12 Myoblasts through Feedback Inhibition of NF-ΚB Signaling Activity. Biochem. Biophys. Res. Commun. 2015, 464, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Chiusa, M.; Cadar, A.G.; Lin, A.; Samaras, S.; Davidson, J.M.; Lim, C.C. Targeted Inhibition of ANKRD1 Disrupts Sarcomeric ERK-GATA4 Signal Transduction and Abrogates Phenylephrine-Induced Cardiomyocyte Hypertrophy. Cardiovasc. Res. 2015, 106, 261–271. [Google Scholar] [CrossRef]
- Torrado, M.; Nespereira, B.; López, E.; Centeno, A.; Castro-Beiras, A.; Mikhailov, A.T. ANKRD1 Specifically Binds CASQ2 in Heart Extracts and Both Proteins Are Co-Enriched in Piglet Cardiac Purkinje Cells. J. Mol. Cell. Cardiol. 2005, 38, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Franklin, J.M.; Ghosh, R.P.; Shi, Q.; Reddick, M.P.; Liphardt, J.T. Concerted Localization-Resets Precede YAP-Dependent Transcription. Nat. Commun. 2020, 11, 4581. [Google Scholar] [CrossRef]
- Jiménez, A.P.; Traum, A.; Boettger, T.; Hackstein, H.; Richter, A.M.; Dammann, R.H. The Tumor Suppressor RASSF1A Induces the YAP1 Target Gene ANKRD1 That Is Epigenetically Inactivated in Human Cancers and Inhibits Tumor Growth. Oncotarget 2017, 8, 88437–88452. [Google Scholar] [CrossRef]
- Moon, S.; Kim, W.; Kim, S.; Kim, Y.; Song, Y.; Bilousov, O.; Kim, J.; Lee, T.; Cha, B.; Kim, M.; et al. Phosphorylation by NLK Inhibits YAP-14-3-3-Interactions and Induces Its Nuclear Localization. EMBO Rep. 2017, 18, 61–71. [Google Scholar] [CrossRef]
- Lange, S.; Gehmlich, K.; Lun, A.S.; Blondelle, J.; Hooper, C.; Dalton, N.D.; Alvarez, E.A.; Zhang, X.; Bang, M.-L.; Abassi, Y.A.; et al. MLP and CARP Are Linked to Chronic PKCα Signalling in Dilated Cardiomyopathy. Nat. Commun. 2016, 7, 12120. [Google Scholar] [CrossRef]
- Murphy, N.P.; Lubbers, E.R.; Mohler, P.J. Advancing Our Understanding of AnkRD1 in Cardiac Development and Disease. Cardiovasc. Res. 2020, 116, 1402–1404. [Google Scholar] [CrossRef]
- Francisco, J.; Zhang, Y.; Jeong, J.I.; Mizushima, W.; Ikeda, S.; Ivessa, A.; Oka, S.; Zhai, P.; Tallquist, M.D.; Del Re, D.P. Blockade of Fibroblast YAP Attenuates Cardiac Fibrosis and Dysfunction Through MRTF-A Inhibition. JACC Basic Transl. Sci. 2020, 5, 931–945. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Xu, J.; Li, Y.; Jia, C.; Ma, X.; Zhang, L.; Xie, X.; Zhang, Y.; Gao, X.; Zhang, Y.; et al. Cardiac Ankyrin Repeat Protein Attenuates Cardiac Hypertrophy by Inhibition of ERK1/2 and TGF-β Signaling Pathways. PLoS ONE 2012, 7, e50436. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Lu, D.; Yuan, J.; Wang, L.; Wang, J.; Lei, Z.; Liu, S.; Wu, J.; Wang, J.; Huang, L. Storax Attenuates Cardiac Fibrosis Following Acute Myocardial Infarction in Rats via Suppression of AT1R-Ankrd1-P53 Signaling Pathway. Int. J. Mol. Sci. 2022, 23, 13161. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Wang, Z.; Chen, K.; Shah, A.M.; Tan, W.; Duan, L.; Sanchez-Ortiz, E.; Li, H.; Xu, L.; Liu, N.; et al. Dynamic Transcriptional Responses to Injury of Regenerative and Non-Regenerative Cardiomyocytes Revealed by Single-Nucleus RNA Sequencing. Dev. Cell 2020, 53, 102–116.e8. [Google Scholar] [CrossRef]
- Lee, M.J.; Kwak, Y.K.; You, K.R.; Lee, B.H.; Kim, D.G. Involvement of GADD153 and Cardiac Ankyrin Repeat Protein in Cardiac Ischemia-Reperfusion Injury. Exp. Mol. Med. 2009, 41, 243–252. [Google Scholar] [CrossRef]
- Bogomolovas, J.; Brohm, K.; Čelutkienė, J.; Balčiūnaitė, G.; Bironaitė, D.; Bukelskienė, V.; Daunoravičus, D.; Witt, C.C.; Fielitz, J.; Grabauskienė, V.; et al. Induction of Ankrd1 in Dilated Cardiomyopathy Correlates with the Heart Failure Progression. BioMed Res. Int. 2015, 2015, 273936. [Google Scholar] [CrossRef]
- Jensen, N.F.; Stenvang, J.; Beck, M.K.; Hanakova, B.; Belling, K.C.; Do, K.N.; Viuff, B.; Nygard, S.B.; Gupta, R.; Rasmussen, M.H.; et al. Establishment and Characterization of Models of Chemotherapy Resistance in Colorectal Cancer: Towards a Predictive Signature of Chemoresistance. Mol. Oncol. 2015, 9, 1169–1185. [Google Scholar] [CrossRef]
- Lei, Y.; Henderson, B.R.; Emmanuel, C.; Harnett, P.R.; deFazio, A. Inhibition of ANKRD1 Sensitizes Human Ovarian Cancer Cells to Endoplasmic Reticulum Stress-Induced Apoptosis. Oncogene 2015, 34, 485–495. [Google Scholar] [CrossRef]
- Scurr, L.L.; Guminski, A.D.; Chiew, Y.E.; Balleine, R.L.; Sharma, R.; Lei, Y.; Pryor, K.; Wain, G.V.; Brand, A.; Byth, K.; et al. Ankyrin Repeat Domain 1, ANKRD1, a Novel Determinant of Cisplatin Sensitivity Expressed in Ovarian Cancer. Clin. Cancer Res. 2008, 14, 6924–6932. [Google Scholar] [CrossRef]
- Rakha, E.A.; Tse, G.M.; Quinn, C.M. An Update on the Pathological Classification of Breast Cancer. Histopathology 2023, 82, 5–16. [Google Scholar] [CrossRef]
- Florke Gee, R.R.; Chen, H.; Lee, A.K.; Daly, C.A.; Wilander, B.A.; Fon Tacer, K.; Potts, P.R. Emerging Roles of the MAGE Protein Family in Stress Response Pathways. J. Biol. Chem. 2020, 295, 16121–16155. [Google Scholar] [CrossRef] [PubMed]
- Almodovar-Garcia, K.; Kwon, M.; Samaras, S.E.; Davidson, J.M. ANKRD1 Acts as a Transcriptional Repressor of MMP13 via the AP-1 Site. Mol. Cell. Biol. 2014, 34, 1500–1511. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.; Chen, J.; Ruiz-Lozano, P.; Zou, Y.; Nemer, M.; Chien, K.R. Control of Segmental Expression of the Cardiac-Restricted Ankyrin Repeat Protein Gene by Distinct Regulatory Pathways in Murine Cardiogenesis. Development 1999, 126, 4223–4234. [Google Scholar] [CrossRef]
- Shi, Y.; Reitmaier, B.; Regenbogen, J.; Slowey, R.M.; Opalenik, S.R.; Wolf, E.; Goppelt, A.; Davidson, J.M. CARP, a Cardiac Ankyrin Repeat Protein, Is up-Regulated during Wound Healing and Induces Angiogenesis in Experimental Granulation Tissue. Am. J. Pathol. 2005, 166, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Jeyaseelan, R.; Poizat, C.; Baker, R.K.; Abdishoo, S.; Isterabadi, L.B.; Lyons, G.E.; Kedes, L. A Novel Cardiac-Restricted Target for Doxorubicin. CARP, a Nuclear Modulator of Gene Expression in Cardiac Progenitor Cells and Cardiomyocytes. J. Biol. Chem. 1997, 272, 22800–22808. [Google Scholar] [CrossRef]
- Henson, D.E.; Ries, L.; Freedman, L.S.; Carriaga, M. Relationship among Outcome, Stage of Disease, and Histologic Grade for 22,616 Cases of Breast Cancer. The Basis for a Prognostic Index. Cancer 1991, 68, 2142–2149. [Google Scholar] [CrossRef]
- Samaras, S.E.; Almodovar-Garcia, K.; Wu, N.; Yu, F.; Davidson, J.M. Global Deletion of Ankrd1 Results in a Wound-Healing Phenotype Associated with Dermal Fibroblast Dysfunction. Am. J. Pathol. 2015, 185, 96–109. [Google Scholar] [CrossRef]
- Elkin, M.; Vlodavsky, I. Tail Vein Assay of Cancer Metastasis. Curr. Protoc. Cell Biol. 2001, 12, 19.2.1–19.2.7. [Google Scholar] [CrossRef]
- Werbeck, J.L.; Thudi, N.K.; Martin, C.K.; Premanandan, C.; Yu, L.; Ostrowksi, M.C.; Rosol, T.J. Tumor Microenvironment Regulates Metastasis and Metastasis Genes of Mouse MMTV-PymT Mammary Cancer Cells in Vivo. Vet. Pathol. 2014, 51, 868–881. [Google Scholar] [CrossRef]
- Cheng, Y.; Hou, T.; Ping, J.; Chen, T.; Yin, B. LMO3 Promotes Hepatocellular Carcinoma Invasion, Metastasis and Anoikis Inhibition by Directly Interacting with LATS1 and Suppressing Hippo Signaling. J. Exp. Clin. Cancer Res. 2018, 37, 228. [Google Scholar] [CrossRef]
- Janse van Rensburg, H.J.; Yang, X. The Roles of the Hippo Pathway in Cancer Metastasis. Cell. Signal. 2016, 28, 1761–1772. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhang, L.; Purohit, V.; Shukla, S.K.; Chen, X.; Yu, F.; Fu, K.; Chen, Y.; Solheim, J.; Singh, P.K.; et al. Active YAP Promotes Pancreatic Cancer Cell Motility, Invasion and Tumorigenesis in a Mitotic Phosphorylation-Dependent Manner through LPAR3. Oncotarget 2015, 6, 36019–36031. [Google Scholar] [CrossRef]
- Barker, P.A.; Salehi, A. The MAGE Proteins: Emerging Roles in Cell Cycle Progression, Apoptosis, and Neurogenetic Disease. J. Neurosci. Res. 2002, 67, 705–712. [Google Scholar] [CrossRef]
- Simpson, A.J.G.; Caballero, O.L.; Jungbluth, A.; Chen, Y.-T.; Old, L.J. Cancer/Testis Antigens, Gametogenesis and Cancer. Nat. Rev. Cancer 2005, 5, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Nardiello, T.; Jungbluth, A.A.; Mei, A.; Diliberto, M.; Huang, X.; Dabrowski, A.; Andrade, V.C.C.; Wasserstrum, R.; Ely, S.; Niesvizky, R.; et al. MAGE-A Inhibits Apoptosis in Proliferating Myeloma Cells through Repression of Bax and Maintenance of Survivin. Clin. Cancer. Res. 2011, 17, 4309–4319. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Cheng, S.; Asa, S.L.; Ezzat, S. The Melanoma-Associated Antigen A3 Mediates Fibronectin-Controlled Cancer Progression and Metastasis. Cancer Res. 2008, 68, 8104–8112. [Google Scholar] [CrossRef]
- Yang, F.; Zhou, X.; Miao, X.; Zhang, T.; Hang, X.; Tie, R.; Liu, N.; Tian, F.; Wang, F.; Yuan, J. MAGEC2, an Epithelial-Mesenchymal Transition Inducer, Is Associated with Breast Cancer Metastasis. Breast Cancer Res. Treat. 2014, 145, 23–32. [Google Scholar] [CrossRef]
- Brasseur, F.; Rimoldi, D.; Liénard, D.; Lethé, B.; Carrel, S.; Arienti, F.; Suter, L.; Vanwijck, R.; Bourlond, A.; Humblet, Y. Expression of MAGE Genes in Primary and Metastatic Cutaneous Melanoma. Int. J. Cancer 1995, 63, 375–380. [Google Scholar] [CrossRef]
- Ye, X.; Xie, J.; Huang, H.; Deng, Z. Knockdown of MAGEA6 Activates AMP-Activated Protein Kinase (AMPK) Signaling to Inhibit Human Renal Cell Carcinoma Cells. Cell. Physiol. Biochem. 2018, 45, 1205–1218. [Google Scholar] [CrossRef]
- Pineda, C.T.; Ramanathan, S.; Fon Tacer, K.; Weon, J.L.; Potts, M.B.; Ou, Y.H.; White, M.A.; Potts, P.R. Degradation of AMPK by a Cancer-Specific Ubiquitin Ligase. Cell 2015, 160, 715–728. [Google Scholar] [CrossRef] [PubMed]
- Pineda, C.T.; Potts, P.R. Oncogenic MAGEA-TRIM28 Ubiquitin Ligase Downregulates Autophagy by Ubiquitinating and Degrading AMPK in Cancer. Autophagy 2015, 11, 844–846. [Google Scholar] [CrossRef]
- van Tongelen, A.; Loriot, A.; de Smet, C. Oncogenic Roles of DNA Hypomethylation through the Activation of Cancer-Germline Genes. Cancer Lett. 2017, 396, 130–137. [Google Scholar] [CrossRef]
- Tajima, K.; Obata, Y.; Tamaki, H.; Yoshida, M.; Chen, Y.-T.; Scanlan, M.J.; Old, L.J.; Kuwano, H.; Takahashi, T.; Takahashi, T.; et al. Expression of Cancer/Testis (CT) Antigens in Lung Cancer. Lung Cancer 2003, 42, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Ayyoub, M.; Scarlata, C.-M.; Hamaï, A.; Pignon, P.; Valmori, D. Expression of MAGE-A3/6 in Primary Breast Cancer Is Associated with Hormone Receptor Negative Status, High Histologic Grade, and Poor Survival. J. Immunother. 2014, 37, 73–76. [Google Scholar] [CrossRef]
- Otte, M.; Zafrakas, M.; Riethdorf, L.; Pichlmeier, U.; Löning, T.; Jänicke, F.; Pantel, K. MAGE-A Gene Expression Pattern in Primary Breast Cancer. Cancer Res. 2001, 61, 6682–6687. [Google Scholar] [PubMed]
- Andrade, V.C.C.; Vettore, A.L.; Felix, R.S.; Almeida, M.S.S.; Carvalho, F.; Oliveira, J.S.R.; Chauffaille, M.L.L.F.; Andriolo, A.; Caballero, O.L.; Zago, M.A.; et al. Prognostic Impact of Cancer/Testis Antigen Expression in Advanced Stage Multiple Myeloma Patients. Cancer Immun. 2008, 8, 2. [Google Scholar] [CrossRef]
- Zhao, L.; Jiang, L.; Zhang, M.; Zhang, Q.; Guan, Q.; Li, Y.; He, M.; Zhang, J.; Wei, M. NF-ΚB-Activated SPRY4-IT1 Promotes Cancer Cell Metastasis by Downregulating TCEB1 MRNA via Staufen1-Mediated MRNA Decay. Oncogene 2021, 40, 4919–4929. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Ma, C.; Zhang, Z.; Zhang, H.; Hu, H. NF-ΚB Signaling in Inflammation and Cancer. MedComm 2021, 2, 618–653. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Nag, S.A.; Zhang, R. Targeting the NFκB Signaling Pathways for Breast Cancer Prevention and Therapy. Curr. Med. Chem. 2015, 22, 264–289. [Google Scholar] [CrossRef]
- Harrington, B.S.; Annunziata, C.M. NF-ΚB Signaling in Ovarian Cancer. Cancers 2019, 11, 1182. [Google Scholar] [CrossRef]
- Pan, S.-J.; Ren, J.; Jiang, H.; Liu, W.; Hu, L.-Y.; Pan, Y.-X.; Sun, B.; Sun, Q.-F.; Bian, L.-G. MAGEA6 Promotes Human Glioma Cell Survival via Targeting AMPKα1. Cancer Lett. 2018, 412, 21–29. [Google Scholar] [CrossRef]
- Liu, M.; Li, J.; Wang, Y.; Ghaffar, M.; Yang, Y.; Wang, M.; Li, C. MAGEA6 Positively Regulates MSMO1 and Promotes the Migration and Invasion of Oesophageal Cancer Cells. Exp. Ther. Med. 2022, 23, 204. [Google Scholar] [CrossRef]
- Kendall, S.E.; Battelli, C.; Irwin, S.; Mitchell, J.G.; Glackin, C.A.; Verdi, J.M. NRAGE Mediates P38 Activation and Neural Progenitor Apoptosis via the Bone Morphogenetic Protein Signaling Cascade. Mol. Cell. Biol. 2005, 25, 7711–7724. [Google Scholar] [CrossRef]
- Matluk, N.; Rochira, J.A.; Karaczyn, A.; Adams, T.; Verdi, J.M. A Role for NRAGE in NF-KappaB Activation through the Non-Canonical BMP Pathway. BMC Biol. 2010, 8, 7. [Google Scholar] [CrossRef]
- Rochira, J.A.; Matluk, N.N.; Adams, T.L.; Karaczyn, A.A.; Oxburgh, L.; Hess, S.T.; Verdi, J.M. A Small Peptide Modeled after the NRAGE Repeat Domain Inhibits XIAP-TAB1-TAK1 Signaling for NF-ΚB Activation and Apoptosis in P19 Cells. PLoS ONE 2011, 6, e20659. [Google Scholar] [CrossRef]
- Dolcet, X.; Llobet, D.; Pallares, J.; Matias-Guiu, X. NF-KB in Development and Progression of Human Cancer. Virchows Arch. 2005, 446, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Gaptulbarova, K.A.; Tsyganov, M.M.; Pevzner, A.M.; Ibragimova, M.K.; Litviakov, N. V NF-KB as a Potential Prognostic Marker and a Candidate for Targeted Therapy of Cancer. Exp. Oncol. 2020, 42, 263–269. [Google Scholar] [CrossRef]
- Pires, B.R.B.; Mencalha, A.L.; Ferreira, G.M.; de Souza, W.F.; Morgado-Díaz, J.A.; Maia, A.M.; Corrêa, S.; Abdelhay, E.S.F.W. NF-KappaB Is Involved in the Regulation of EMT Genes in Breast Cancer Cells. PLoS ONE 2017, 12, e0169622. [Google Scholar] [CrossRef]
- Qian, Z.; Zhang, G.; Song, G.; Shi, J.; Gong, L.; Mou, Y.; Han, Y. Integrated Analysis of Genes Associated with Poor Prognosis of Patients with Colorectal Cancer Liver Metastasis. Oncotarget 2017, 8, 25500–25512. [Google Scholar] [CrossRef]
- Laban, S.; Giebel, G.; Klümper, N.; Schröck, A.; Doescher, J.; Spagnoli, G.; Thierauf, J.; Theodoraki, M.-N.; Remark, R.; Gnjatic, S.; et al. MAGE Expression in Head and Neck Squamous Cell Carcinoma Primary Tumors, Lymph Node Metastases and Respective Recurrences-Implications for Immunotherapy. Oncotarget 2017, 8, 14719–14735. [Google Scholar] [CrossRef]
- Alsalloum, A.; Shevchenko, J.A.; Sennikov, S. The Melanoma-Associated Antigen Family A (MAGE-A): A Promising Target for Cancer Immunotherapy? Cancers 2023, 15, 1779. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Sang, M.; Yin, D.; Liu, F.; Wu, Y.; Liu, S.; Huang, W.; Shan, B. MAGE-A Gene Expression in Peripheral Blood Serves as a Poor Prognostic Marker for Patients with Lung Cancer. Thorac. Cancer 2018, 9, 431–438. [Google Scholar] [CrossRef]
- Doyle, J.M.; Gao, J.; Wang, J.; Yang, M.; Potts, P.R. MAGE-RING Protein Complexes Comprise a Family of E3 Ubiquitin Ligases. Mol. Cell 2010, 39, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Powell, E.; Piwnica-Worms, D.; Piwnica-Worms, H. Contribution of P53 to Metastasis. Cancer Discov. 2014, 4, 405–414. [Google Scholar] [CrossRef]
- Kwon, S.; Kang, S.H.; Ro, J.; Jeon, C.-H.; Park, J.-W.; Lee, E.S. The Melanoma Antigen Gene as a Surveillance Marker for the Detection of Circulating Tumor Cells in Patients with Breast Carcinoma. Cancer 2005, 104, 251–256. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brisam, M.; Rauthe, S.; Hartmann, S.; Linz, C.; Brands, R.C.; Kübler, A.C.; Rosenwald, A.; Müller-Richter, U.D. Expression of MAGE-A1-A12 Subgroups in the Invasive Tumor Front and Tumor Center in Oral Squamous Cell Carcinoma. Oncol. Rep. 2016, 35, 1979–1986. [Google Scholar] [CrossRef] [PubMed]
- Mei, A.H.-C.; Tung, K.; Han, J.; Perumal, D.; Laganà, A.; Keats, J.; Auclair, D.; Chari, A.; Jagannath, S.; Parekh, S.; et al. MAGE-A Inhibit Apoptosis and Promote Proliferation in Multiple Myeloma through Regulation of BIM and P21Cip1. Oncotarget 2020, 11, 727–739. [Google Scholar] [CrossRef]
- Deng, L.; Meng, T.; Chen, L.; Wei, W.; Wang, P. The Role of Ubiquitination in Tumorigenesis and Targeted Drug Discovery. Signal Transduct. Target. Ther. 2020, 5, 11. [Google Scholar] [CrossRef]
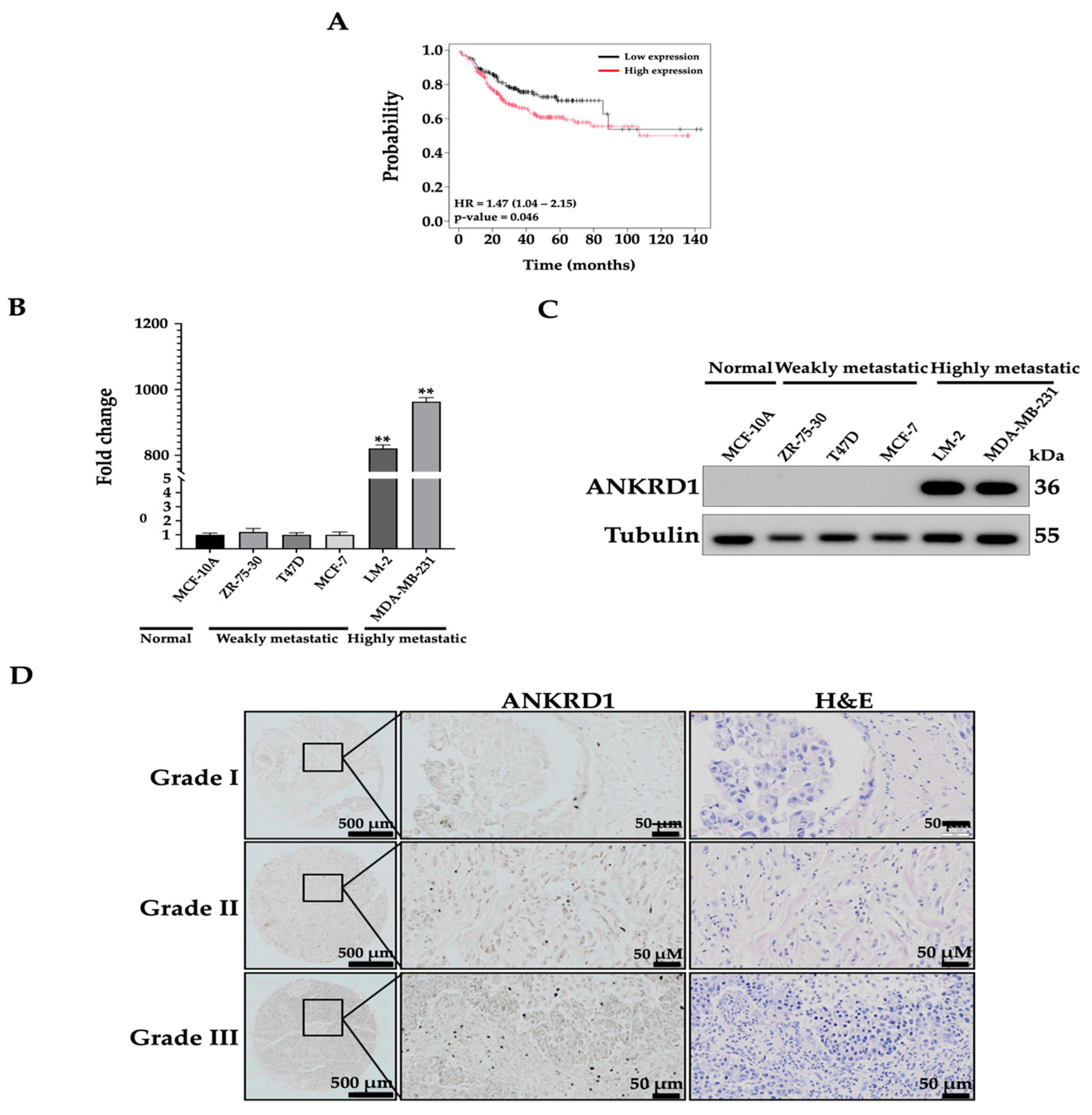

| Grade 1 (Number of Samples = 7) | Grade 2 (Number of Samples = 58) | Grade 3 (Number of Samples = 35) | p-Value | ||||
|---|---|---|---|---|---|---|---|
| Pathologists | ImageJ | Pathologists | ImageJ | Pathologists | ImageJ | ||
| Staining intensity | + | 0.19 ± 0.001 | ++ | 0.65 ± 0.185 | +++ | 16.93 ± 2.523 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diskul-Na-Ayudthaya, P.; Bae, S.J.; Bae, Y.-U.; Van, N.T.; Kim, W.; Ryu, S. ANKRD1 Promotes Breast Cancer Metastasis by Activating NF-κB-MAGE-A6 Pathway. Cancers 2024, 16, 3306. https://doi.org/10.3390/cancers16193306
Diskul-Na-Ayudthaya P, Bae SJ, Bae Y-U, Van NT, Kim W, Ryu S. ANKRD1 Promotes Breast Cancer Metastasis by Activating NF-κB-MAGE-A6 Pathway. Cancers. 2024; 16(19):3306. https://doi.org/10.3390/cancers16193306
Chicago/Turabian StyleDiskul-Na-Ayudthaya, Penchatr, Seon Joo Bae, Yun-Ui Bae, Ngu Trinh Van, Wootae Kim, and Seongho Ryu. 2024. "ANKRD1 Promotes Breast Cancer Metastasis by Activating NF-κB-MAGE-A6 Pathway" Cancers 16, no. 19: 3306. https://doi.org/10.3390/cancers16193306
APA StyleDiskul-Na-Ayudthaya, P., Bae, S. J., Bae, Y.-U., Van, N. T., Kim, W., & Ryu, S. (2024). ANKRD1 Promotes Breast Cancer Metastasis by Activating NF-κB-MAGE-A6 Pathway. Cancers, 16(19), 3306. https://doi.org/10.3390/cancers16193306







