Long Non-Coding RNAs as Regulators for Targeting Breast Cancer Stem Cells and Tumor Immune Microenvironment: Biological Properties and Therapeutic Potential
Abstract
:Simple Summary
Abstract
1. Introduction
2. BCSC and TIME
2.1. BCSC Markers
2.2. Role of TIME in CSCs
2.2.1. Hypoxia
2.2.2. Stromal Cell Interactions
3. Biological Characteristics and Mechanisms of LncRNAs
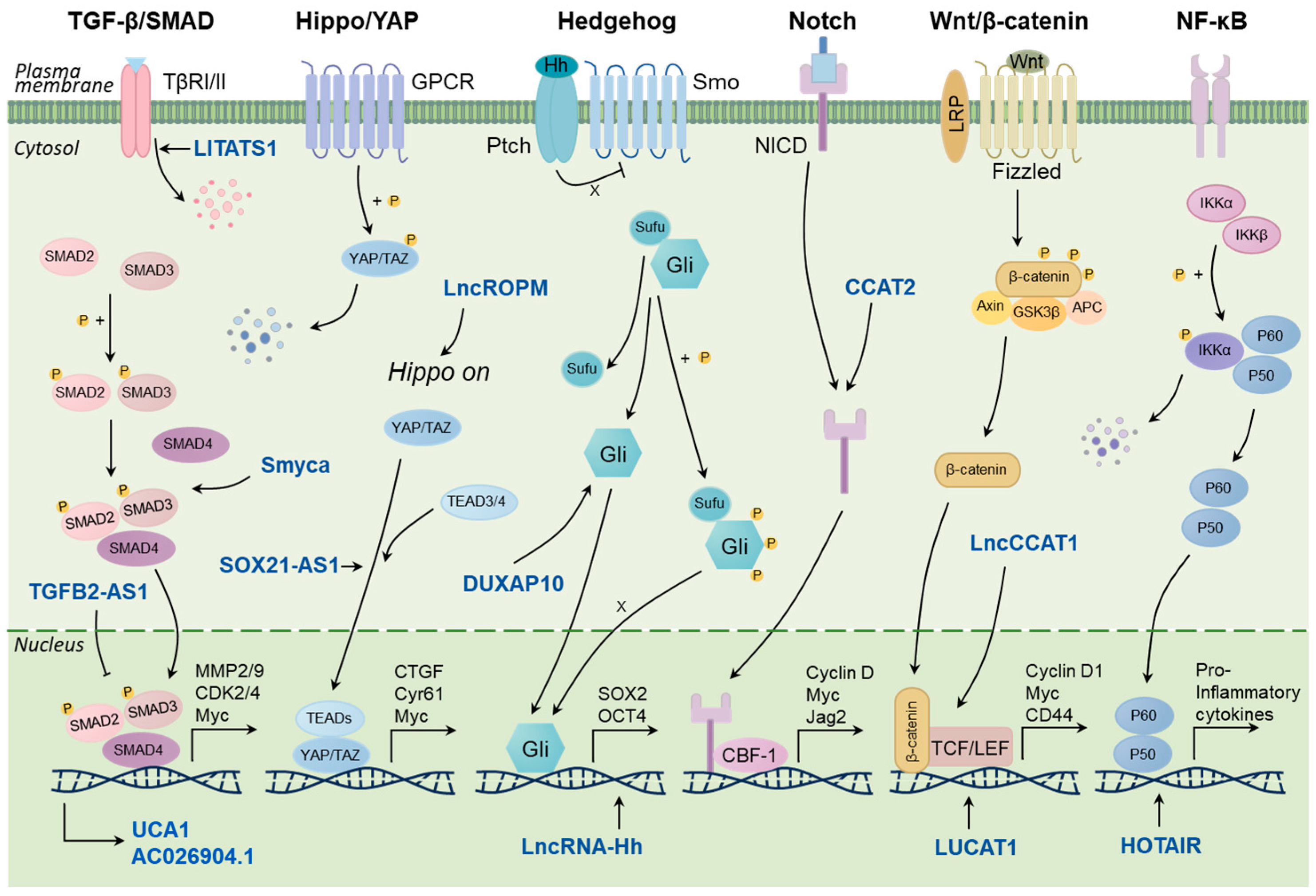
4. LncRNAs and Regulatory Pathways Associated with BCSC
4.1. TGF-β/Smad Signaling Pathway
4.2. Hippo/YAP Signaling Pathway
4.3. Hedgehog Signaling Pathway
4.4. Other Signaling Pathways
5. Immune-Specific LncRNAs in the TIME
5.1. CAFs
5.2. TAMs
5.3. Myeloid-Derived Suppressor Cells (MDSCs)
5.4. Regulatory T Cells (Tregs)
5.5. Natural Killer Cells (NKs)
6. Clinical Implications of lncRNAs in BCSC and TIME
6.1. Prognostic Value of Stemness-Related LncRNAs
6.2. LncRNAs Impact Sensitivity to Immunotherapy
6.3. Strategies of Eliminating CSCs and Regulating TIME by Targeting LncRNAs
7. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Korkaya, H.; Kim, G.I.; Davis, A.; Malik, F.; Henry, N.L.; Ithimakin, S.; Quraishi, A.A.; Tawakkol, N.; D’Angelo, R.; Paulson, A.K.; et al. Activation of an IL6 inflammatory loop mediates trastuzumab resistance in HER2+ breast cancer by expanding the cancer stem cell population. Mol. Cell 2012, 47, 570–584. [Google Scholar] [CrossRef]
- Beziaud, L.; Young, C.M.; Alonso, A.M.; Norkin, M.; Minafra, A.R.; Huelsken, J. IFNgamma-induced stem-like state of cancer cells as a driver of metastatic progression following immunotherapy. Cell Stem Cell 2023, 30, 818–831.e6. [Google Scholar] [CrossRef] [PubMed]
- Postepska-Igielska, A.; Giwojna, A.; Gasri-Plotnitsky, L.; Schmitt, N.; Dold, A.; Ginsberg, D.; Grummt, I. LncRNA Khps1 Regulates Expression of the Proto-oncogene SPHK1 via Triplex-Mediated Changes in Chromatin Structure. Mol. Cell 2015, 60, 626–636. [Google Scholar] [CrossRef] [PubMed]
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016, 539, 452–455. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Hu, J.; Zou, K.; Ye, M.; Chen, Y.; Wu, C.; Chen, X.; Han, M. Activation of LncRNA TINCR by H3K27 acetylation promotes Trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast Cancer. Mol. Cancer 2019, 18, 3. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hou, P.; Fan, D.; Dong, M.; Ma, M.; Li, H.; Yao, R.; Li, Y.; Wang, G.; Geng, P.; et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017, 24, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Yang, L.; Zou, Y.; Liang, J.Y.; Liu, P.; Gao, G.; Yang, A.; Tang, H.; Xie, X. Long non-coding RNA HUMT hypomethylation promotes lymphangiogenesis and metastasis via activating FOXK1 transcription in triple-negative breast cancer. J. Hematol. Oncol. 2020, 13, 17. [Google Scholar] [CrossRef]
- Jin, X.; Ge, L.P.; Li, D.Q.; Shao, Z.M.; Di, G.H.; Xu, X.E.; Jiang, Y.Z. LncRNA TROJAN promotes proliferation and resistance to CDK4/6 inhibitor via CDK2 transcriptional activation in ER+ breast cancer. Mol. Cancer 2020, 19, 87. [Google Scholar] [CrossRef]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef]
- Wright, M.H.; Calcagno, A.M.; Salcido, C.D.; Carlson, M.D.; Ambudkar, S.V.; Varticovski, L. Brca1 breast tumors contain distinct CD44+/CD24− and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008, 10, R10. [Google Scholar] [CrossRef] [PubMed]
- Vassilopoulos, A.; Chisholm, C.; Lahusen, T.; Zheng, H.; Deng, C.X. A critical role of CD29 and CD49f in mediating metastasis for cancer-initiating cells isolated from a Brca1-associated mouse model of breast cancer. Oncogene 2014, 33, 5477–5482. [Google Scholar] [CrossRef] [PubMed]
- Vaillant, F.; Asselin-Labat, M.L.; Shackleton, M.; Forrest, N.C.; Lindeman, G.J.; Visvader, J.E. The mammary progenitor marker CD61/beta3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 2008, 68, 7711–7717. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Xu, J.; Tang, L.; Guan, X. Breast cancer stem cell: The roles and therapeutic implications. Cell. Mol. Life Sci. 2017, 74, 951–966. [Google Scholar] [CrossRef] [PubMed]
- Badve, S.; Nakshatri, H. Breast-cancer stem cells-beyond semantics. Lancet Oncol. 2012, 13, e43–e48. [Google Scholar] [CrossRef] [PubMed]
- Heddleston, J.M.; Li, Z.; McLendon, R.E.; Hjelmeland, A.B.; Rich, J.N. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle 2009, 8, 3274–3284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Samanta, D.; Lu, H.; Bullen, J.W.; Zhang, H.; Chen, I.; He, X.; Semenza, G.L. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. USA 2016, 113, E2047–E2056. [Google Scholar] [CrossRef]
- van den Beucken, T.; Koch, E.; Chu, K.; Rupaimoole, R.; Prickaerts, P.; Adriaens, M.; Voncken, J.W.; Harris, A.L.; Buffa, F.M.; Haider, S.; et al. Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nat. Commun. 2014, 5, 5203. [Google Scholar] [CrossRef]
- Samanta, D.; Gilkes, D.M.; Chaturvedi, P.; Xiang, L.; Semenza, G.L. Hypoxia-inducible factors are required for chemotherapy resistance of breast cancer stem cells. Proc. Natl. Acad. Sci. USA 2014, 111, E5429–E5438. [Google Scholar] [CrossRef]
- Su, S.; Chen, J.; Yao, H.; Liu, J.; Yu, S.; Lao, L.; Wang, M.; Luo, M.; Xing, Y.; Chen, F.; et al. CD10(+)GPR77(+) Cancer-Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness. Cell 2018, 172, 841–856.e16. [Google Scholar] [CrossRef]
- Yu, Y.; Xiao, C.H.; Tan, L.D.; Wang, Q.S.; Li, X.Q.; Feng, Y.M. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of breast cancer cells through paracrine TGF-beta signalling. Br. J. Cancer 2014, 110, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Pelon, F.; Bourachot, B.; Kieffer, Y.; Magagna, I.; Mermet-Meillon, F.; Bonnet, I.; Costa, A.; Givel, A.M.; Attieh, Y.; Barbazan, J.; et al. Cancer-associated fibroblast heterogeneity in axillary lymph nodes drives metastases in breast cancer through complementary mechanisms. Nat. Commun. 2020, 11, 404. [Google Scholar] [CrossRef] [PubMed]
- Valenti, G.; Quinn, H.M.; Heynen, G.; Lan, L.; Holland, J.D.; Vogel, R.; Wulf-Goldenberg, A.; Birchmeier, W. Cancer Stem Cells Regulate Cancer-Associated Fibroblasts via Activation of Hedgehog Signaling in Mammary Gland Tumors. Cancer Res. 2017, 77, 2134–2147. [Google Scholar] [CrossRef] [PubMed]
- Bartoschek, M.; Oskolkov, N.; Bocci, M.; Lovrot, J.; Larsson, C.; Sommarin, M.; Madsen, C.D.; Lindgren, D.; Pekar, G.; Karlsson, G.; et al. Spatially and functionally distinct subclasses of breast cancer-associated fibroblasts revealed by single cell RNA sequencing. Nat. Commun. 2018, 9, 5150. [Google Scholar] [CrossRef]
- Costa, A.; Kieffer, Y.; Scholer-Dahirel, A.; Pelon, F.; Bourachot, B.; Cardon, M.; Sirven, P.; Magagna, I.; Fuhrmann, L.; Bernard, C.; et al. Fibroblast Heterogeneity and Immunosuppressive Environment in Human Breast Cancer. Cancer Cell 2018, 33, 463–479.e410. [Google Scholar] [CrossRef]
- Okuda, H.; Kobayashi, A.; Xia, B.; Watabe, M.; Pai, S.K.; Hirota, S.; Xing, F.; Liu, W.; Pandey, P.R.; Fukuda, K.; et al. Hyaluronan synthase HAS2 promotes tumor progression in bone by stimulating the interaction of breast cancer stem-like cells with macrophages and stromal cells. Cancer Res. 2012, 72, 537–547. [Google Scholar] [CrossRef]
- Radharani, N.N.V.; Yadav, A.S.; Nimma, R.; Kumar, T.V.S.; Bulbule, A.; Chanukuppa, V.; Kumar, D.; Patnaik, S.; Rapole, S.; Kundu, G.C. Tumor-associated macrophage derived IL-6 enriches cancer stem cell population and promotes breast tumor progression via Stat-3 pathway. Cancer Cell Int. 2022, 22, 122. [Google Scholar] [CrossRef]
- Chen, X.; Yang, M.; Yin, J.; Li, P.; Zeng, S.; Zheng, G.; He, Z.; Liu, H.; Wang, Q.; Zhang, F.; et al. Tumor-associated macrophages promote epithelial-mesenchymal transition and the cancer stem cell properties in triple-negative breast cancer through CCL2/AKT/beta-catenin signaling. Cell Commun. Signal. 2022, 20, 92. [Google Scholar] [CrossRef]
- Chin, A.R.; Wang, S.E. Cytokines driving breast cancer stemness. Mol. Cell. Endocrinol. 2014, 382, 598–602. [Google Scholar] [CrossRef]
- Li, C.; Qiu, S.; Liu, X.; Guo, F.; Zhai, J.; Li, Z.; Deng, L.; Ge, L.; Qian, H.; Yang, L.; et al. Extracellular matrix-derived mechanical force governs breast cancer cell stemness and quiescence transition through integrin-DDR signaling. Signal Transduct. Target. Ther. 2023, 8, 247. [Google Scholar] [CrossRef]
- Wu, Y.; Liang, H.; Luo, A.; Li, Y.; Liu, Z.; Li, X.; Li, W.; Liang, K.; Li, J.; Liu, Z.; et al. Gelatin-based 3D biomimetic scaffolds platform potentiates culture of cancer stem cells in esophageal squamous cell carcinoma. Biomaterials 2023, 302, 122323. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Hainer, S.J.; Pruneski, J.A.; Mitchell, R.D.; Monteverde, R.M.; Martens, J.A. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 2011, 25, 29–40. [Google Scholar] [CrossRef]
- Zhou, C.; Wang, D.; Li, J.; Wang, Q.; Wo, L.; Zhang, X.; Hu, Z.; Wang, Z.; Zhan, M.; He, M.; et al. TGFB2-AS1 inhibits triple-negative breast cancer progression via interaction with SMARCA4 and regulating its targets TGFB2 and SOX2. Proc. Natl. Acad. Sci. USA 2022, 119, e2117988119. [Google Scholar] [CrossRef]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Wu, F.; Li, Y.; Qin, C.; Zhao, Y.; Xie, M.; Dai, H.; Yao, H.; Cai, H.; Wang, Q.; et al. MALAT1 modulates alternative splicing by cooperating with the splicing factors PTBP1 and PSF. Sci. Adv. 2022, 8, eabq7289. [Google Scholar] [CrossRef]
- Maida, Y.; Yasukawa, M.; Furuuchi, M.; Lassmann, T.; Possemato, R.; Okamoto, N.; Kasim, V.; Hayashizaki, Y.; Hahn, W.C.; Masutomi, K. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature 2009, 461, 230–235. [Google Scholar] [CrossRef]
- Smola, M.J.; Christy, T.W.; Inoue, K.; Nicholson, C.O.; Friedersdorf, M.; Keene, J.D.; Lee, D.M.; Calabrese, J.M.; Weeks, K.M. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc. Natl. Acad. Sci. USA 2016, 113, 10322–10327. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Cui, X.; Tan, Y.; Wang, Q.; Wang, Y.; Xu, C.; Fang, C.; Kang, C. LncRNA PRADX-mediated recruitment of PRC2/DDX5 complex suppresses UBXN1 expression and activates NF-kappaB activity, promoting tumorigenesis. Theranostics 2021, 11, 4516–4530. [Google Scholar] [CrossRef] [PubMed]
- Michelini, F.; Pitchiaya, S.; Vitelli, V.; Sharma, S.; Gioia, U.; Pessina, F.; Cabrini, M.; Wang, Y.; Capozzo, I.; Iannelli, F.; et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 2017, 19, 1400–1411. [Google Scholar] [CrossRef] [PubMed]
- Yeh, H.W.; Hsu, E.C.; Lee, S.S.; Lang, Y.D.; Lin, Y.C.; Chang, C.Y.; Lee, S.Y.; Gu, D.L.; Shih, J.H.; Ho, C.M.; et al. PSPC1 mediates TGF-beta1 autocrine signalling and Smad2/3 target switching to promote EMT, stemness and metastasis. Nat. Cell Biol. 2018, 20, 479–491. [Google Scholar] [CrossRef]
- Hou, L.; Tu, J.; Cheng, F.; Yang, H.; Yu, F.; Wang, M.; Liu, J.; Fan, J.; Zhou, G. Long noncoding RNA ROR promotes breast cancer by regulating the TGF-beta pathway. Cancer Cell Int. 2018, 18, 142. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Chan, S.J.; Liu, X.; Wei, A.C.; Jian, R.I.; Huang, K.W.; Lang, Y.D.; Shih, J.H.; Liao, C.C.; Luan, C.L.; et al. Long noncoding RNA Smyca coactivates TGF-beta/Smad and Myc pathways to drive tumor progression. J. Hematol. Oncol. 2022, 15, 85. [Google Scholar] [CrossRef]
- Li, G.Y.; Wang, W.; Sun, J.Y.; Xin, B.; Zhang, X.; Wang, T.; Zhang, Q.F.; Yao, L.B.; Han, H.; Fan, D.M.; et al. Long non-coding RNAs AC026904.1 and UCA1: A “one-two punch” for TGF-beta-induced SNAI2 activation and epithelial-mesenchymal transition in breast cancer. Theranostics 2018, 8, 2846–2861. [Google Scholar] [CrossRef]
- Fan, C.; Wang, Q.; Kuipers, T.B.; Cats, D.; Iyengar, P.V.; Hagenaars, S.C.; Mesker, W.E.; Devilee, P.; Tollenaar, R.; Mei, H.; et al. LncRNA LITATS1 suppresses TGF-beta-induced EMT and cancer cell plasticity by potentiating TbetaRI degradation. EMBO J. 2023, 42, e112806. [Google Scholar] [CrossRef]
- Maugeri-Sacca, M.; De Maria, R. Hippo pathway and breast cancer stem cells. Crit. Rev. Oncol. Hematol. 2016, 99, 115–122. [Google Scholar] [CrossRef]
- Cordenonsi, M.; Zanconato, F.; Azzolin, L.; Forcato, M.; Rosato, A.; Frasson, C.; Inui, M.; Montagner, M.; Parenti, A.R.; Poletti, A.; et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 2011, 147, 759–772. [Google Scholar] [CrossRef]
- Cheng, Z.; Liang, X.; Zhang, C.; Wang, R.; Wei, T.; Ning, B.; Poreba, E.; Li, L.; Wang, H.; Ding, J. SOX9-transactived long non-coding RNA NEAT1 promotes the self-renewal of liver cancer stem cells through PKA/Hippo signaling. Signal Transduct. Target. Ther. 2021, 6, 87. [Google Scholar] [CrossRef]
- Liu, S.; Sun, Y.; Hou, Y.; Yang, L.; Wan, X.; Qin, Y.; Liu, Y.; Wang, R.; Zhu, P.; Teng, Y.; et al. A novel lncRNA ROPM-mediated lipid metabolism governs breast cancer stem cell properties. J. Hematol. Oncol. 2021, 14, 178. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Meng, D.; Wang, R. Long non-coding RNA SOX21-AS1 enhances the stemness of breast cancer cells via the Hippo pathway. FEBS Open Bio 2021, 11, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.P.; Wang, Z.; Yang, C. LncRNA DUXAP10 Upregulation and the Hedgehog Pathway Activation Are Critically Involved in Chronic Cadmium Exposure-Induced Cancer Stem Cell-Like Property. Toxicol. Sci. 2021, 184, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Hou, Y.; Yang, G.; Zhang, H.; Tu, G.; Du, Y.E.; Wen, S.; Xu, L.; Tang, X.; Tang, S.; et al. LncRNA-Hh Strengthen Cancer Stem Cells Generation in Twist-Positive Breast Cancer via Activation of Hedgehog Signaling Pathway. Stem Cells 2016, 34, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Takebe, N.; Harris, P.J.; Warren, R.Q.; Ivy, S.P. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat. Rev. Clin. Oncol. 2011, 8, 97–106. [Google Scholar] [CrossRef]
- D’Angelo, R.C.; Ouzounova, M.; Davis, A.; Choi, D.; Tchuenkam, S.M.; Kim, G.; Luther, T.; Quraishi, A.A.; Senbabaoglu, Y.; Conley, S.J.; et al. Notch reporter activity in breast cancer cell lines identifies a subset of cells with stem cell activity. Mol. Cancer Ther. 2015, 14, 779–787. [Google Scholar] [CrossRef]
- Xu, Z.; Liu, C.; Zhao, Q.; Lu, J.; Ding, X.; Luo, A.; He, J.; Wang, G.; Li, Y.; Cai, Z.; et al. Long non-coding RNA CCAT2 promotes oncogenesis in triple-negative breast cancer by regulating stemness of cancer cells. Pharmacol. Res. 2020, 152, 104628. [Google Scholar] [CrossRef]
- Tang, T.; Guo, C.; Xia, T.; Zhang, R.; Zen, K.; Pan, Y.; Jin, L. LncCCAT1 Promotes Breast Cancer Stem Cell Function through Activating WNT/beta-catenin Signaling. Theranostics 2019, 9, 7384–7402. [Google Scholar] [CrossRef]
- Zheng, A.; Song, X.; Zhang, L.; Zhao, L.; Mao, X.; Wei, M.; Jin, F. Long non-coding RNA LUCAT1/miR-5582-3p/TCF7L2 axis regulates breast cancer stemness via Wnt/beta-catenin pathway. J. Exp. Clin. Cancer Res. 2019, 38, 305. [Google Scholar] [CrossRef]
- Raju, G.S.R.; Pavitra, E.; Bandaru, S.S.; Varaprasad, G.L.; Nagaraju, G.P.; Malla, R.R.; Huh, Y.S.; Han, Y.K. HOTAIR: A potential metastatic, drug-resistant and prognostic regulator of breast cancer. Mol. Cancer 2023, 22, 65. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Li, P.; Wang, J.; Shu, Y.; Zhong, X.; Gao, Z.; Yang, J.; Jiang, Y.; Zhou, X.; et al. Long noncoding RNA HOTAIR regulates the stemness of breast cancer cells via activation of the NF-kappaB signaling pathway. J. Biol. Chem. 2022, 298, 102630. [Google Scholar] [CrossRef]
- Fiori, M.E.; Di Franco, S.; Villanova, L.; Bianca, P.; Stassi, G.; De Maria, R. Cancer-associated fibroblasts as abettors of tumor progression at the crossroads of EMT and therapy resistance. Mol. Cancer 2019, 18, 70. [Google Scholar] [CrossRef] [PubMed]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Yang, C.; Chen, X.; Sun, H.; He, X.; Wang, W. Breast cancer-derived exosomal lncRNA SNHG14 induces normal fibroblast activation to cancer-associated fibroblasts via the EBF1/FAM171A1 axis. Breast Cancer 2023, 30, 1028–1040. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yu, J.; Lv, C.; Luo, Z. Cancer-associated fibroblasts-derived lncRNA signature as a putative biomarker in breast cancer. Front. Oncol. 2022, 12, 1028664. [Google Scholar] [CrossRef] [PubMed]
- Chanmee, T.; Ontong, P.; Konno, K.; Itano, N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers 2014, 6, 1670–1690. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Song, X.; Li, Y.; Chen, B.; Zhao, W.; Wang, L.; Zhang, H.; Liu, Y.; Han, D.; Zhang, N.; et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol. Cancer 2020, 19, 85. [Google Scholar] [CrossRef]
- Zhao, Y.; Yu, Z.; Ma, R.; Zhang, Y.; Zhao, L.; Yan, Y.; Lv, X.; Zhang, L.; Su, P.; Bi, J.; et al. lncRNA-Xist/miR-101-3p/KLF6/C/EBPalpha axis promotes TAM polarization to regulate cancer cell proliferation and migration. Mol. Ther. Nucleic Acids 2021, 23, 536–551. [Google Scholar] [CrossRef]
- Liu, S.Q.; Zhou, Z.Y.; Dong, X.; Guo, L.; Zhang, K.J. LncRNA GNAS-AS1 facilitates ER+ breast cancer cells progression by promoting M2 macrophage polarization via regulating miR-433-3p/GATA3 axis. Biosci. Rep. 2020, 40, BSR20200626. [Google Scholar] [CrossRef]
- Tao, S.; Chen, Q.; Lin, C.; Dong, H. Linc00514 promotes breast cancer metastasis and M2 polarization of tumor-associated macrophages via Jagged1-mediated notch signaling pathway. J. Exp. Clin. Cancer Res. 2020, 39, 191. [Google Scholar] [CrossRef]
- Marvel, D.; Gabrilovich, D.I. Myeloid-derived suppressor cells in the tumor microenvironment: Expect the unexpected. J. Clin. Investig. 2015, 125, 3356–3364. [Google Scholar] [CrossRef] [PubMed]
- Di Mitri, D.; Toso, A.; Alimonti, A. Molecular Pathways: Targeting Tumor-Infiltrating Myeloid-Derived Suppressor Cells for Cancer Therapy. Clin. Cancer Res. 2015, 21, 3108–3112. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Sun, W.; Shang, W.; Li, Y.; Zhang, D.; Wang, T.; Zhang, X.; Zhang, S.; Zhang, Y.; Yang, R. Lnc-C/EBPbeta Negatively Regulates the Suppressive Function of Myeloid-Derived Suppressor Cells. Cancer Immunol. Res. 2018, 6, 1352–1363. [Google Scholar] [CrossRef] [PubMed]
- Adewunmi, O.; Shen, Y.; Zhang, X.H.; Rosen, J.M. Targeted inhibition of lncRNA Malat1 alters the tumor immune microenvironment in preclinical syngeneic mouse models of triple negative breast cancer. Cancer Immunol. Res. 2023, 11, 1462–1479. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, Y.; Nishikawa, H. Roles of regulatory T cells in cancer immunity. Int. Immunol. 2016, 28, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Moallemi-Rad, L.; Ghorbani, A.; Dadyar, M.; Hussen, B.M.; Rasul, M.F.; Eslami, S.; Taheri, M.; Jamali, E.; Ghafouri-Fard, S. Expression of Treg-associated lncRNAs in breast cancer. Pathol. Res. Pract. 2023, 241, 154270. [Google Scholar] [CrossRef]
- Qiu, P.; Guo, Q.; Lin, J.; Pan, K.; Chen, J.; Ding, M. An exosome-related long non-coding RNAs risk model could predict survival outcomes in patients with breast cancer. Sci. Rep. 2022, 12, 22322. [Google Scholar] [CrossRef]
- Ni, C.; Fang, Q.Q.; Chen, W.Z.; Jiang, J.X.; Jiang, Z.; Ye, J.; Zhang, T.; Yang, L.; Meng, F.B.; Xia, W.J.; et al. Breast cancer-derived exosomes transmit lncRNA SNHG16 to induce CD73+γδ1 Treg cells. Signal Transduct. Target. Ther. 2020, 5, 41. [Google Scholar] [CrossRef]
- Shimasaki, N.; Jain, A.; Campana, D. NK cells for cancer immunotherapy. Nat. Rev. Drug Discov. 2020, 19, 200–218. [Google Scholar] [CrossRef]
- Zhang, K.J.; Tan, X.L.; Guo, L. LncRNA TYMSOS facilitates breast cancer metastasis and immune escape through downregulating ULBP3. iScience 2023, 26, 107556. [Google Scholar] [CrossRef]
- Mekky, R.Y.; Ragab, M.F.; Manie, T.; Attia, A.A.; Youness, R.A. MALAT-1: Immunomodulatory lncRNA hampering the innate and the adaptive immune arms in triple negative breast cancer. Transl. Oncol. 2023, 31, 101653. [Google Scholar] [CrossRef]
- Yin, J.Y.; Zhou, Y.; Ding, X.M.; Gong, R.Z.; Zhou, Y.; Hu, H.Y.; Liu, Y.; Lv, X.B.; Zhang, B. UCA1 inhibits NKG2D-mediated cytotoxicity of NK cells to breast cancer. Curr. Cancer Drug Targets 2023, 24, 204–219. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Liu, X.; Zhou, S.; Li, W.; Liu, C.; Chadwick, M.; Qian, C. Long non-coding RNA FGF13-AS1 inhibits glycolysis and stemness properties of breast cancer cells through FGF13-AS1/IGF2BPs/Myc feedback loop. Cancer Lett. 2019, 450, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; He, F.; Hou, Y.; Tu, G.; Li, Q.; Jin, T.; Zeng, H.; Qin, Y.; Wan, X.; Qiao, Y.; et al. A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene 2021, 40, 1609–1627. [Google Scholar] [CrossRef]
- Qin, Y.; Hou, Y.; Liu, S.; Zhu, P.; Wan, X.; Zhao, M.; Peng, M.; Zeng, H.; Li, Q.; Jin, T.; et al. A Novel Long Non-Coding RNA lnc030 Maintains Breast Cancer Stem Cell Stemness by Stabilizing SQLE mRNA and Increasing Cholesterol Synthesis. Adv. Sci. 2021, 8, 2002232. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Li, Y.; Ma, Y.; Lu, J.; Chen, Y.; Jiang, Q.; Qin, Q.; Zhao, L.; Huang, Q.; Luo, Z.; et al. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J. Exp. Clin. Cancer Res. 2018, 37, 289. [Google Scholar] [CrossRef]
- Ma, Y.; Zhu, Y.; Shang, L.; Qiu, Y.; Shen, N.; Wang, J.; Adam, T.; Wei, W.; Song, Q.; Li, J.; et al. LncRNA XIST regulates breast cancer stem cells by activating proinflammatory IL-6/STAT3 signaling. Oncogene 2023, 42, 1419–1437. [Google Scholar] [CrossRef]
- Schouten, P.C.; Vollebergh, M.A.; Opdam, M.; Jonkers, M.; Loden, M.; Wesseling, J.; Hauptmann, M.; Linn, S.C. High XIST and Low 53BP1 Expression Predict Poor Outcome after High-Dose Alkylating Chemotherapy in Patients with a BRCA1-like Breast Cancer. Mol. Cancer Ther. 2016, 15, 190–198. [Google Scholar] [CrossRef]
- Li, Z.H.; Yu, N.S.; Deng, Q.; Zhang, Y.; Hu, Y.Y.; Liu, G.; Huang, K. LncRNA SNHG7 Mediates the Chemoresistance and Stemness of Breast Cancer by Sponging miR-34a. Front. Oncol. 2020, 10, 592757. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Yu, X.; Jin, F. Identification and validation of stemness-related lncRNA prognostic signature for breast cancer. J. Transl. Med. 2020, 18, 331. [Google Scholar] [CrossRef]
- Liu, L.; Wang, Q.; Qiu, Z.; Kang, Y.; Liu, J.; Ning, S.; Yin, Y.; Pang, D.; Xu, S. Noncoding RNAs: The shot callers in tumor immune escape. Signal Transduct. Target. Ther. 2020, 5, 102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, N.; Song, P.; Fu, Y.; Ren, Y.; Li, Z.; Wang, J. LncRNA GATA3-AS1 facilitates tumour progression and immune escape in triple-negative breast cancer through destabilization of GATA3 but stabilization of PD-L1. Cell Prolif. 2020, 53, e12855. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, G.; Ma, X.; Liu, L.; Liu, J.; Yin, Y.; Li, H.; Chen, Y.; Zhang, X.; Zhang, L.; et al. LncRNA TINCR impairs the efficacy of immunotherapy against breast cancer by recruiting DNMT1 and downregulating MiR-199a-5p via the STAT1-TINCR-USP20-PD-L1 axis. Cell Death Dis. 2023, 14, 76. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Ye, Y.; Chan, L.C.; Li, Y.; Liang, K.; Lin, A.; Egranov, S.D.; Zhang, Y.; Xia, W.; Gong, J.; et al. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat. Immunol. 2019, 20, 835–851. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Wang, J.H.; Fan, W.J.; Meng, Y.T.; Li, M.M.; Li, T.T.; Cui, B.; Wang, H.F.; Zhao, Y.; An, F.; et al. Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene 2018, 37, 1062–1074. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, L.; Fan, P.; Bauer, N.; Gladkich, J.; Ryschich, E.; Bazhin, A.V.; Giese, N.A.; Strobel, O.; Hackert, T.; et al. Aspirin counteracts cancer stem cell features, desmoplasia and gemcitabine resistance in pancreatic cancer. Oncotarget 2015, 6, 9999–10015. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Liu, J.; Ben, Q.; Qu, Y.; Li, M.; Wang, Y.; Chen, W.; Zhang, J. The aspirin-induced long non-coding RNA OLA1P2 blocks phosphorylated STAT3 homodimer formation. Genome Biol. 2016, 17, 24. [Google Scholar] [CrossRef]
- Ko, Y.C.; Choi, H.S.; Liu, R.; Kim, J.H.; Kim, S.L.; Yun, B.S.; Lee, D.S. Inhibitory Effects of Tangeretin, A Citrus Peel-Derived Flavonoid, on Breast Cancer Stem Cell Formation through Suppression of Stat3 Signaling. Molecules 2020, 25, 2599. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, F.; Chen, L.; Yang, Y.; Cao, S.; Ye, Y.; Wang, X.; Mu, J.; Li, Z.; Li, L. Blockage of TGFbeta-SMAD2 by demethylation-activated miR-148a is involved in caffeic acid-induced inhibition of cancer stem cell-like properties in vitro and in vivo. FEBS Open Bio 2015, 5, 466–475. [Google Scholar] [CrossRef]
- Jiang, F.; Li, Y.; Mu, J.; Hu, C.; Zhou, M.; Wang, X.; Si, L.; Ning, S.; Li, Z. Glabridin inhibits cancer stem cell-like properties of human breast cancer cells: An epigenetic regulation of miR-148a/SMAd2 signaling. Mol. Carcinog. 2016, 55, 929–940. [Google Scholar] [CrossRef]
- Sakunrangsit, N.; Ketchart, W. Plumbagin inhibits cancer stem-like cells, angiogenesis and suppresses cell proliferation and invasion by targeting Wnt/beta-catenin pathway in endocrine resistant breast cancer. Pharmacol. Res. 2019, 150, 104517. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, R.; Yokota, T. Knocking Down Long Noncoding RNAs Using Antisense Oligonucleotide Gapmers. Methods Mol. Biol. 2020, 2176, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Roux, B.T.; Lindsay, M.A.; Heward, J.A. Knockdown of Nuclear-Located Enhancer RNAs and Long ncRNAs Using Locked Nucleic Acid GapmeRs. Methods Mol. Biol. 2017, 1468, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, M.; Yanagawa, N. Knockdown of Nuclear lncRNAs by Locked Nucleic Acid (LNA) Gapmers in Nephron Progenitor Cells. Methods Mol. Biol. 2020, 2161, 29–36. [Google Scholar] [CrossRef]
- He, Y.; Jiang, X.; Duan, L.; Xiong, Q.; Yuan, Y.; Liu, P.; Jiang, L.; Shen, Q.; Zhao, S.; Yang, C.; et al. LncRNA PKMYT1AR promotes cancer stem cell maintenance in non-small cell lung cancer via activating Wnt signaling pathway. Mol. Cancer 2021, 20, 156. [Google Scholar] [CrossRef]
- Park, E.G.; Pyo, S.J.; Cui, Y.; Yoon, S.H.; Nam, J.W. Tumor immune microenvironment lncRNAs. Brief Bioinform. 2022, 23, bbab504. [Google Scholar] [CrossRef]

| LncRNA | Alterations | Targets | Functions | Prognostic Value |
|---|---|---|---|---|
| TGFB2-AS1 [36] | Downregulated in MDA-231-LM2 cells (showing enhanced lung metastatic activity compared to the parental MDA-231 cells) | TGFβ2, SOX2 | Attenuate BCSC self-renewal activity and inhibit the malignant character | Correlated with better outcome |
| FGF13-AS1 [83] | Downregulated in breast cancer cells | IGF2BPs, MYC | Inhibit breast cancer cells proliferation, invasion, metastasis, glycolysis and stemness features | Correlated with better outcome |
| LncROPM [51] | Upregulated in BCSCs | PLA2G16 | Promote BCSC stemness by regulating PLA2G16-mediated phospholipid metabolism | Correlated with poor outcome |
| KB-1980E6.3 [84] | Upregulated in hypoxic breast cancer cells | IGF2BP1, c-MYC | Maintain the stemness of BCSCs | Correlated with poor outcome |
| Lnc030 [85] | Upregulated in BCSCs | SQLE | Maintain BCSC stemness by stabilizing SQLE mRNA and increasing cholesterol synthesis | Correlated with poor outcome |
| LINC00511 [86] | Upregulated in breast cancer cells | miR-185-3p/E2F1/NANOG | Promote proliferation, invasion, maintenance of BCSC features | Correlated with poor outcome |
| XIST [87,88] | Downregulated in breast cancer cells | let-7a-2-3p/IL-6/STAT3 | Promote BCSCs by activating proinflammatory IL-6/STAT3 signaling | Correlated with poor outcome |
| SNHG7 [89] | Upregulated in chemoresistant breast cancer tissues and cells | miR-34a | Modulate chemoresistance and stemness partially via MiR-34a | Correlated with chemoresistance |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, F.; Yang, Y.; Qiu, Y.; Tang, L.; Xie, L.; Guan, X. Long Non-Coding RNAs as Regulators for Targeting Breast Cancer Stem Cells and Tumor Immune Microenvironment: Biological Properties and Therapeutic Potential. Cancers 2024, 16, 290. https://doi.org/10.3390/cancers16020290
Yang F, Yang Y, Qiu Y, Tang L, Xie L, Guan X. Long Non-Coding RNAs as Regulators for Targeting Breast Cancer Stem Cells and Tumor Immune Microenvironment: Biological Properties and Therapeutic Potential. Cancers. 2024; 16(2):290. https://doi.org/10.3390/cancers16020290
Chicago/Turabian StyleYang, Fang, Yiqi Yang, Yuling Qiu, Lin Tang, Li Xie, and Xiaoxiang Guan. 2024. "Long Non-Coding RNAs as Regulators for Targeting Breast Cancer Stem Cells and Tumor Immune Microenvironment: Biological Properties and Therapeutic Potential" Cancers 16, no. 2: 290. https://doi.org/10.3390/cancers16020290
APA StyleYang, F., Yang, Y., Qiu, Y., Tang, L., Xie, L., & Guan, X. (2024). Long Non-Coding RNAs as Regulators for Targeting Breast Cancer Stem Cells and Tumor Immune Microenvironment: Biological Properties and Therapeutic Potential. Cancers, 16(2), 290. https://doi.org/10.3390/cancers16020290






