Sirt6-Mediated Cell Death Associated with Sirt1 Suppression in Gastric Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Samples and Data Collection
2.2. Cell Lines and Chemicals
2.3. Western Blot Analysis
2.4. Flow Cytometric DNA Analysis
2.5. ROS Measurement
2.6. IHC Analysis of Sirt6 and Evaluation of IHC Reactions
2.7. ICC-Immunofluorescence (IF) for Sirt1 and Sirt6 Double Staining
2.8. Immunoprecipitation (IP) Assay
2.9. Preparation of Adenovirus and Transduction
2.10. Short Hairpin RNA (shRNA)-Mediated Silencing of Sirt6 and MDM2
2.11. Preparation of Nuclear and Cytosolic Extracts
2.12. Xenograft Mouse Model
2.13. Statistics
3. Results
3.1. Sirt6 Is Downregulated in Gastric Cancer, and Its Higher Expression Levels Are Linked to Improved Patient Survival
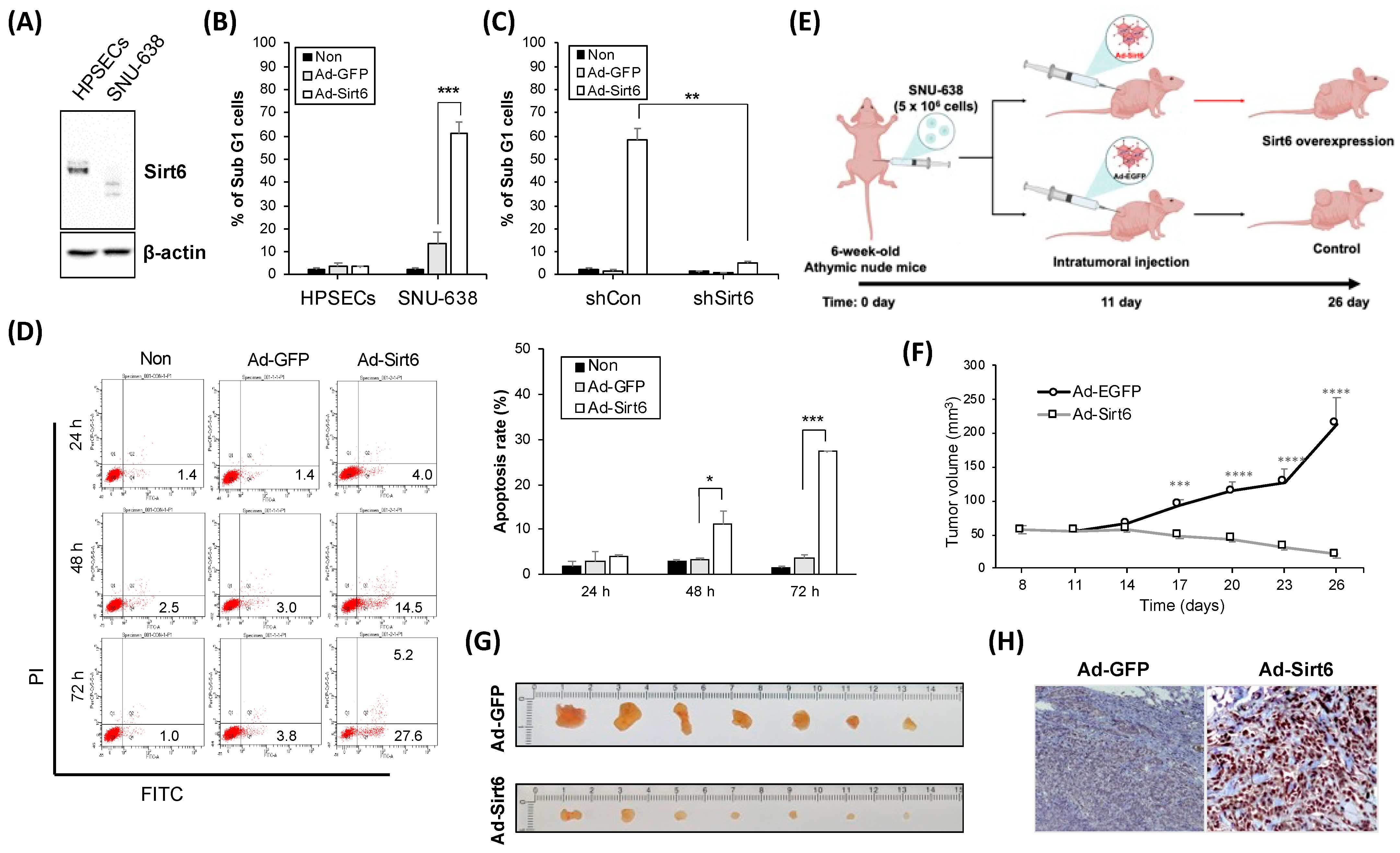
3.2. Manipulation of Sirt6 Levels Affects Cell Death Rates and Tumor Volume in Gastric Cancer Models
3.3. Modulation of Sirt6 Influences ROS Production and Apoptotic Responses in Gastric Cancer Cells
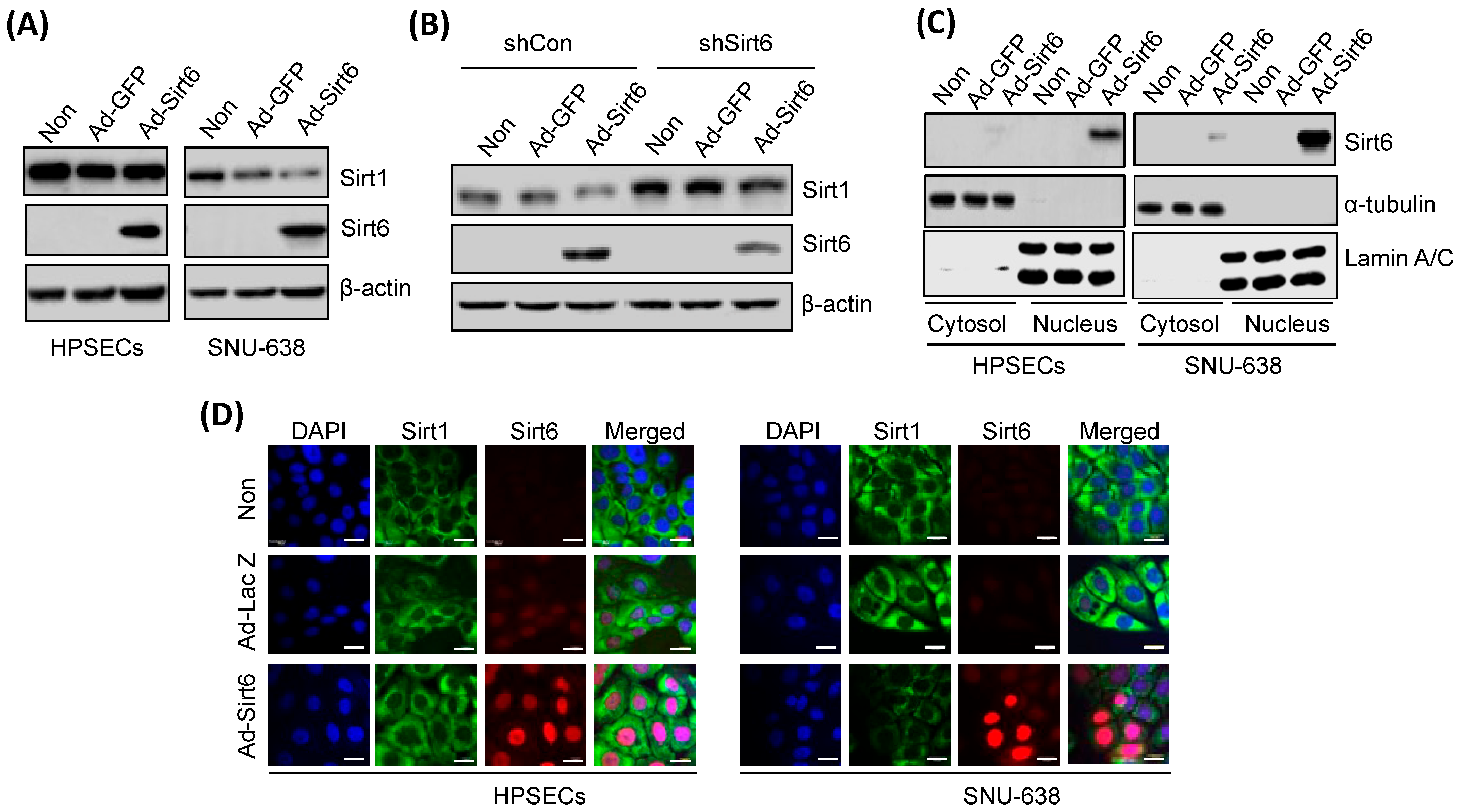
3.4. Changes in Sirt6 Expression Alter Sirt1 Levels, Affecting Gastric Cancer Cell Apoptosis
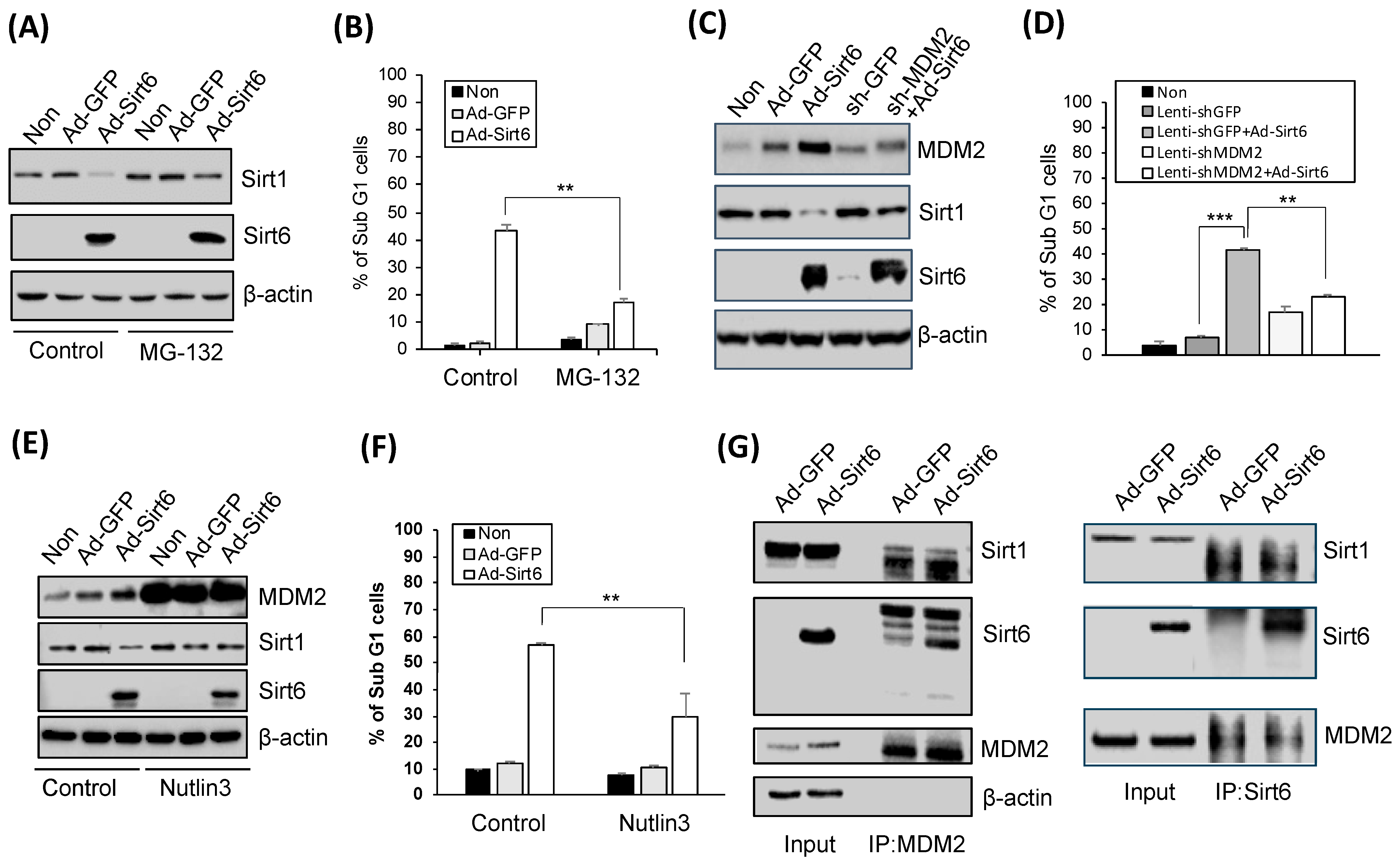
3.5. Proteasome Inhibition and MDM2 Modulation Significantly Affect Sirt1 and Sirt6 Levels
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yamamoto, H.; Schoonjans, K.; Auwerx, J. Sirtuin functions in health and disease. Mol. Endocrinol. 2007, 21, 1745–1755. [Google Scholar] [CrossRef] [PubMed]
- Vassilopoulos, A.; Fritz, K.S.; Petersen, D.R.; Gius, D. The human sirtuin family: Evolutionary divergences and functions. Hum. Genom. 2011, 5, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Chalkiadaki, A.; Guarente, L. The multifaceted functions of sirtuins in cancer. Nat. Rev. Cancer 2015, 15, 608–624. [Google Scholar] [CrossRef]
- Bosch-Presegue, L.; Vaquero, A. The dual role of sirtuins in cancer. Genes Cancer 2011, 2, 648–662. [Google Scholar] [CrossRef] [PubMed]
- Zhao, E.; Hou, J.; Ke, X.; Abbas, M.N.; Kausar, S.; Zhang, L.; Cui, H. The Roles of Sirtuin Family Proteins in Cancer Progression. Cancers 2019, 11, 1949. [Google Scholar] [CrossRef]
- Park, J.J.; Hah, Y.S.; Ryu, S.; Cheon, S.Y.; Won, S.J.; Lee, J.S.; Hwa, J.S.; Seo, J.H.; Chang, H.W.; Kim, S.W.; et al. MDM2-dependent Sirt1 degradation is a prerequisite for Sirt6-mediated cell death in head and neck cancers. Exp. Mol. Med. 2021, 53, 422–431. [Google Scholar] [CrossRef]
- Meng, F.; Qian, M.; Peng, B.; Peng, L.; Wang, X.; Zheng, K.; Liu, Z.; Tang, X.; Zhang, S.; Sun, S.; et al. Synergy between SIRT1 and SIRT6 helps recognize DNA breaks and potentiates the DNA damage response and repair in humans and mice. eLife 2020, 15, 9. [Google Scholar] [CrossRef]
- Zhao, B.; Li, X.; Zhou, L.; Wang, Y.; Shang, P. SIRT1: A potential tumour biomarker and therapeutic target. J. Drug Target. 2019, 27, 1046–1052. [Google Scholar] [CrossRef]
- Li, Y.; Chen, X.; Cui, Y.; Wei, Q.; Chen, S.; Wang, X. Effects of SIRT1 silencing on viability, invasion and metastasis of human glioma cell lines. Oncol. Lett. 2019, 17, 3701–3708. [Google Scholar] [CrossRef]
- Poniewierska-Baran, A.; Warias, P.; Zgutka, K. Sirtuins (SIRTs) As a Novel Target in Gastric Cancer. Int. J. Mol. Sci. 2022, 23, 15119. [Google Scholar] [CrossRef]
- Yan, H.; Qiu, C.; Sun, W.; Gu, M.; Xiao, F.; Zou, J.; Zhang, L. Yap regulates gastric cancer survival and migration via SIRT1/Mfn2/mitophagy. Oncol. Rep. 2018, 39, 1671–1681. [Google Scholar] [CrossRef]
- Mohammadi Saravle, S.; Ahmadi Hedayati, M.; Mohammadi, E.; Sheikhesmaeili, F.; Nikkhoo, B. Sirt1 Gene Expression and Gastric Epithelial Cells Tumor Stage in Patients with Helicobacter pylori Infection. Asian Pac. J. Cancer Prev. 2018, 19, 913–916. [Google Scholar]
- Liu, H.; Liu, N.; Zhao, Y.; Zhu, X.; Wang, C.; Liu, Q.; Gao, C.; Zhao, X.; Li, J. Oncogenic USP22 supports gastric cancer growth and metastasis by activating c-Myc/NAMPT/SIRT1-dependent FOXO1 and YAP signaling. Aging 2019, 11, 9643–9660. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Ko, Y.S.; Park, J.; Choi, Y.; Park, J.W.; Kim, Y.; Pyo, J.S.; Yoo, Y.B.; Lee, J.S.; Lee, B.L. Forkhead Transcription Factor FOXO1 Inhibits Angiogenesis in Gastric Cancer in Relation to SIRT1. Cancer Res. Treat. 2016, 48, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Huang, S.; Deng, C.; Cao, Y.; Yang, J.; Chen, G.; Zhang, B.; Duan, C.; Shi, J.; Kong, B.; et al. Co-ordinated overexpression of SIRT1 and STAT3 is associated with poor survival outcome in gastric cancer patients. Oncotarget 2017, 8, 18848–18860. [Google Scholar] [CrossRef] [PubMed]
- Qiu, G.; Li, X.; Che, X.; Wei, C.; He, S.; Lu, J.; Jia, Z.; Pang, K.; Fan, L. SIRT1 is a regulator of autophagy: Implications in gastric cancer progression and treatment. FEBS Lett. 2015, 589, 2034–2042. [Google Scholar] [CrossRef]
- Li, Q.K.; Liu, Y.K.; Li, J.W.; Liu, Y.T.; Li, Y.F.; Li, B.H. Circ-sirt1 inhibits growth and invasion of gastric cancer by sponging miR-132-3p/miR-212-3p and upregulating sirt1 expression. Neoplasma 2021, 68, 780–787. [Google Scholar] [CrossRef]
- Zhu, M.; Wei, C.; Wang, H.; Han, S.; Cai, L.; Li, X.; Liao, X.; Che, X.; Li, X.; Fan, L.; et al. SIRT1 mediated gastric cancer progression under glucose deprivation through the FoxO1-Rab7-autophagy axis. Front. Oncol. 2023, 13, 1175151. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, B.; Gao, W.; Huang, S.; Liu, Z.; Li, W.; Jia, J. SIRT1 is downregulated in gastric cancer and leads to G1-phase arrest via NF-kappaB/Cyclin D1 signaling. Mol. Cancer Res. 2013, 11, 1497–1507. [Google Scholar] [CrossRef]
- Xu, J.; Wang, X.; Wang, W.; Zhang, L.; Huang, P. Candidate oncogene circularNOP10 mediates gastric cancer progression by regulating miR-204/SIRT1 pathway. J. Gastrointest. Oncol. 2021, 12, 1428–1443. [Google Scholar] [CrossRef]
- Fu, L.; Han, B.K.; Meng, F.F.; Wang, J.W.; Wang, T.Y.; Li, H.J.; Sun, Y.Y.; Zou, G.N.; Li, X.R.; Li, W.; et al. Jaridon 6, a new diterpene from Rabdosia rubescens (Hemsl.) Hara, can display anti-gastric cancer resistance by inhibiting SIRT1 and inducing autophagy. Phytother. Res. 2021, 35, 5720–5733. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, X.; Chen, P. MiR-204 down regulates SIRT1 and reverts SIRT1-induced epithelial-mesenchymal transition, anoikis resistance and invasion in gastric cancer cells. BMC Cancer 2013, 13, 290. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, A.; Kikuchi, K.; Zheng, H.; Takahashi, H.; Miyagi, Y.; Aoki, I.; Takano, Y. SIRT1 expression is associated with a poor prognosis, whereas DBC1 is associated with favorable outcomes in gastric cancer. Cancer Med. 2014, 3, 1553–1561. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Xia, L.; Zhang, Y.; Wang, H.; Xu, W.; Hu, H.; Wang, J.; Xin, J.; Gang, Y.; Sha, S.; et al. Activating transcription factor 4 confers a multidrug resistance phenotype to gastric cancer cells through transactivation of SIRT1 expression. PLoS ONE 2012, 7, e31431. [Google Scholar] [CrossRef]
- Li, H.J.; Che, X.M.; Zhao, W.; He, S.C.; Zhang, Z.L.; Chen, R.; Fan, L.; Jia, Z.L. Diet-induced obesity promotes murine gastric cancer growth through a nampt/sirt1/c-myc positive feedback loop. Oncol. Rep. 2013, 30, 2153–2160. [Google Scholar] [CrossRef]
- Li, J.; Dong, G.; Wang, B.; Gao, W.; Yang, Q. miR-543 promotes gastric cancer cell proliferation by targeting SIRT1. Biochem. Biophys. Res. Commun. 2016, 469, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Ding, Y.; Zhang, L. SIRT1/APE1 promotes the viability of gastric cancer cells by inhibiting p53 to suppress ferroptosis. Open Med. 2023, 18, 20220620. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liao, K.; Liu, D. MiRNA-12129 Suppresses Cell Proliferation and Block Cell Cycle Progression by Targeting SIRT1 in GASTRIC Cancer. Technol. Cancer Res. Treat. 2020, 19, 1533033820928144. [Google Scholar] [CrossRef]
- Deng, X.J.; Zheng, H.L.; Ke, X.Q.; Deng, M.; Ma, Z.Z.; Zhu, Y.; Cui, Y.Y. Hsa-miR-34a-5p reverses multidrug resistance in gastric cancer cells by targeting the 3′-UTR of SIRT1 and inhibiting its expression. Cell Signal. 2021, 84, 110016. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, X.; Zhang, D.; Fan, Y.; Qin, L.; Dong, S.; Zhang, L. Upregulated miR-132 in Lgr5+ gastric cancer stem cell-like cells contributes to cisplatin-resistance via SIRT1/CREB/ABCG2 signaling pathway. Mol. Carcinog. 2017, 56, 2022–2034. [Google Scholar] [CrossRef]
- Shi, Y.; Yang, Z.; Zhang, T.; Shen, L.; Li, Y.; Ding, S. SIRT1-targeted miR-543 autophagy inhibition and epithelial-mesenchymal transition promotion in Helicobacter pylori CagA-associated gastric cancer. Cell Death Dis. 2019, 10, 625. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; He, C.; Wang, X.; Wang, H.; Nan, G.; Fang, L. MicroRNA-183 affects the development of gastric cancer by regulating autophagy via MALAT1-miR-183-SIRT1 axis and PI3K/AKT/mTOR signals. Artif. Cells Nanomed. Biotechnol. 2019, 47, 3163–3171. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Wang, B.; An, Y.; Li, J.; Wang, X.; Jia, J.; Yang, Q. SIRT1 suppresses the migration and invasion of gastric cancer by regulating ARHGAP5 expression. Cell Death Dis. 2018, 9, 977. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; Wang, B.; Wang, X.; Dong, G.; Jia, J.; Yang, Q. SIRT1 inhibits chemoresistance and cancer stemness of gastric cancer by initiating an AMPK/FOXO3 positive feedback loop. Cell Death Dis. 2020, 11, 115. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, Y.; Huang, S.; Deng, C.; Zhou, S.; Yang, J.; Ca, Y.; Xu, L.; Yuan, Y.; Yang, J.; et al. SIRT1 inhibits gastric cancer proliferation and metastasis via STAT3/MMP-13 signaling. J. Cell. Physiol. 2019, 234, 15395–15406. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Zhang, L.; Chen, X.; Lu, Q.; Yang, Y.; Liu, J.; Ma, X. SIRT1 counteracted the activation of STAT3 and NF-kappaB to repress the gastric cancer growth. Int. J. Clin. Exp. Med. 2014, 7, 5050–5058. [Google Scholar]
- Yang, Q.; Wang, B.; Zang, W.; Wang, X.; Liu, Z.; Li, W.; Jia, J. Resveratrol inhibits the growth of gastric cancer by inducing G1 phase arrest and senescence in a Sirt1-dependent manner. PLoS ONE 2013, 8, e70627. [Google Scholar] [CrossRef]
- Zhou, J.; Wu, A.; Yu, X.; Zhu, J.; Dai, H. SIRT6 inhibits growth of gastric cancer by inhibiting JAK2/STAT3 pathway. Oncol. Rep. 2017, 38, 1059–1066. [Google Scholar] [CrossRef]
- Cai, S.; Fu, S.; Zhang, W.; Yuan, X.; Cheng, Y.; Fang, J. SIRT6 silencing overcomes resistance to sorafenib by promoting ferroptosis in gastric cancer. Biochem. Biophys. Res. Commun. 2021, 577, 158–164. [Google Scholar] [CrossRef]
- Weinberg, F.; Ramnath, N.; Nagrath, D. Reactive Oxygen Species in the Tumor Microenvironment: An Overview. Cancers 2019, 11, 1191. [Google Scholar] [CrossRef]
- Smith, M.G.; Hold, G.L.; Tahara, E.; El-Omar, E.M. Cellular and molecular aspects of gastric cancer. World J. Gastroenterol. 2006, 12, 2979–2990. [Google Scholar] [CrossRef] [PubMed]
- Kountouras, J.; Zavos, C.; Chatzopoulos, D.; Katsinelos, P. New aspects of Helicobacter pylori infection involvement in gastric oncogenesis. J. Surg. Res. 2008, 146, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Polk, D.B.; Peek, R.M., Jr. Helicobacter pylori: Gastric cancer and beyond. Nat. Rev. Cancer 2010, 10, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Singh, C.K.; Chhabra, G.; Ndiaye, M.A.; Garcia-Peterson, L.M.; Mack, N.J.; Ahmad, N. The Role of Sirtuins in Antioxidant and Redox Signaling. Antioxid. Redox Signal. 2018, 28, 643–661. [Google Scholar] [CrossRef]
- You, Y.; Liang, W. SIRT1 and SIRT6: The role in aging-related diseases. Biochim. Biophys. Acta. Mol. Basis Dis. 2023, 1869, 166815. [Google Scholar] [CrossRef]
- Park, E.Y.; Woo, Y.W.; Kim, S.J.; Kim, D.H.; Lee, E.K.; De, U.; Kim, K.S.; Lee, J.W.; Jung, J.H.; Ha, K.-T.; et al. Anticancer effects of a new SIRT inhibitor, MHY2256, against human breast cancer MCF-7 cells via regulation of MDM2-p53 binding. Int. J. Biol. Sci. 2016, 12, 1555–1567. [Google Scholar] [CrossRef]
- Grabowska, W.; Sikora, E.; Bielak-Zmijewska, A. Sirtuins, a promising target in slowing down the ageing process. Biogerontology 2017, 18, 447–476. [Google Scholar]



| Name | Function | Mechanism | References |
|---|---|---|---|
| Sirt1 | Pro-tumorigenic | Yap signaling pathway | [11,12] |
| Modulation of angiogenesis via FOXO1 | [13] | ||
| Deacetylation of histone substrates, transcription factors, and cofactors | [14] | ||
| FOXO1 and YAP signaling pathway modulated by USP22 | [15] | ||
| Regulating autophagy through the deacetylation of ATGs | [16] | ||
| Targeting p53 to suppress ferroptosis | [17] | ||
| Mediated by candidate oncogene circNOP10 | [18] | ||
| FoxO1-Rab7–autophagy axis | [19] | ||
| Deacetylation of Beclin-1 and other autophagy mediators | [20] | ||
| Inhibiting PI3K/AKT pathway | [21] | ||
| Sirt1 downregulation by MiR-204 | [22] | ||
| By the results of DBC1, H4K16Ac, and H3K9Ac | [23] | ||
| Sirt1 transactivation by ATF4 | [24] | ||
| Nampt/sirt1/c-myc positive feedback loop | [25] | ||
| Anti-tumorigenic | Sirt1 targeted by micro-RNAs (miR-543, miR-132-3p/miR-212-3p, miRNA-12129, miR-1301-3p, Has-miR-34a-5p, miR-132, miR-543, miR-183) | [15,26,27,28,29,30,31,32] | |
| Regulating ARHGAP5 expression | [33] | ||
| Initiating an AMPK/FOXO3 positive feedback loop | [34] | ||
| Via STAT3/MMP-12 signaling | [35] | ||
| G1-phase arrest via NF-κB/Cyclin D1 signaling | [19] | ||
| Repression of activation of STAT3 and NF-κB proteins via deacetylation | [36] | ||
| Inducing G1 phase arrest and senescence by resveratrol | [37] | ||
| Sirt6 | Anti-tumorigenic | Inhibition of JAK2/STAT3 pathway | [38] |
| Promoting ferroptosis | [39] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, J.H.; Ryu, S.; Cheon, S.Y.; Lee, S.-J.; Won, S.-J.; Yim, C.D.; Lee, H.-J.; Hah, Y.-S.; Park, J.J. Sirt6-Mediated Cell Death Associated with Sirt1 Suppression in Gastric Cancer. Cancers 2024, 16, 387. https://doi.org/10.3390/cancers16020387
Seo JH, Ryu S, Cheon SY, Lee S-J, Won S-J, Yim CD, Lee H-J, Hah Y-S, Park JJ. Sirt6-Mediated Cell Death Associated with Sirt1 Suppression in Gastric Cancer. Cancers. 2024; 16(2):387. https://doi.org/10.3390/cancers16020387
Chicago/Turabian StyleSeo, Ji Hyun, Somi Ryu, So Young Cheon, Seong-Jun Lee, Seong-Jun Won, Chae Dong Yim, Hyun-Jin Lee, Young-Sool Hah, and Jung Je Park. 2024. "Sirt6-Mediated Cell Death Associated with Sirt1 Suppression in Gastric Cancer" Cancers 16, no. 2: 387. https://doi.org/10.3390/cancers16020387
APA StyleSeo, J. H., Ryu, S., Cheon, S. Y., Lee, S. -J., Won, S. -J., Yim, C. D., Lee, H. -J., Hah, Y. -S., & Park, J. J. (2024). Sirt6-Mediated Cell Death Associated with Sirt1 Suppression in Gastric Cancer. Cancers, 16(2), 387. https://doi.org/10.3390/cancers16020387






