The Diversity of Methylation Patterns in Serous Borderline Ovarian Tumors and Serous Ovarian Carcinomas
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Clinicopathological Parameters
2.2. DNA Isolation and Quality Assessment
2.3. DNA Bisulfite Conversion
2.4. Microarray Profiling
2.5. Methylation-Specific PCR and Sanger Sequencing
2.6. Bioinformatic and Statistical Analyses
3. Results
3.1. The Analysis of the MDM2/TP53/CDKN1A (p21) Axis
3.2. Differences in Methylation Patterns Between Groups
3.3. CpG Sites with the Most Differentiated Methylation
3.4. Ontological Analyses
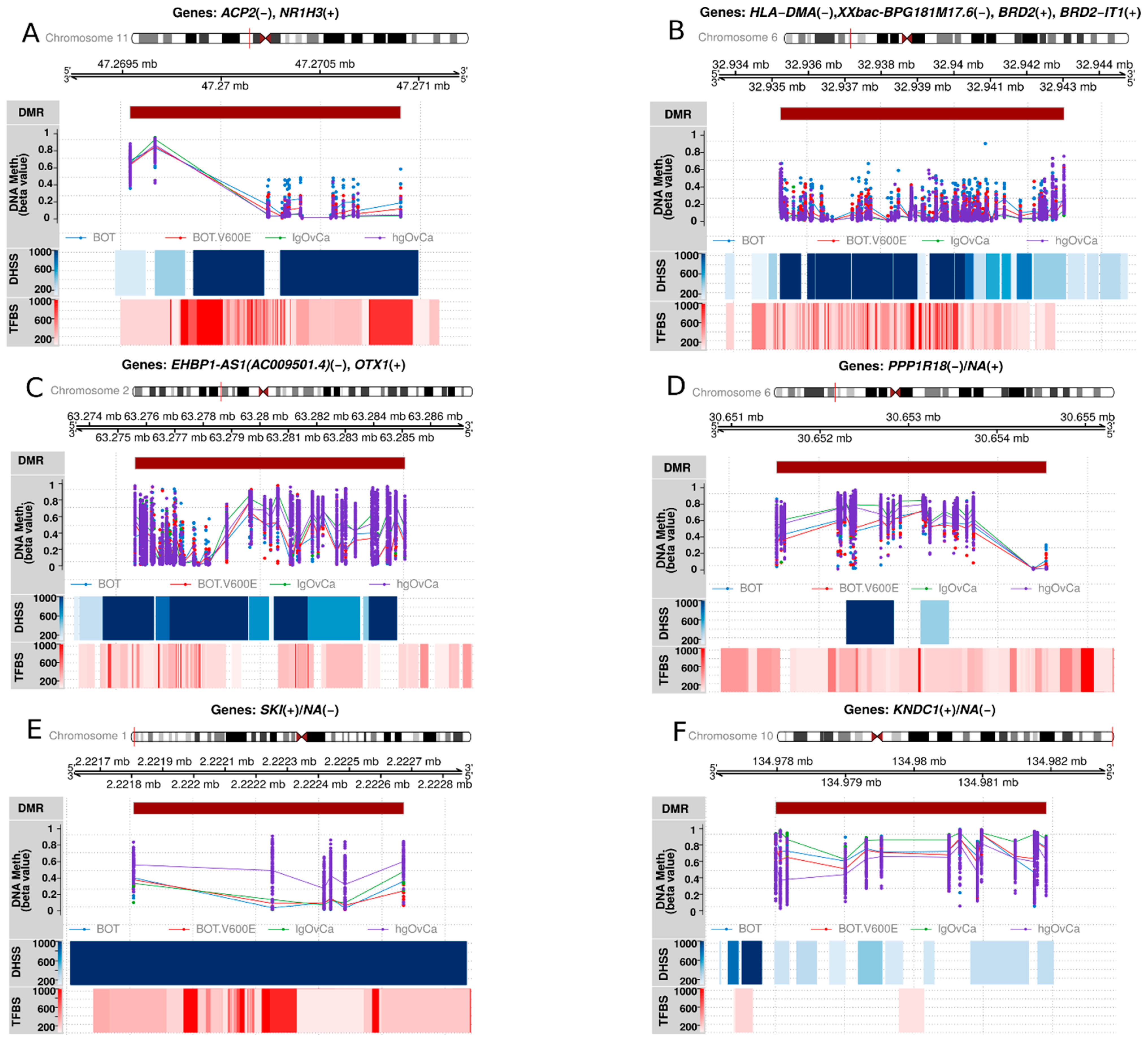
3.5. The Most Statistically Significant DMRs
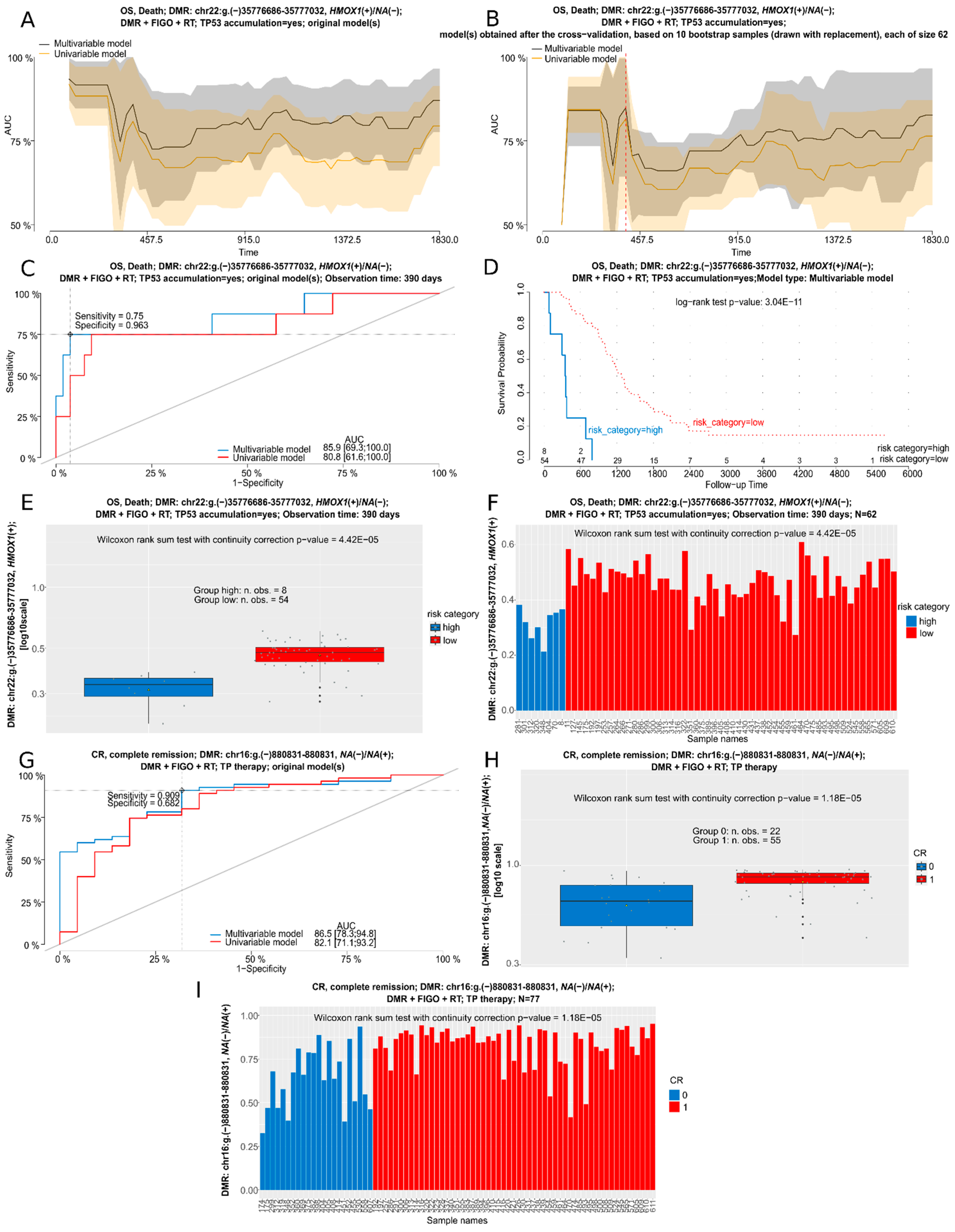
3.6. Cox and Logistic Regression Analyses for DMRs in BOTS and hgOvCa
4. Discussion
5. Limitations of the Study
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Heery, R.; Schaefer, M.H. DNA Methylation Variation along the Cancer Epigenome and the Identification of Novel Epigenetic Driver Events. Nucleic Acids Res. 2021, 49, 12692–12705. [Google Scholar] [CrossRef] [PubMed]
- Hanson, H.E.; Liebl, A.L. The Mutagenic Consequences of DNA Methylation within and across Generations. Epigenomes 2022, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- Bourdel, N.; Huchon, C.; Abdel Wahab, C.; Azaïs, H.; Bendifallah, S.; Bolze, P.-A.; Brun, J.-L.; Canlorbe, G.; Chauvet, P.; Chereau, E.; et al. Borderline Ovarian Tumors: French Guidelines from the CNGOF. Part 2. Surgical Management, Follow-up, Hormone Replacement Therapy, Fertility Management and Preservation. J. Gynecol. Obstet. Hum. Reprod. 2021, 50, 101966. [Google Scholar] [CrossRef] [PubMed]
- Timor-Tritsch, I.E.; Foley, C.E.; Brandon, C.; Yoon, E.; Ciaffarrano, J.; Monteagudo, A.; Mittal, K.; Boyd, L. New Sonographic Marker of Borderline Ovarian Tumor: Microcystic Pattern of Papillae and Solid Components. Ultrasound Obstet. Gynecol. Off. J. Int. Soc. Ultrasound Obstet. Gynecol. 2019, 54, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Niu, L.; Tian, H.; Xu, Y.; Cao, J.; Zhang, X.; Zhang, J.; Hou, J.; Lv, W.; Wang, J.; Xin, L.; et al. Recurrence Characteristics and Clinicopathological Results of Borderline Ovarian Tumors. BMC Womens Health 2021, 21, 134. [Google Scholar] [CrossRef]
- Shih, K.; Zhou, Q.; Huh, J.; Morgan, J.; Iasonos, A.; Aghajanian, C.; Chi, D.; Barakat, R.; Abu-Rustum, N. Risk Factors for Recurrence of Ovarian Borderline Tumors. Gynecol. Oncol. 2011, 120, 480–484. [Google Scholar] [CrossRef]
- Hauptmann, S.; Friedrich, K.; Redline, R.; Avril, S. Ovarian Borderline Tumors in the 2014 WHO Classification: Evolving Concepts and Diagnostic Criteria. Virchows Arch. 2017, 470, 125–142. [Google Scholar] [CrossRef]
- Gershenson, D.M.; Silva, E.G.; Levy, L.; Burke, T.W.; Wolf, J.K.; Tornos, C. Ovarian Serous Borderline Tumors with Invasive Peritoneal Implants. Cancer 1998, 82, 1096–1103. [Google Scholar] [CrossRef]
- Vang, R.; Hannibal, C.G.; Junge, J.; Frederiksen, K.; Kjaer, S.K.; Kurman, R.J. Long-Term Behavior of Serous Borderline Tumors Subdivided Into Atypical Proliferative Tumors and Noninvasive Low-Grade Carcinomas: A Population-Based Clinicopathologic Study of 942 Cases. Am. J. Surg. Pathol. 2017, 41, 725–737. [Google Scholar] [CrossRef]
- Moujaber, T.; Balleine, R.L.; Gao, B.; Madsen, I.; Harnett, P.R.; DeFazio, A. New Therapeutic Opportunities for Women with Low-Grade Serous Ovarian Cancer. Endocr. Relat. Cancer 2022, 29, R1–R16. [Google Scholar] [CrossRef]
- Shih, I.-M.; Chen, L.; Wang, C.; Gu, J.; Davidson, B.; Cope, L.; Kurman, R.J.; Xuan, J.; Wang, T.-L. Distinct DNA Methylation Profiles in Ovarian Serous Neoplasms and Their Implications in Ovarian Carcinogenesis. Am. J. Obstet. Gynecol. 2010, 203, 584.e1–584.e22. [Google Scholar] [CrossRef] [PubMed]
- Zeller, C.; Dai, W.; Curry, E.; Siddiq, A.; Walley, A.; Masrour, N.; Kitsou-Mylona, I.; Anderson, G.; Ghaem-Maghami, S.; Brown, R.; et al. The DNA Methylomes of Serous Borderline Tumors Reveal Subgroups with Malignant- or Benign-like Profiles. Am. J. Pathol. 2013, 182, 668–677. [Google Scholar] [CrossRef] [PubMed]
- Reyes, H.D.; Devor, E.J.; Warrier, A.; Newtson, A.M.; Mattson, J.; Wagner, V.; Duncan, G.N.; Leslie, K.K.; Gonzalez-Bosquet, J. Differential DNA Methylation in High-Grade Serous Ovarian Cancer (HGSOC) Is Associated with Tumor Behavior. Sci. Rep. 2019, 9, 17996. [Google Scholar] [CrossRef] [PubMed]
- Ferris, J.S.; Wang, T.; Wang, S.; Hibshoosh, H.; Su, T.; Wang, X.; Chen, X.; Yu, A.; Santella, R.M.; Wright, J.D.; et al. Identifying DNA Methylation Signatures in High-Grade Serous Ovarian Cancer: Results Vary by Control Tissue Type. J. Clin. Oncol. 2022, 40, e17559. [Google Scholar] [CrossRef]
- Kupryjańczyk, J.; Szymańska, T.; Madry, R.; Timorek, A.; Stelmachów, J.; Karpińska, G.; Rembiszewska, A.; Ziółkowska, I.; Kraszewska, E.; Debniak, J.; et al. Evaluation of Clinical Significance of TP53, BCL-2, BAX and MEK1 Expression in 229 Ovarian Carcinomas Treated with Platinum-Based Regimen. Br. J. Cancer 2003, 88, 848–854. [Google Scholar] [CrossRef]
- Kupryjańczyk, J.; Madry, R.; Plisiecka-Hałasa, J.; Bar, J.; Kraszewska, E.; Ziółkowska, I.; Timorek, A.; Stelmachów, J.; Emerich, J.; Jedryka, M.; et al. TP53 Status Determines Clinical Significance of ERBB2 Expression in Ovarian Cancer. Br. J. Cancer 2004, 91, 1916–1923. [Google Scholar] [CrossRef]
- Mayr, D.; Hirschmann, A.; Löhrs, U.; Diebold, J. KRAS and BRAF Mutations in Ovarian Tumors: A Comprehensive Study of Invasive Carcinomas, Borderline Tumors and Extraovarian Implants. Gynecol. Oncol. 2006, 103, 883–887. [Google Scholar] [CrossRef]
- Wong, K.-K.; Tsai, C.-C.; Gershenson, D.M. BRAF Mutational Analysis in Ovarian Tumors: Recent Perspectives. Pathol. Lab. Med. Int. 2015, 7, 75–82. [Google Scholar] [CrossRef]
- Szafron, L.A.; Iwanicka-Nowicka, R.; Podgorska, A.; Bonna, A.M.; Sobiczewski, P.; Kupryjanczyk, J.; Szafron, L.M. The Clinical Significance of CRNDE Gene Methylation, Polymorphisms, and CRNDEP Micropeptide Expression in Ovarian Tumors. Int. J. Mol. Sci. 2024, 25, 7531. [Google Scholar] [CrossRef]
- Mehra, P.; Aditi, S.; Prasad, K.M.; Bariar, N. k Histomorphological Analysis of Ovarian Neoplasms According to the 2020 WHO Classification of Ovarian Tumors: A Distribution Pattern in a Tertiary Care Center. Cureus 2023, 15, e38273. [Google Scholar] [CrossRef]
- Szafron, L.A.; Sobiczewski, P.; Dansonka-Mieszkowska, A.; Kupryjanczyk, J.; Szafron, L.M. An Analysis of Genetic Polymorphisms in 76 Genes Related to the Development of Ovarian Tumors of Different Aggressiveness. International Journal of Molecular Sciences 2024, 25, 10876. [Google Scholar] [CrossRef]
- Woroniecka, R.; Rymkiewicz, G.; Szafron, L.M.; Blachnio, K.; Szafron, L.A.; Bystydzienski, Z.; Pienkowska-Grela, B.; Borkowska, K.; Rygier, J.; Kotyl, A.; et al. Cryptic MYC Insertions in Burkitt Lymphoma: New Data and a Review of the Literature. PLoS ONE 2022, 17, e0263980. [Google Scholar] [CrossRef] [PubMed]
- Maksimovic, J.; Phipson, B.; Oshlack, A. A Cross-Package Bioconductor Workflow for Analysing Methylation Array Data. F1000Research 2016, 5, 1281. [Google Scholar] [CrossRef] [PubMed]
- Gerds, T.A.; Cai, T.; Schumacher, M. The Performance of Risk Prediction Models. Biom. J. 2008, 50, 457–479. [Google Scholar] [CrossRef]
- Lotfi Garavand, A.; Mohammadi, M.; Mohammadzadeh, S. Evaluation of TP53 Codon 72, P21 Codon 31, and MDM2 SNP309 Polymorphisms in Iranian Patients with Acute Lymphocytic Leukemia. Rep. Biochem. Mol. Biol. 2020, 9, 26–32. [Google Scholar] [CrossRef]
- Li, S.; Zhang, J.; Huang, S.; He, X. Genome-Wide Analysis Reveals That Exon Methylation Facilitates Its Selective Usage in the Human Transcriptome. Brief. Bioinform. 2018, 19, 754–764. [Google Scholar] [CrossRef]
- Lakshminarasimhan, R.; Liang, G. The Role of DNA Methylation in Cancer. Adv. Exp. Med. Biol. 2016, 945, 151–172. [Google Scholar] [CrossRef]
- Haerinck, J.; Goossens, S.; Berx, G. The Epithelial–Mesenchymal Plasticity Landscape: Principles of Design and Mechanisms of Regulation. Nat. Rev. Genet. 2023, 24, 590–609. [Google Scholar] [CrossRef]
- Hentze, J.L.; Høgdall, C.K.; Høgdall, E.V. Methylation and Ovarian Cancer: Can DNA Methylation Be of Diagnostic Use? Mol. Clin. Oncol. 2019, 10, 323–330. [Google Scholar] [CrossRef]
- Jain, S.; Chang, T.-T.; Hamilton, J.P.; Lin, S.Y.; Lin, Y.-J.; Evans, A.A.; Selaru, F.M.; Lin, P.-W.; Chen, S.-H.; Block, T.M.; et al. Methylation of the CpG Sites Only on the Sense Strand of the APC Gene Is Specific for Hepatocellular Carcinoma. PLoS ONE 2011, 6, e26799. [Google Scholar] [CrossRef]
- Chmelarova, M.; Krepinska, E.; Spacek, J.; Laco, J.; Beranek, M.; Palicka, V. Methylation in the P53 Promoter in Epithelial Ovarian Cancer. Clin. Transl. Oncol. Off. Publ. Fed. Span. Oncol. Soc. Natl. Cancer Inst. Mex. 2013, 15, 160–163. [Google Scholar] [CrossRef] [PubMed]
- Singer, M.; Kosti, I.; Pachter, L.; Mandel-Gutfreund, Y. A Diverse Epigenetic Landscape at Human Exons with Implication for Expression. Nucleic Acids Res. 2015, 43, 3498–3508. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadi, D.; Esteve-Codina, A.; Piferrer, F. Consistent Inverse Correlation between DNA Methylation of the First Intron and Gene Expression across Tissues and Species. Epigenetics Chromatin 2018, 11, 37. [Google Scholar] [CrossRef] [PubMed]
- McGuire, M.H.; Herbrich, S.M.; Dasari, S.K.; Wu, S.Y.; Wang, Y.; Rupaimoole, R.; Lopez-Berestein, G.; Baggerly, K.A.; Sood, A.K. Pan-Cancer Genomic Analysis Links 3’UTR DNA Methylation with Increased Gene Expression in T Cells. EBioMedicine 2019, 43, 127–137. [Google Scholar] [CrossRef]
- Buitrago, D.; Labrador, M.; Arcon, J.P.; Lema, R.; Flores, O.; Esteve-Codina, A.; Blanc, J.; Villegas, N.; Bellido, D.; Gut, M.; et al. Impact of DNA Methylation on 3D Genome Structure. Nat. Commun. 2021, 12, 3243. [Google Scholar] [CrossRef]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef]
- Kulis, M.; Esteller, M. 2—DNA Methylation and Cancer. In Advances in Genetics; Herceg, Z., Ushijima, T., Eds.; Epigenetics and Cancer, Part A; Academic Press: Cambridge, MA, USA, 2010; Volume 70, pp. 27–56. [Google Scholar]
- Shi, D.; Gu, W. Dual Roles of MDM2 in the Regulation of P53. Genes Cancer 2012, 3, 240–248. [Google Scholar] [CrossRef]
- Manousakis, E.; Miralles, C.M.; Esquerda, M.G.; Wright, R.H.G. CDKN1A/P21 in Breast Cancer: Part of the Problem, or Part of the Solution? Int. J. Mol. Sci. 2023, 24, 17488. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef]
- Roszkowska, K.A.; Piecuch, A.; Sady, M.; Gajewski, Z.; Flis, S. Gain of Function (GOF) Mutant P53 in Cancer-Current Therapeutic Approaches. Int. J. Mol. Sci. 2022, 23, 13287. [Google Scholar] [CrossRef]
- Parreno, V.; Martinez, A.-M.; Cavalli, G. Mechanisms of Polycomb Group Protein Function in Cancer. Cell Res. 2022, 32, 231–253. [Google Scholar] [CrossRef] [PubMed]
- Gendronneau, G.; Boucherat, O.; Aubin, J.; Lemieux, M.; Jeannotte, L. The Loss of Hoxa5 Function Causes Estrous Acyclicity and Ovarian Epithelial Inclusion Cysts. Endocrinology 2012, 153, 1484–1497. [Google Scholar] [CrossRef] [PubMed]
- Jeannotte, L.; Gotti, F.; Landry-Truchon, K. Hoxa5: A Key Player in Development and Disease. J. Dev. Biol. 2016, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- Bracken, A.P.; Dietrich, N.; Pasini, D.; Hansen, K.H.; Helin, K. Genome-Wide Mapping of Polycomb Target Genes Unravels Their Roles in Cell Fate Transitions. Genes Dev. 2006, 20, 1123–1136. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Qian, Q.; Cao, D.; Yang, J.; Gui, T.; Shen, K. Role of BMI1 in Epithelial Ovarian Cancer: Investigated via the CRISPR/Cas9 System and RNA Sequencing. J. Ovarian Res. 2018, 11, 31. [Google Scholar] [CrossRef]
- Kielbik, M.; Przygodzka, P.; Szulc-Kielbik, I.; Klink, M. Snail Transcription Factors as Key Regulators of Chemoresistance, Stemness and Metastasis of Ovarian Cancer Cells. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 189003. [Google Scholar] [CrossRef]
- Curry, E.W.; Stronach, E.A.; Rama, N.R.; Wang, Y.Y.; Gabra, H.; El-Bahrawy, M.A. Molecular Subtypes of Serous Borderline Ovarian Tumor Show Distinct Expression Patterns of Benign Tumor and Malignant Tumor-Associated Signatures. Mod. Pathol. 2014, 27, 433–442. [Google Scholar] [CrossRef]
- Donninger, H.; Bonome, T.; Li, J.-Y.; Park, D.-C.; Radonovich, M.; Pise-Masison, C.; Brady, J.; Barrett, J.C.; Mok, S.C.; Birrer, M.J. Expression Profiling of Microdissected Papillary Serous Ovarian Epithelial Cancers Identifies Genes Describing the Unique Phenotypes of Borderline and Malignant Tumors. J. Clin. Oncol. 2005, 23, 5029. [Google Scholar] [CrossRef]
- Yang, L.; Bie, L.; Sun, L.; Yue, Y. Neural Activities Are Unfavorable for the Prognosis of Ovarian Cancer through mRNA Expression Analysis. Biomark. Med. 2019, 13, 663–673. [Google Scholar] [CrossRef]
- Ahmed, N.; Latifi, A.; Riley, C.B.; Findlay, J.K.; Quinn, M.A. Neuronal Transcription Factor Brn-3a(l) Is over Expressed in High-Grade Ovarian Carcinomas and Tumor Cells from Ascites of Patients with Advanced-Stage Ovarian Cancer. J. Ovarian Res. 2010, 3, 17. [Google Scholar] [CrossRef]
- Huang, R.-L.; Gu, F.; Kirma, N.B.; Ruan, J.; Chen, C.-L.; Wang, H.-C.; Liao, Y.-P.; Chang, C.-C.; Yu, M.-H.; Pilrose, J.M.; et al. Comprehensive Methylome Analysis of Ovarian Tumors Reveals Hedgehog Signaling Pathway Regulators as Prognostic DNA Methylation Biomarkers. Epigenetics 2013, 8, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M.; Corn, P.G.; Baylin, S.B.; Herman, J.G. A Gene Hypermethylation Profile of Human Cancer. Cancer Res. 2001, 61, 3225–3229. [Google Scholar] [PubMed]
- Bhandari, N.; Acharya, D.; Chatterjee, A.; Mandve, L.; Kumar, P.; Pratap, S.; Malakar, P.; Shukla, S.K. Pan-Cancer Integrated Bioinformatic Analysis of RNA Polymerase Subunits Reveal RNA Pol I Member CD3EAP Regulates Cell Growth by Modulating Autophagy. Cell Cycle 2023, 22, 1986–2002. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wu, Y.; Guo, Q.; Wang, S.; Wu, X. RNA-Binding Proteins in Ovarian Cancer: A Novel Avenue of Their Roles in Diagnosis and Treatment. J. Transl. Med. 2022, 20, 37. [Google Scholar] [CrossRef] [PubMed]
- Piqué, L.; Martinez de Paz, A.; Piñeyro, D.; Martínez-Cardús, A.; Castro de Moura, M.; Llinàs-Arias, P.; Setien, F.; Gomez-Miragaya, J.; Gonzalez-Suarez, E.; Sigurdsson, S.; et al. Epigenetic Inactivation of the Splicing RNA-Binding Protein CELF2 in Human Breast Cancer. Oncogene 2019, 38, 7106–7112. [Google Scholar] [CrossRef]
- Guo, Q.; Wu, Y.; Guo, X.; Cao, L.; Xu, F.; Zhao, H.; Zhu, J.; Wen, H.; Ju, X.; Wu, X. The RNA-Binding Protein CELF2 Inhibits Ovarian Cancer Progression by Stabilizing FAM198B. Mol. Ther. Nucleic Acids 2021, 23, 169–184. [Google Scholar] [CrossRef]
- Bae, G.; Berezhnoy, G.; Koch, A.; Cannet, C.; Schäfer, H.; Kommoss, S.; Brucker, S.; Beziere, N.; Trautwein, C. Stratification of Ovarian Cancer Borderline from High-Grade Serous Carcinoma Patients by Quantitative Serum NMR Spectroscopy of Metabolites, Lipoproteins, and Inflammatory Markers. Front. Mol. Biosci. 2023, 10, 1158330. [Google Scholar] [CrossRef]
- Huang, Y.; Du, Y.; Zheng, Y.; Wen, C.; Zou, H.; Huang, J.; Zhou, H.; Zhao, H.; Wu, L. Ct-OATP1B3 Promotes High-Grade Serous Ovarian Cancer Metastasis by Regulation of Fatty Acid Beta-Oxidation and Oxidative Phosphorylation. Cell Death Dis. 2022, 13, 556. [Google Scholar] [CrossRef]
- Xue, Y.; Yang, Y.; Tian, H.; Quan, H.; Liu, S.; Zhang, L.; Yang, L.; Zhu, H.; Wu, H.; Gao, Y.Q. Computational Characterization of Domain-Segregated 3D Chromatin Structure and Segmented DNA Methylation Status in Carcinogenesis. Mol. Oncol. 2022, 16, 699–716. [Google Scholar] [CrossRef]
- Lie, B.A.; Thorsby, E. Several Genes in the Extended Human MHC Contribute to Predisposition to Autoimmune Diseases. Curr. Opin. Immunol. 2005, 17, 526–531. [Google Scholar] [CrossRef]
- Palmer, R.E.; Lee, S.B.; Wong, J.C.; Reynolds, P.A.; Zhang, H.; Truong, V.; Oliner, J.D.; Gerald, W.L.; Haber, D.A. Induction of BAIAP3 by the EWS-WT1 Chimeric Fusion Implicates Regulated Exocytosis in Tumorigenesis. Cancer Cell 2002, 2, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Franzè, E.; Stolfi, C.; Troncone, E.; Scarozza, P.; Monteleone, G. Role of Interleukin-34 in Cancer. Cancers 2020, 12, 252. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.-J.; Abudukeyoumu, A.; Zhang, X.; Liu, L.-B.; Li, M.-Q.; Xie, F. Heme Oxygenase 1: A Novel Oncogene in Multiple Gynecological Cancers. Int. J. Biol. Sci. 2021, 17, 2252–2261. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Li, L.; Yan, J.; Huang, T. Prioritization of Drug Targets for Thyroid Cancer: A Multi-Omics Mendelian Randomization Study. Endocrine 2024. [Google Scholar] [CrossRef]
- Li, Y.-Z.; Zhang, C.; Pei, J.-P.; Zhang, W.-C.; Zhang, C.-D.; Dai, D.-Q. The Functional Role of Pescadillo Ribosomal Biogenesis Factor 1 in Cancer. J. Cancer 2022, 13, 268–277. [Google Scholar] [CrossRef]
- Li, J.; Zhuang, Q.; Lan, X.; Zeng, G.; Jiang, X.; Huang, Z. PES1 Differentially Regulates the Expression of ERα and ERβ in Ovarian Cancer. IUBMB Life 2013, 65, 1017–1025. [Google Scholar] [CrossRef]
- Namasu, C.Y.; Katzerke, C.; Bräuer-Hartmann, D.; Wurm, A.A.; Gerloff, D.; Hartmann, J.-U.; Schwind, S.; Müller-Tidow, C.; Hilger, N.; Fricke, S.; et al. ABR, a Novel Inducer of Transcription Factor C/EBPα, Contributes to Myeloid Differentiation and Is a Favorable Prognostic Factor in Acute Myeloid Leukemia. Oncotarget 2017, 8, 103626–103639. [Google Scholar] [CrossRef]

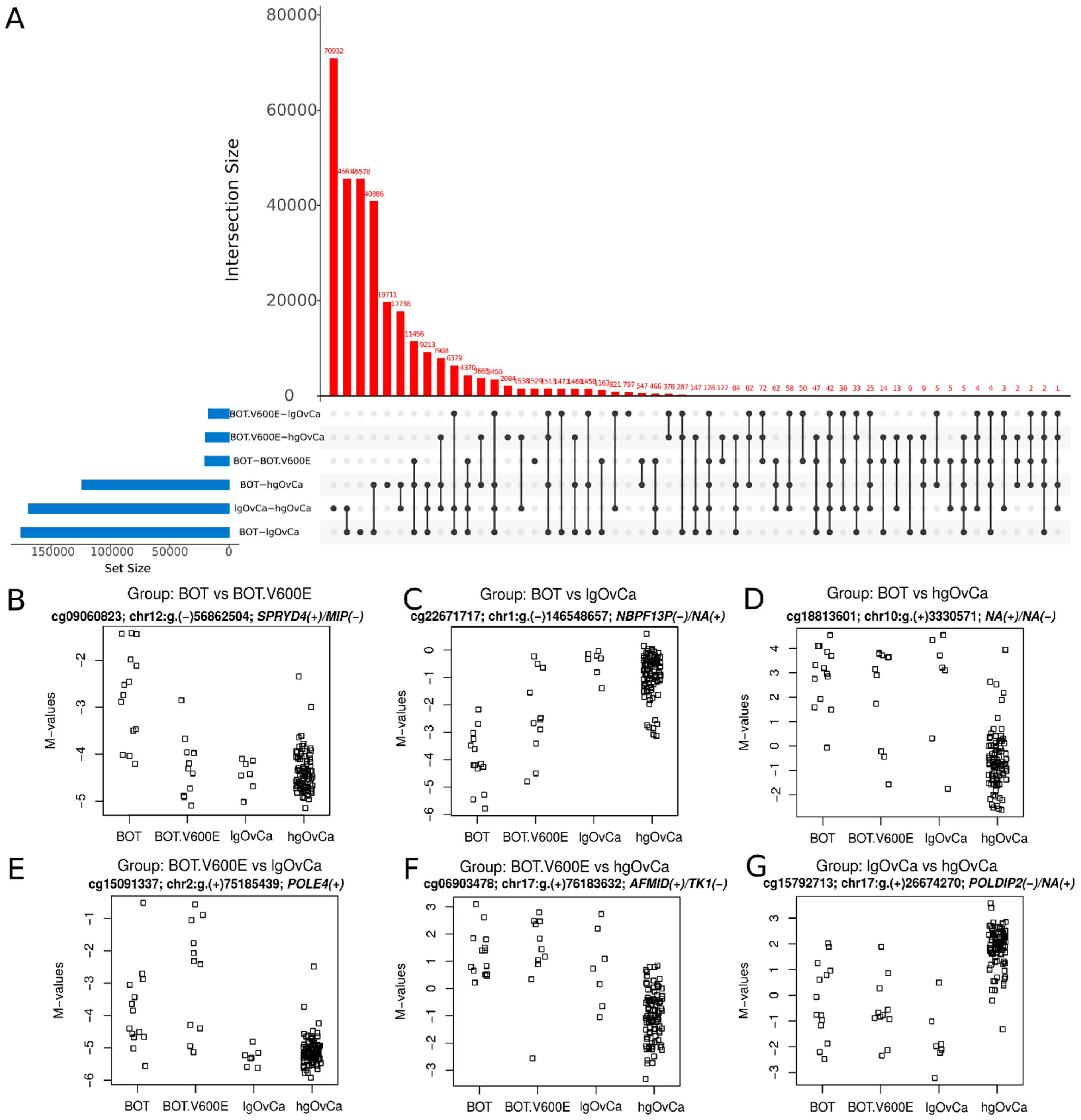
| CpGs | ||||||
| DM CpGs | BOT vs. BOT V600E | BOT vs. lgOvCa | BOT vs. hgOvCa | BOT V600E vs. lgOvCa | BOT V600E vs. hgOvCa | lgOvCa vs. hgOvCa |
| Upmethylated | 16,108 | 86,834 | 93,667 | 5438 | 12,170 | 136,293 |
| Downmethylated | 4035 | 88,467 | 30,227 | 11,665 | 7369 | 32,832 |
| Sum of DM CpGs | 20,143 | 175,301 | 123,894 | 17,103 | 19,539 | 169,125 |
| NS | 579,360 | 424,202 | 475,609 | 582,400 | 579,964 | 430,378 |
| Up/Down ratio | 3.99 | 0.98 | 3.1 | 0.47 | 1.65 | 4.15 |
| DMRs | ||||||
| DMRs | BOT vs. BOT V600E | BOT vs. lgOvCa | BOT vs. hgOvCa | BOT V600E vs. lgOvCa | BOT V600E vs. hgOvCa | lgOvCa vs. hgOvCa |
| Upmethylated | 1837 | 12,438 | 11,442 | 1062 | 2127 | 21,555 |
| Downmethylated | 25 | 7646 | 1979 | 869 | 1385 | 5759 |
| Sum of DMRs | 1862 | 20,084 | 13,421 | 1931 | 3512 | 27,314 |
| Up/Down ratio | 73.48 | 1.63 | 5.78 | 1.22 | 1.54 | 3.74 |
| BOT vs. BOT.V600E | BOT vs. lgOvCa | BOT vs. hgOvCa |
| cg09060823; chr12:g.(−)56862504 SPRYD4(+)/MIP(−) | cg22671717; chr1:g.(−)146548657 NBPF13P(−)/NA(+) | cg18813601; chr10:g.(+)3330571 NA(+)/NA(−) |
| cg00598858; chr19:g.(−)11545966 PRKCSH(+)/ODAD3 (CCDC151)(−) | cg06869971; chr15:g.(−)69706519 KIF23(+)/RP11-253M7.1 (KIF23-AS1)(−) | cg25977528; chr13:g.(+)100633444 ZIC2(+) |
| cg24443198; chr6:g.(−)84569302 CYB5R4(+)/NA(−) | cg22011361; chr14:g.(−)70821355 COX16(−)/SYNJ2BP-COX16(−) | cg00614081; chr4:g.(−)1233439 CTBP1(−) |
| cg10664618; chr12:g.(+)57579466 LRP1(+) | cg25977528; chr13:g.(+)100633444 ZIC2(+) | cg06903478; chr17:g.(+)76183632 AFMID(+)/TK1(−) |
| cg15086746; chr1:g.(−)44084965 PTPRF(+)/NA(−) | cg03751813; chr19:g.(−)37701393 ZNF585B(−) | cg02608914; chr2:g.(−)171784720 GORASP2(+)/NA(−) |
| cg00500457; chr22:g.(−)39055589 CBY1(+)/FAM227A(−) | cg23639257; chr17:g.(−)73663270 RECQL5(−)/SAP30BP(+) | cg22671717; chr1:g.(−)146548657 NBPF13P(−)/NA(+) |
| cg08427970; chr10:g.(−)99122398 RRP12(−) | cg10479053; chr17:g.(−)38136919 PSMD3(+)/NA(−) | cg11704490; chr2:g.(−)162284894 NA(−)/SLC4A10(+)/AC009487.5(+) |
| cg02608656; chr12:g.(+)56090830 ITGA7(−)/NA(+) | cg17908846; chr20:g.(+)32320553 ZNF341(+) | cg10659805; chr7:g.(+)96631680 DLX6(+)/DLX6-AS1(−) |
| cg02901790; chr8:g.(+)144391601 TOP1MT(−)/NA(+) | cg22437020; chr17:g.(−)41623744 ETV4(−)/RP11-392O1.4(+) | cg02215357; chr13:g.(−)53191046 NA(−)/HNRNPA1L2(+)/MRPS31P4(+) |
| cg19623237; chr17:g.(+)77818582 NA(+)/NA(−) | cg00528793; chr22:g.(−)19842837 GNB1L(−)/RTL10 (C22Orf29)(−) | cg25899337; chr19:g.(−)1970441 CSNK1G2(+)/NA(−) |
| BOT.V600E vs. lgOvCa | BOT.V600E vs. hgOvCa | lgOvCa vs. hgOvCa |
| cg15091337; chr2:g.(+)75185439 POLE4(+) | cg06903478; chr17:g.(+)76183632 AFMID(+)/TK1(−) | cg15792713; chr17:g.(+)26674270 POLDIP2(−)/NA(+) |
| cg13518540; chr17:g.(+)72781248 TMEM104(+) | cg27641801; chr4:g.(−)4429265 STX18(−) | cg11610925; chr10:g.(−)134978049 KNDC1(+)/NA(−) |
| cg00376288; chr19:g.(+)3656580 PIP5K1C(−)/NA(+) | cg08271229; chr1:g.(+)2222674 SKI(+) | cg00454305; chr16:g.(−)1429905 UNKL(−) |
| cg10168722; chr14:g.(−)38068608 FOXA1(−)/TTC6(+) | cg18813601; chr10:g.(+)3330571 NA(+)/NA(−) | cg18468569; chr8:g.(+)125984720 ZNF572(+) |
| cg11199810; chr1:g.(−)150123146 PLEKHO1(+)/NA(−) | cg17026391; chr11:g.(+)61159442 TMEM216(+) | cg14636714; chr10:g.(−)135018298 KNDC1(+)/NA(−) |
| cg18656829; chr13:g.(−)100632250 NA(−)/ZIC2(+) | cg00614081; chr4:g.(−)1233439 CTBP1(−) | cg07570470; chr8:g.(+)142318841 NA(+)/SLC45A4(−) |
| cg02941008; chr1:g.(+)20810527 CAMK2N1(−)/NA(+) | cg00817355; chr2:g.(−)85073409 TRABD2A(−) | cg19823504; chr19:g.(+)4556982 SEMA6B(−)/NA(+) |
| cg27641801; chr4:g.(−)4429265 STX18(−) | cg15792713; chr17:g.(+)26674270 POLDIP2(−)/NA(+) | cg21633143; chr7:g.(−)154862021 HTR5A(+)/HTR5A-AS1(−) |
| cg07819108; chr8:g.(+)128921817 PVT1(+) | cg05222982; chr13:g.(+)28545214 NA(+)/CDX2(−) | cg05640731; chr10:g.(−)135018226 KNDC1(+)/NA(−) |
| cg17707487; chr13(+)114261869 TFDP1(+) | cg19875936; chr12:g.(−)7858848 NA(−)/NA(+) | cg19307500; chr19:g.(−)1083193 HMHA1 (ARHGAP45)(+)/NA(−) |
| BOT vs. BOT.V600E | BOT vs. lgOvCa | BOT vs. hgOvCa |
| chr11:g.both 47269539–47270908; NR1H3(+)/ACP2(−) | chr6:g.both 32935236–32943025; BRD2(+)/BRD2-IT1(+)/XXbac-BPG181M17.6(−)/HLA-DMA(−) | chr2:g.both 63275602–63285097; EHBP1-AS1(AC009501.4)(−)/OTX1(+) |
| chr6:g.both 31762409–31763873; VARS1(−)/NA(+) | chr6:g.both 30684340–30690844; TUBB(+)/MDC1(−) | chr6:g.both 30683787–30690844; TUBB(+)/MDC1(−) |
| chr15:g.(−)69706375–69707291; KIF23(+)/RP11-253M7.1(KIF23-AS1)(−) | chr6:g.both 31626915–31634890; C6orf47(−)/C6orf47-AS1(+)/CSNK2B(+)/GPANK1(−)/LY6G5B(+) | chr10:g.both 119291766–119296942; EMX2OS(−)/NA(+) |
| chr6:g.(−)31762409–31763873; VARS1(−)/NA(+) | chr6:g.both 30874989–30886161; GTF2H4(+)/VARS2(+)/NA(−) | chr4:g.both 1232112–1236678; CTBP1(−)/NA(+) |
| chr20:g.both 33459881–33461321; ACSS2(+)/GGT7(−) | chr6:g.(+)32935236–32943025; BRD2(+)/BRD2-IT1(+)/XXbac-BPG181M17.6(−)/HLA-DMA(−) | chr22:g.both 22899991–22902665; IGL locus (+): LL22NC03-63E9.3(+)/PRAME(−) |
| chr12:g.both 7282081–7283890; CLSTN3(+)/RBP5(−)/RP11-273B20.1(−) | chr6:g.both 31850189–31857100; SLC44A4(−)/EHMT2(−)/EHMT2-AS1(+) | chr15:g.both 37391121–37395115; MEIS2(−)/RP11-128A17.1(+) |
| chr7:g.both 137686266–137687260; AKR1D1(+)/CREB3L2(−) | chr6:g.both 33279563–33287809; TAPBP(−)/ZBTB22(−)/DAXX(−)/ NA(+) | chr6:g.both 32094845–32098253; ATF6B(−)/FKBPL(−)/NA(+) |
| chr6:g.both 32861863–32862953; LOC100294145(+)/HLA-Z(+)NA(−) | chr6:g.both 30519312–30525976; GNL1(−)/PRR3(+) | chr22:g.both 51016386–51017723; CPT1B(−)/CHKB-CPT1B(−)/CHKB-DT(+)/CHKB(−) |
| chr11:g.both 9595191–9596475; WEE1(+)/NA(−) | chr6:g.both 30651511–30659692; PPP1R18(−)/NRM(−)/NA(+) | chr2:g.both 171784610–171786316; GORASP2(+)/NA(−) |
| chr11:g.(+)47269539–47270669; NR1H3(+)/ACP2(−) | chr6:g.both 33381680–33387205; PHF1(+)/SYNGAP1(+)/CUTA(−) | chr5:g.both 134362967–134369605; PITX1(−)/PITX1-AS1(+) |
| BOT.V600E vs. lgOvCa | BOT.V600E vs. hgOvCa | lgOvCa vs. hgOvCa |
| chr6:g.both 30651511–30654559; PPP1R18(−)/NA(+) | chr1:g.both 2221807–2222674; SKI(+)/NA(−) | chr10:g.both 134977981–134981930; KNDC1(+)/NA(−) |
| chr6:g.both 31733434–31734580; VWA7(−)/SAPCD1-AS1(−)/NA(+) | chr19:g.both 58220080–58220818; ZNF551(+)/AC003006.7(+)/ZNF154(−) | chr6:g.both 32044869–32057846; TNXB(−)/RNA5SP206(−)/NA(+) |
| chr1:g.both 19664276–19665757; CAPZB(−)/NA(+) | chr1:g.both 1102276–1106175; MIR200B(+)/MIR200A(+)/MIR429(+)/TTLL10(+)/RP11-465B22.8(+)/NA(−) | chr6:g.both 30127760–30132715; TRIM15(+)/TRIM10(−) |
| chr6:g.both 152127812–152129791; ESR1(+)/ NA(−) | chr17:g.both 78865087–78866579; RPTOR(+)/NA(−) | chr19:g.both 405795–409510; C2CD4C(−)/NA(+) |
| chr7:g.both 964629–967277; ADAP1(−)/NA(+) | chr22:g.both 51016386–51017723; CPT1B(−)/CHKB-CPT1B(−)/CHKB-DT(+)/CHKB(−) | chr10:g.both 119291766–119297716; EMX2OS(−)/EMX2(+) |
| chr11:g.61521905–61523045; MYRF(+)/MYRF-AS1(−)/RP11-467L20.10(−) | chr16:g.2082689–2083393; NHERF2(SLC9A3R2)(+)/NA(−) | chr12:g.both 132686912–132689907; GALNT9(−)/NA(+) |
| chr3:g.both 129692836–129694665; TRH(+)/NA(−) | chr19:g.(+)58220080–58220818; ZNF551(+)/AC003006.7(+)/ZNF154(−) | chr12:g.both 132847907–132856142; LOC100130238(+)/GALNT9(−)/RP13-895J2.3(+) |
| chr12:g.both 6483708–6487080; LTBR(+)/SCNN1A(−) | chr3:g.both 185911208–185912486; DGKG(−)/NA(+) | chr4:g.both 100571622–100574653; NA(+)/C4orf54(−) |
| chr3:g.both 188664632–188666540; TPRG1(+)/TPRG1-AS1(−) | chr16:g.(−)2082745–2083178; NHERF2(SLC9A3R2)(+)/NA(−) | chr16:g.both 1127792–1132709; SSTR5(+)/SSTR5-AS1(−) |
| chr3:g.(+)129692836–129694665; TRH(+)/NA(−) | chr1:g.(+)1102276–1106175; MIR200B(+)/MIR200A(+)/MIR429(+)/TTLL10(+)/RP11-465B22.8(+)/NA(−) | chr16:g.both 1428639–1430367; UNKL(−)/NA(+) |
| hgOvCa | ||||||
| Cox Regression (alpha = 0.0005) | Mean beta Value (%) for DMR | |||||
| OS in the TP53 Accumulation = Yes Subgroup | HR [95% Cl] | p-Value | BOT | BOT V600E | lgOvCa | hgOvCa |
| HMOX1(+)/NA(−): chr22:g.(−)35776686–35777032 a | 8.4 × 10−5 [0–0.005] | 4.11 × 10−6 | 51.05 | 54.64 | 49.59 | 45.18 |
| Residual tumor > 2 cm vs. 0 cm | 6.24 [2.315–16.823] | 0.0003 | ||||
| OS in the TP therapy and TP53 accumulation = yes subgroup | ||||||
| HMOX1(+)/NA(−): chr22:g.(−)35775959–35777032 b | 3.71 × 10−6 [0–0.001] | 4.33 × 10−6 | 63.24 | 66.81 | 65.14 | 60.05 |
| Residual tumor > 2 cm vs. 0 cm | 8.3 [2.525–27.269] | 0.0005 | ||||
| TCN2(+)/PES1(−)/RP1-56J10.8(+): chr22:g.(−)31002067–31003655 c | 1.13 × 10−7 [0–0] | 5.26 × 10−6 | 36.66 | 32.48 | 31.57 | 26.55 |
| TCN2(+)/PES1(−)/RP1-56J10.8(+): chr22:g.both 31002067–31003655 c | 4.06 × 10−11 [0–0] | 6.35 × 10−6 | 27.65 | 23.92 | 22.3 | 18.88 |
| TCN2(+)/PES1(−)/RP1-56J10.8(+): chr22:g.both 31002362–31004367 c | 1.15 × 10−9 [0–0] | 7.31 × 10−6 | 31.29 | 27.29 | 25.84 | 22.48 |
| Logistic regression (alpha = 0.005) | Mean beta value (%) for DMR | |||||
| CR in the TP therapy subgroup | OR [95% Cl] | p-value | BOT | BOT V600E | lgOvCa | hgOvCa |
| NA(−)/NA(+): chr16:g.(−)880831–880831 | 5.14 [2.207–11.957] | 0.00015 | 83.21 | 85.29 | 92.53 | 77.47 |
| CR in the whole group (full table) | ||||||
| ABR(−)/NA(+): chr17:g.(−)1131424–1131781 d | 7.86 [2.566–24.063] | 0.00031 | 31.71 | 26.14 | 38 | 21.59 |
| NA(−)/NA(+): chr16:g.(−)880831–880831 | 3.4 [1.72–6.707] | 0.00043 | 83.21 | 85.29 | 92.53 | 77.47 |
| NCAM1(+)/RP11-629G13.1(−): chr11:g.(−)112831728–112832249 c | 4.77 [1.975–11.535] | 0.00052 | 28.82 | 19.77 | 17.35 | 13.42 |
| AC006372.4 (+)/NA(−): chr7:g.(−)157258854–157259343 c | 5.54 [2.104–14.596] | 0.00053 | 57.89 | 60.4 | 65.37 | 42.17 |
| PS in the whole group (full table) | ||||||
| NPTXR(−)/NA(+): chr.22:g.(+)39240094–39240424 | 4.04 [1.81–9.03] | 0.00066 | 13.97 | 8.24 | 3.42 | 3.86 |
| Residual tumor > 2 cm vs. 0 cm | 0.042 [0.006–0.294] | 0.0014 | ||||
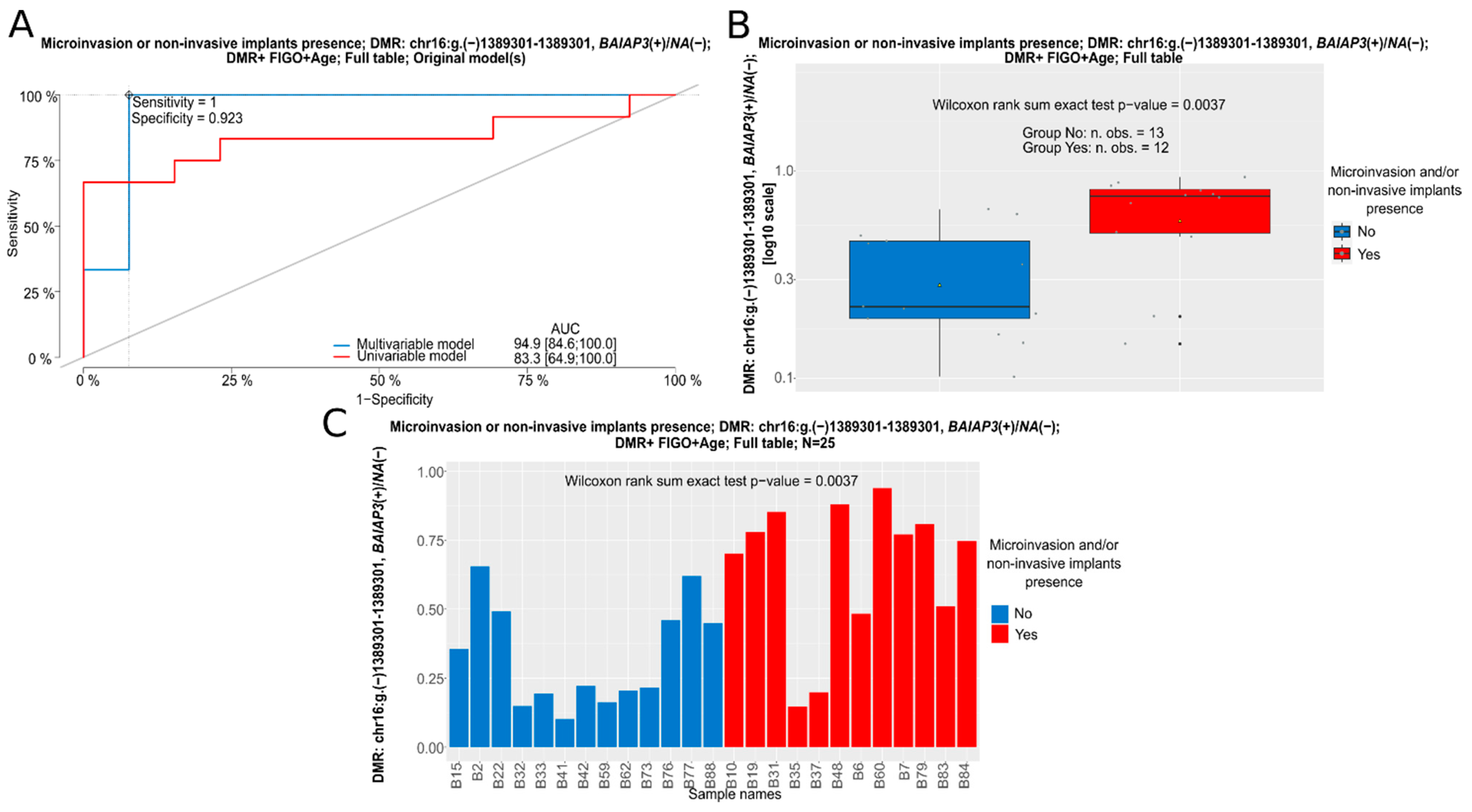
| BOTS | ||||||
| Logistic regression (alpha = 0.05) | Mean beta value (%) for DMR | |||||
| The presence of microinvasion and/or non-invasive implants in the whole group (full table) | OR [95% Cl] | p-value | BOT | BOT V600E | lgOvCa | hgOvCa |
| BAIAP3(+)/NA(−): chr.16:g.(−)1389301–1389301 | 49.04 [1.863–1290.778] | 0.02 | 45.4 | 52.22 | 63.23 | 38.27 |
| IL34(+)/NA(−): chr16:g.both 70613332–70613944 | 0.168 [0.037–0.769] | 0.022 | 52.68 | 49.89 | 42.6 | 47.9 |
| FIGO II/III vs FIGO IA/IB | 185.5 [2.166–15883.94] | 0.021 | ||||
| IL34(+)/NA(−): chr16:g.(−)70613332–70613944 | 0.139 [0.025–0.759] | 0.023 | 54.96 | 51.69 | 47.07 | 50.71 |
| FIGO II/III vs. FIGO IA/IB | 117.39 [1.936–7116.43] | 0.023 | ||||
| WNT10A(+)/NA(−): chr2:g.(+)219748780–219748780 | 0.14 [0.025–0.762] | 0.023 | 41.98 | 36.23 | 44.2 | 30.48 |
| FIGO II/III vs. FIGO IA/IB | 157.11 [1.691–14593.4] | 0.029 | ||||
| NEU1(−)/SLC44A4(−)/NA(+): chr.6:g.(+)31827414–31834178 | 0.022 [0.001–0.601] | 0.024 | 53.63 | 53.38 | 53.2 | 53.14 |
| FIGO II/III vs. FIGO IA/IB | 569.6 [1.093–296737.5] | 0.047 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szafron, L.A.; Iwanicka-Nowicka, R.; Sobiczewski, P.; Koblowska, M.; Dansonka-Mieszkowska, A.; Kupryjanczyk, J.; Szafron, L.M. The Diversity of Methylation Patterns in Serous Borderline Ovarian Tumors and Serous Ovarian Carcinomas. Cancers 2024, 16, 3524. https://doi.org/10.3390/cancers16203524
Szafron LA, Iwanicka-Nowicka R, Sobiczewski P, Koblowska M, Dansonka-Mieszkowska A, Kupryjanczyk J, Szafron LM. The Diversity of Methylation Patterns in Serous Borderline Ovarian Tumors and Serous Ovarian Carcinomas. Cancers. 2024; 16(20):3524. https://doi.org/10.3390/cancers16203524
Chicago/Turabian StyleSzafron, Laura A., Roksana Iwanicka-Nowicka, Piotr Sobiczewski, Marta Koblowska, Agnieszka Dansonka-Mieszkowska, Jolanta Kupryjanczyk, and Lukasz M. Szafron. 2024. "The Diversity of Methylation Patterns in Serous Borderline Ovarian Tumors and Serous Ovarian Carcinomas" Cancers 16, no. 20: 3524. https://doi.org/10.3390/cancers16203524
APA StyleSzafron, L. A., Iwanicka-Nowicka, R., Sobiczewski, P., Koblowska, M., Dansonka-Mieszkowska, A., Kupryjanczyk, J., & Szafron, L. M. (2024). The Diversity of Methylation Patterns in Serous Borderline Ovarian Tumors and Serous Ovarian Carcinomas. Cancers, 16(20), 3524. https://doi.org/10.3390/cancers16203524






