Predicting Biochemical Recurrence of Prostate Cancer Post-Prostatectomy Using Artificial Intelligence: A Systematic Review
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Literature Search Strategy
2.2. Eligibility Criteria
2.3. Screening and Study Selection
2.4. Quality and Risk of Bias Assessment
3. Results
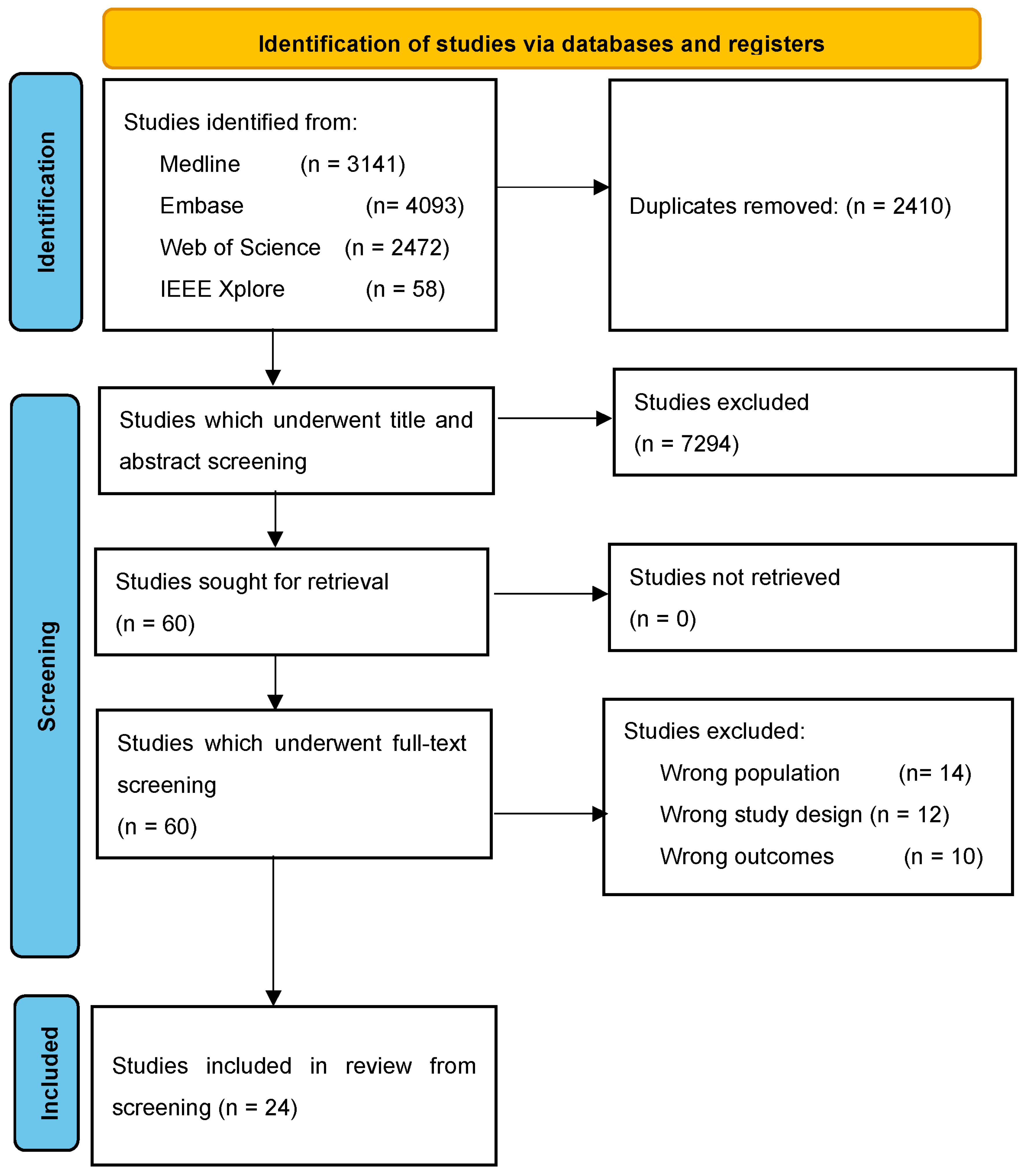
3.1. Screening Process
3.2. Characteristics of Included Studies
3.3. Characteristics of Patients in Included Studies
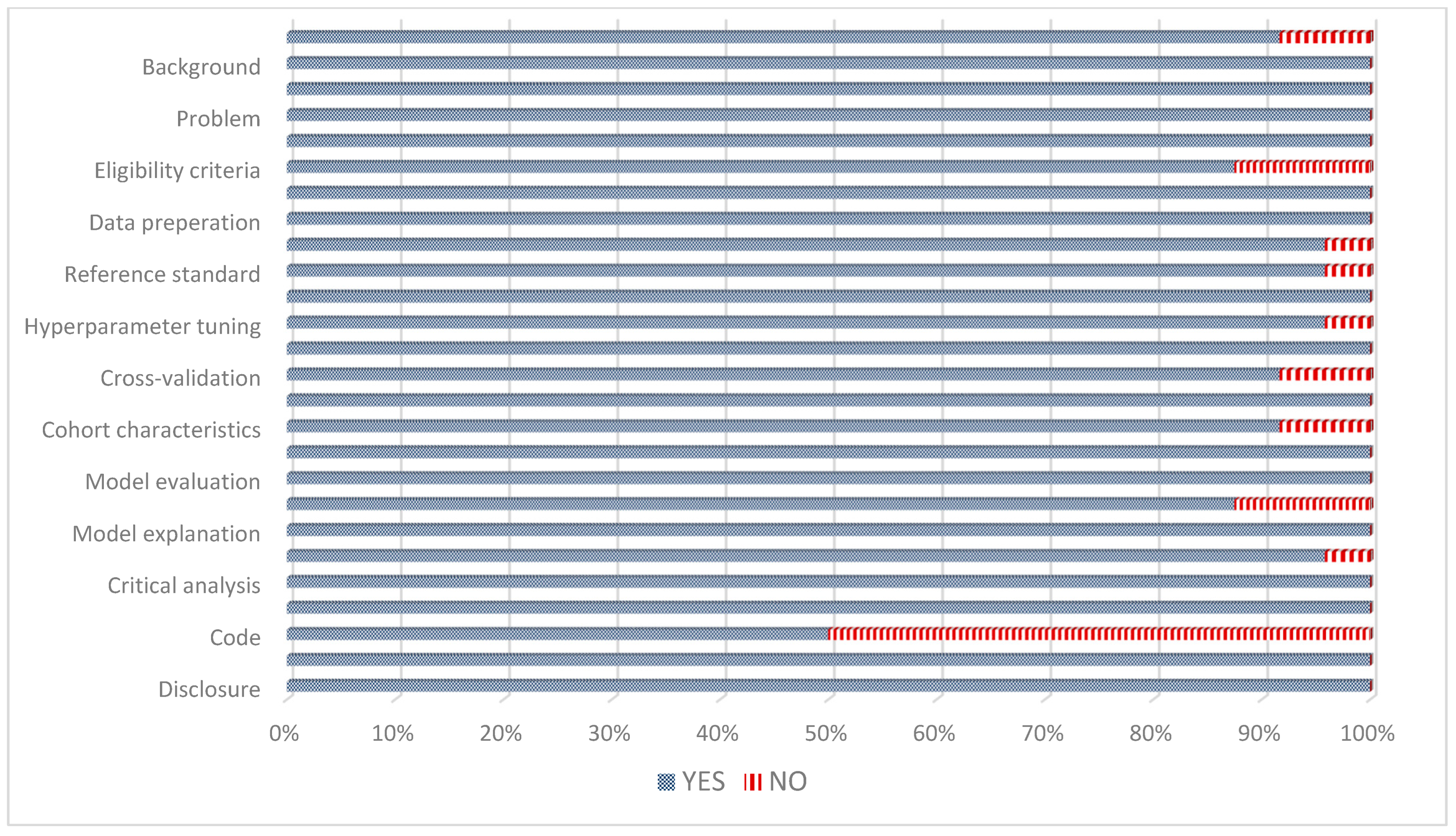
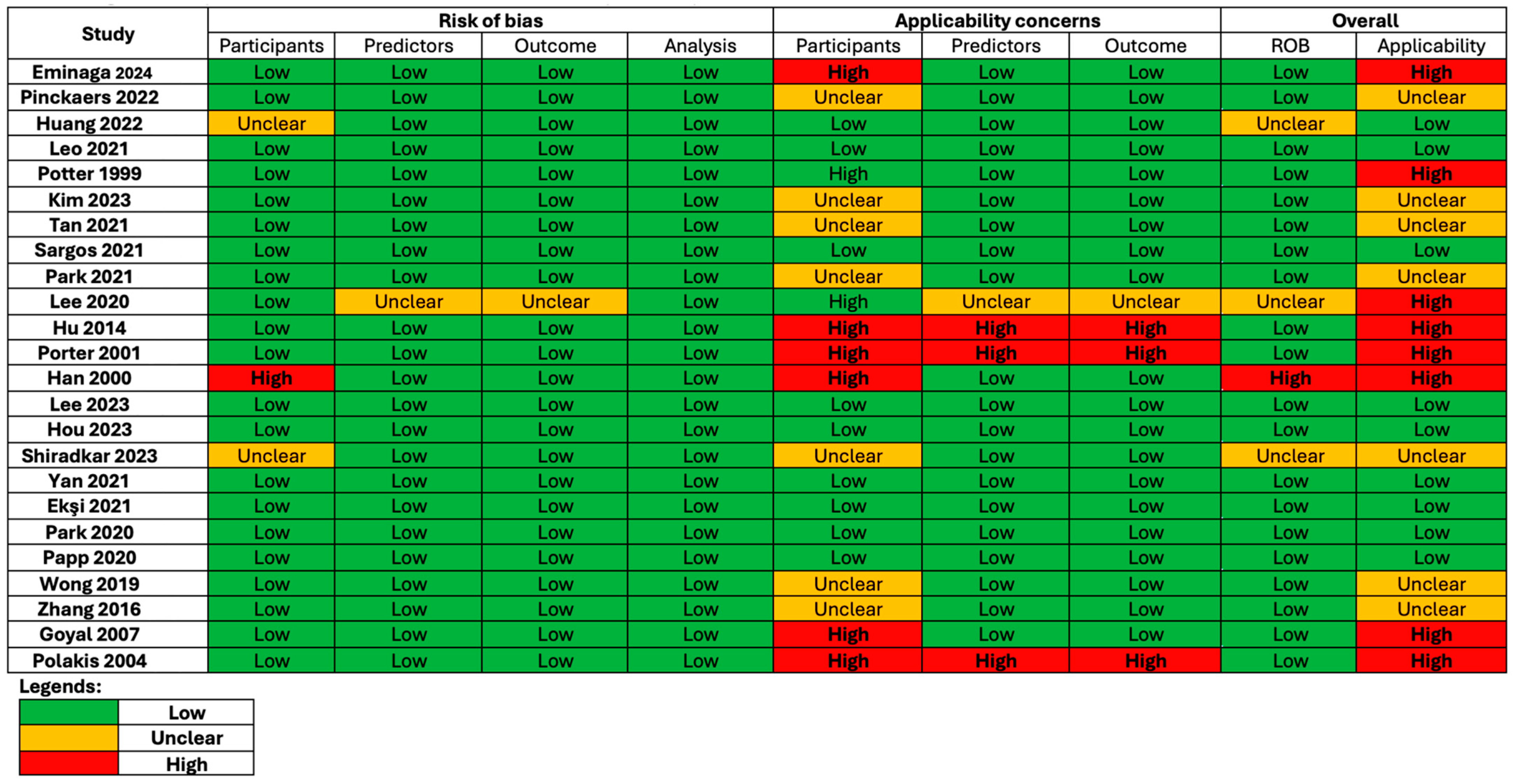
3.4. Quality and Risk of Bias Assessment of Included Studies
3.5. AI Developed Using Histological Variables Only
3.6. AI Developed Using Clinical and Histological Variables
3.7. AI Developed Using Radiological Variables
3.8. Comparing AI Models
3.9. Comparing AI against Traditional Methods of Predicting BCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cookson, M.S.; Aus, G.; Burnett, A.L.; Canby-Hagino, E.D.; D’Amico, A.V.; Dmochowski, R.R.; Eton, D.T.; Forman, J.D.; Goldenberg, S.L.; Hernandez, J.; et al. Variation in the definition of biochemical recurrence in patients treated for localized prostate cancer: The American Urological Association Prostate Guidelines for Localized Prostate Cancer Update Panel report and recommendations for a standard in the reporting of surgical outcomes. J. Urol. 2007, 177, 540–545. [Google Scholar] [PubMed]
- Carroll, P.R.; Parsons, J.K.; Andriole, G.; Bahnson, R.R.; Barocas, D.A.; Castle, E.P.; Catalona, W.J.; Dahl, D.M.; Davis, J.W.; Epstein, J.I.; et al. NCCN Clinical Practice Guidelines Prostate Cancer Early Detection, Version 2.2015. J. Natl. Compr. Cancer Netw. 2015, 13, 1534–1561. [Google Scholar] [CrossRef] [PubMed]
- Van den Broeck, T.; van den Bergh, R.C.N.; Arfi, N.; Gross, T.; Moris, L.; Briers, E.; Cumberbatch, M.; De Santis, M.; Tilki, D.; Fanti, S.; et al. Prognostic Value of Biochemical Recurrence Following Treatment with Curative Intent for Prostate Cancer: A Systematic Review. Eur. Urol. 2019, 75, 967–987. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, A.J.; Scardino, P.T.; Eastham, J.A.; Bianco, F.J., Jr.; Dotan, Z.A.; DiBlasio, C.J.; Reuther, A.; Klein, E.A.; Kattan, M.W. Postoperative nomogram predicting the 10-year probability of prostate cancer recurrence after radical prostatectomy. J. Clin. Oncol. 2005, 23, 7005–7012. [Google Scholar] [CrossRef]
- Cooperberg, M.R.; Hilton, J.F.; Carroll, P.R. The CAPRA-S score: A straightforward tool for improved prediction of outcomes after radical prostatectomy. Cancer 2011, 117, 5039–5046. [Google Scholar] [CrossRef]
- Stephenson, A.J.; Eggener, S.E.; Hernandez, A.V.; Klein, E.A.; Kattan, M.W.; Wood, D.P., Jr.; Rabah, D.M.; Eastham, J.A.; Scardino, P.T. Do margins matter? The influence of positive surgical margins on prostate cancer-specific mortality. Eur. Urol. 2014, 65, 675–680. [Google Scholar] [CrossRef]
- Pound, C.R.; Partin, A.W.; Eisenberger, M.A.; Chan, D.W.; Pearson, J.D.; Walsh, P.C. Natural History of Progression After PSA Elevation Following Radical Prostatectomy. JAMA 1999, 281, 1591–1597. [Google Scholar] [CrossRef]
- Tourinho-Barbosa, R.; Srougi, V.; Nunes-Silva, I.; Baghdadi, M.; Rembeyo, G.; Eiffel, S.S.; Barret, E.; Rozet, F.; Galiano, M.; Cathelineau, X.; et al. Biochemical recurrence after radical prostatectomy: What does it mean? Int. Braz. J. Urol. 2018, 44, 14–21. [Google Scholar] [CrossRef]
- Perera, M.; Papa, N.; Christidis, D.; Wetherell, D.; Hofman, M.S.; Murphy, D.G.; Bolton, D.; Lawrentschuk, N. Sensitivity, Specificity, and Predictors of Positive 68Ga-Prostate-specific Membrane Antigen Positron Emission Tomography in Advanced Prostate Cancer: A Systematic Review and Meta-analysis. Eur. Urol. 2016, 70, 926–937. [Google Scholar] [CrossRef]
- Liu, J.; Cundy, T.P.; Woon, D.T.S.; Desai, N.; Palaniswami, M.; Lawrentschuk, N. A systematic review on artificial intelligence evaluating PSMA PET scan for intraprostatic cancer. BJU Int. 2024. [Google Scholar] [CrossRef]
- Liu, J.; Cundy, T.P.; Woon, D.T.S.; Lawrentschuk, N. A Systematic Review on Artificial Intelligence Evaluating Metastatic Prostatic Cancer and Lymph Nodes on PSMA PET Scans. Cancers 2024, 16, 486. [Google Scholar] [CrossRef] [PubMed]
- Sandeman, K.; Eineluoto, J.T.; Pohjonen, J.; Erickson, A.; Kilpeläinen, T.P.; Järvinen, P.; Santti, H.; Petas, A.; Matikainen, M.; Marjasuo, S.; et al. Prostate MRI added to CAPRA, MSKCC and Partin cancer nomograms significantly enhances the prediction of adverse findings and biochemical recurrence after radical prostatectomy. PLoS ONE 2020, 15, e0235779. [Google Scholar] [CrossRef] [PubMed]
- Kwong, J.C.C.; McLoughlin, L.C.; Haider, M.; Goldenberg, M.G.; Erdman, L.; Rickard, M.; Lorenzo, A.J.; Hung, A.J.; Farcas, M.; Goldenberg, L.; et al. Standardized Reporting of Machine Learning Applications in Urology: The STREAM-URO Framework. Eur. Urol. Focus 2021, 7, 672–682. [Google Scholar] [CrossRef] [PubMed]
- Wolff, R.F.; Moons, K.G.M.; Riley, R.D.; Whiting, P.F.; Westwood, M.; Collins, G.S.; Reitsma, J.B.; Kleijnen, J.; Mallett, S.; for the PROBAST Group. PROBAST: A Tool to Assess the Risk of Bias and Applicability of Prediction Model Studies. Ann. Intern. Med. 2019, 170, 51–58. [Google Scholar] [CrossRef]
- Eminaga, O.; Saad, F.; Tian, Z.; Wolffgang, U.; Karakiewicz, P.I.; Ouellet, V.; Azzi, F.; Spieker, T.; Helmke, B.M.; Graefen, M.; et al. Artificial intelligence unravels interpretable malignancy grades of prostate cancer on histology images. NPJ Imaging 2024, 2, 6. [Google Scholar] [CrossRef]
- Pinckaers, H.; van Ipenburg, J.; Melamed, J.; De Marzo, A.; Platz, E.A.; van Ginneken, B.; van der Laak, J.; Litjens, G. Predicting biochemical recurrence of prostate cancer with artificial intelligence. Commun. Med. 2022, 2, 64. [Google Scholar] [CrossRef]
- Huang, W.; Randhawa, R.; Jain, P.; Hubbard, S.; Eickhoff, J.; Kummar, S.; Wilding, G.; Basu, H.; Roy, R. A Novel Artificial Intelligence-Powered Method for Prediction of Early Recurrence of Prostate Cancer After Prostatectomy and Cancer Drivers. JCO Clin. Cancer Inform. 2022, 6, e2100131. [Google Scholar] [CrossRef]
- Leo, P.; Chandramouli, S.; Farré, X.; Elliott, R.; Janowczyk, A.; Bera, K.; Fu, P.; Janaki, N.; El-Fahmawi, A.; Shahait, M.; et al. Computationally Derived Cribriform Area Index from Prostate Cancer Hematoxylin and Eosin Images Is Associated with Biochemical Recurrence Following Radical Prostatectomy and Is Most Prognostic in Gleason Grade Group 2. Eur. Urol. Focus 2021, 7, 722–732. [Google Scholar] [CrossRef]
- Potter, S.R.; Miller, M.C.; Mangold, L.A.; Jones, K.A.; Epstein, J.I.; Veltri, R.W.; Partin, A.W. Genetically engineered neural networks for predicting prostate cancer progression after radical prostatectomy. Urology 1999, 54, 791–795. [Google Scholar] [CrossRef]
- Kim, J.-K.; Hong, S.-H.; Choi, I.-Y. Partial Correlation Analysis and Neural-Network-Based Prediction Model for Biochemical Recurrence of Prostate Cancer after Radical Prostatectomy. Appl. Sci. 2023, 13, 891. [Google Scholar] [CrossRef]
- Sargos, P.; Leduc, N.; Giraud, N.; Gandaglia, G.; Roumiguié, M.; Ploussard, G.; Rozet, F.; Soulié, M.; Mathieu, R.; Artus, P.M.; et al. Deep Neural Networks Outperform the CAPRA Score in Predicting Biochemical Recurrence after Prostatectomy. Front. Oncol. 2020, 10, 607923. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Rho, M.J.; Moon, H.W.; Kim, J.; Lee, C.; Kim, D.; Kim, C.-S.; Jeon, S.S.; Kang, M.; Lee, J.Y. Answer AI for Prostate Cancer: Predicting Biochemical Recurrence Following Radical Prostatectomy. Technol. Cancer Res. Treat. 2021, 20, 15330338211024660. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Yu, S.H.; Kim, Y.; Kim, J.K.; Hong, J.H.; Kim, C.-S.; Seo, S.I.; Byun, S.-S.; Jeong, C.W.; Lee, J.Y.; et al. Prediction System for Prostate Cancer Recurrence Using Machine Learning. Appl. Sci. 2020, 10, 1333. [Google Scholar] [CrossRef]
- Hu, X.H.; Cammann, H.; Meyer, H.A.; Jung, K.; Lu, H.B.; Leva, N.; Magheli, A.; Stephan, C. Risk prediction models for biochemical recurrence after radical prostatectomy using prostate-specific antigen and Gleason score. Asian J. Androl. 2014, 16, 897–901. [Google Scholar]
- Porter, C.; O’Donnell, C.; Crawford, E.D.; Gamito, E.J.; Errejon, A.; Genega, E.; Sotelo, T.; Tewari, A. Artificial neural network model to predict biochemical failure after radical prostatectomy. Mol. Urol. 2001, 5, 159–162. [Google Scholar] [CrossRef]
- Han, M.; Snow, P.B.; Epstein, J.I.; Chan, T.Y.; Jones, K.A.; Walsh, P.C.; Partin, A.W. A neural network predicts progression for men with gleason score 3+4 versus 4+3 tumors after radical prostatectomy. Urology 2000, 56, 994–999. [Google Scholar] [CrossRef]
- Lee, H.W.; Kim, E.; Na, I.; Kim, C.K.; Seo, S.I.; Park, H. Novel Multiparametric Magnetic Resonance Imaging-Based Deep Learning and Clinical Parameter Integration for the Prediction of Long-Term Biochemical Recurrence-Free Survival in Prostate Cancer after Radical Prostatectomy. Cancers 2023, 15, 3416. [Google Scholar] [CrossRef]
- Hou, Y.; Jiang, K.W.; Wang, L.L.; Zhi, R.; Bao, M.L.; Li, Q.; Zhang, J.; Qu, J.-R.; Zhu, F.-P.; Zhang, Y.-D. Biopsy-free AI-aided precision MRI assessment in prediction of prostate cancer biochemical recurrence. Br. J. Cancer 2023, 129, 1625–1633. [Google Scholar] [CrossRef]
- Shiradkar, R.; Ghose, S.; Mahran, A.; Li, L.; Hubbard, I.; Fu, P.; Tirumani, S.H.; Ponsky, L.; Purysko, A.; Madabhushi, A. Prostate Surface Distension and Tumor Texture Descriptors from Pre-Treatment MRI Are Associated with Biochemical Recurrence Following Radical Prostatectomy: Preliminary Findings. Front. Oncol. 2022, 12, 841801. [Google Scholar] [CrossRef]
- Ekşi, M.; Evren, İ.; Akkaş, F.; Arıkan, Y.; Özdemir, O.; Özlü, D.N.; Ayten, A.; Sahin, S.; Tuğcu, V.; Taşçı, A. Machine learning algorithms can more efficiently predict biochemical recurrence after robot-assisted radical prostatectomy. Prostate 2021, 81, 913–920. [Google Scholar] [CrossRef]
- Park, S.; Byun, J.; Woo, J.Y. A Machine Learning Approach to Predict an Early Biochemical Recurrence after a Radical Prostatectomy. Appl. Sci. 2020, 10, 3854. [Google Scholar] [CrossRef]
- Zhang, Y.D.; Wang, J.; Wu, C.J.; Bao, M.L.; Li, H.; Wang, X.N.; Tao, J.; Shi, H.-B. An imaging-based approach predicts clinical outcomes in prostate cancer through a novel support vector machine classification. Oncotarget 2016, 7, 78140–78151. [Google Scholar] [CrossRef] [PubMed]
- Goyal, N.K.; Kumar, A.; Acharya, R.L.; Dwivedi, U.S.; Trivedi, S.; Singh, P.B.; Singh, T.N. Prediction of biochemical failure in localized carcinoma of prostate after radical prostatectomy by neuro-fuzzy. Indian J. Urol. 2007, 23, 14–17. [Google Scholar] [PubMed]
- Poulakis, V.; Witzsch, U.; de Vries, R.; Emmerlich, V.; Meves, M.; Altmannsberger, H.M.; Becht, E. Preoperative neural network using combined magnetic resonance imaging variables, prostate specific antigen, and Gleason score to predict prostate cancer recurrence after radical prostatectomy. Eur. Urol. 2004, 46, 571–578. [Google Scholar] [CrossRef]
- Yan, Y.; Shao, L.; Liu, Z.; He, W.; Yang, G.; Liu, J.; Xia, H.; Zhang, Y.; Chen, H.; Liu, C.; et al. Deep Learning with Quantitative Features of Magnetic Resonance Images to Predict Biochemical Recurrence of Radical Prostatectomy: A Multi-Center Study. Cancers 2021, 13, 3098. [Google Scholar] [CrossRef]
- Tan, Y.G.; Fang, A.H.S.; Lim, J.K.S.; Khalid, F.; Chen, K.; Ho, H.S.S.; Yuen, J.S.P.; Huang, H.H.; Tay, K.J. Incorporating artificial intelligence in urology: Supervised machine learning algorithms demonstrate comparative advantage over nomograms in predicting biochemical recurrence after prostatectomy. Prostate 2022, 82, 298–305. [Google Scholar] [CrossRef]
- Papp, L.; Spielvogel, C.P.; Grubmüller, B.; Grahovac, M.; Krajnc, D.; Ecsedi, B.; Sareshgi, R.A.; Mohamad, D.; Hamboeck, M.; Rausch, I.; et al. Supervised machine learning enables non-invasive lesion characterization in primary prostate cancer with [68Ga]Ga-PSMA-11 PET/MRI. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 1795–1805. [Google Scholar] [CrossRef]
- Wong, N.C.; Lam, C.; Patterson, L.; Shayegan, B. Use of machine learning to predict early biochemical recurrence after robot-assisted prostatectomy. BJU Int. 2019, 123, 51–57. [Google Scholar] [CrossRef]
- Flach, R.N.; Willemse, P.M.; Suelmann, B.B.M.; Deckers, I.A.G.; Jonges, T.N.; van Dooijeweert, C.; van Diest, P.J.; Meijer, R.P. Significant Inter- and Intralaboratory Variation in Gleason Grading of Prostate Cancer: A Nationwide Study of 35,258 Patients in The Netherlands. Cancers 2021, 13, 5378. [Google Scholar] [CrossRef]
- Annamalai, A.; Fustok, J.N.; Beltran-Perez, J.; Rashad, A.T.; Krane, L.S.; Triche, B.L. Interobserver Agreement and Accuracy in Interpreting mpMRI of the Prostate: A Systematic Review. Curr. Urol. Rep. 2022, 23, 1–10. [Google Scholar] [CrossRef]
- Li, X.T.; Huang, R.Y. Standardization of imaging methods for machine learning in neuro-oncology. Neuro-Oncol. Adv. 2020, 2 (Suppl. S4), iv49–iv55. [Google Scholar] [CrossRef] [PubMed]
- Tilki, D.; van den Bergh, R.C.N.; Briers, E.; Van den Broeck, T.; Brunckhorst, O.; Darraugh, J.; Eberli, D.; De Meerleer, G.; De Santis, M.; Farolfi, A.; et al. EAU-EANM-ESTRO-ESUR-ISUP-SIOG Guidelines on Prostate Cancer. Part II—2024 Update: Treatment of Relapsing and Metastatic Prostate Cancer. Eur. Urol. 2024, 86, 164–182. [Google Scholar] [CrossRef]
- Ho, R.; Siddiqui, M.M.; George, A.K.; Frye, T.; Kilchevsky, A.; Fascelli, M.; Shakir, N.A.; Chelluri, R.; Abboud, S.F.; Walton-Diaz, A.; et al. Preoperative Multiparametric Magnetic Resonance Imaging Predicts Biochemical Recurrence in Prostate Cancer after Radical Prostatectomy. PLoS ONE 2016, 11, e0157313. [Google Scholar] [CrossRef] [PubMed]
- Gandaglia, G.; Ploussard, G.; Valerio, M.; Marra, G.; Moschini, M.; Martini, A.; Roumiguié, M.; Fossati, N.; Stabile, A.; Beauval, J.-B.; et al. Prognostic Implications of Multiparametric Magnetic Resonance Imaging and Concomitant Systematic Biopsy in Predicting Biochemical Recurrence After Radical Prostatectomy in Prostate Cancer Patients Diagnosed with Magnetic Resonance Imaging-targeted Biopsy. Eur. Urol. Oncol. 2020, 3, 739–747. [Google Scholar] [CrossRef] [PubMed]
- Bourbonne, V.; Vallières, M.; Lucia, F.; Doucet, L.; Visvikis, D.; Tissot, V.; Pradier, O.; Hatt, M.; Schick, U. MRI-Derived Radiomics to Guide Post-operative Management for High-Risk Prostate Cancer. Front. Oncol. 2019, 9, 807. [Google Scholar] [CrossRef]
- Bourbonne, V.; Fournier, G.; Vallières, M.; Lucia, F.; Doucet, L.; Tissot, V.; Cuvelier, G.; Hue, S.; Du, H.L.P.; Perdriel, L.; et al. External Validation of an MRI-Derived Radiomics Model to Predict Biochemical Recurrence after Surgery for High-Risk Prostate Cancer. Cancers 2020, 12, 814. [Google Scholar] [CrossRef]
- Hofman, M.S.; Lawrentschuk, N.; Francis, R.J.; Tang, C.; Vela, I.; Thomas, P.; Rutherford, N.; Martin, J.M.; Frydenberg, M.; Shakher, R.; et al. Prostate-specific membrane antigen PET-CT in patients with high-risk prostate cancer before curative-intent surgery or radiotherapy (proPSMA): A prospective, randomised, multicentre study. Lancet 2020, 395, 1208–1216. [Google Scholar] [CrossRef]
- Qiu, X.; Chen, M.; Yin, H.; Zhang, Q.; Li, H.; Guo, S.; Fu, Y.; Zang, S.; Ai, S.; Wang, F.; et al. Prediction of Biochemical Recurrence After Radical Prostatectomy Based on Preoperative (68)Ga-PSMA-11 PET/CT. Front. Oncol. 2021, 11, 745530. [Google Scholar] [CrossRef]
- Baas, D.J.H.; Schilham, M.; Hermsen, R.; de Baaij, J.M.S.; Vrijhof, H.; Hoekstra, R.J.; Sedelaar, J.P.M.; Küsters-Vandevelde, H.V.N.; Gotthardt, M.; Wijers, C.H.W.; et al. Preoperative PSMA-PET/CT as a predictor of biochemical persistence and early recurrence following radical prostatectomy with lymph node dissection. Prostate Cancer Prostatic Dis. 2022, 25, 65–70. [Google Scholar] [CrossRef]
- Coskun, N.; Kartal, M.O.; Erdogan, A.S.; Ozdemir, E. Development and validation of a nomogram for predicting the likelihood of metastasis in prostate cancer patients undergoing Ga-68 PSMA PET/CT due to biochemical recurrence. Nucl. Med. Commun. 2022, 43, 952–958. [Google Scholar] [CrossRef]
- Bodar, Y.J.L.; Veerman, H.; Meijer, D.; de Bie, K.; van Leeuwen, P.J.; Donswijk, M.L.; van Moorselaar, R.J.A.; Hendrikse, N.H.; Boellaard, R.; Oprea-Lager, D.E.; et al. Standardised uptake values as determined on prostate-specific membrane antigen positron emission tomography/computed tomography is associated with oncological outcomes in patients with prostate cancer. BJU Int. 2022, 129, 768–776. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Qiu, X.; Zhang, Q.; Zhang, C.; Zhou, Y.H.; Zhao, X.; Fu, Y.; Wang, F.; Guo, H. PSMA uptake on [68Ga]-PSMA-11-PET/CT positively correlates with prostate cancer aggressiveness. Q. J. Nucl. Med. Mol. Imaging 2022, 66, 67–73. [Google Scholar] [CrossRef]
- Milonas, D.; Venclovas, Z.; Sasnauskas, G.; Ruzgas, T. The Significance of Prostate Specific Antigen Persistence in Prostate Cancer Risk Groups on Long-Term Oncological Outcomes. Cancers 2021, 13, 2453. [Google Scholar] [CrossRef] [PubMed]
- Poon, A.I.F.; Sung, J.J.Y. Opening the black box of AI-Medicine. J Gastroenterol. Hepatol. 2021, 36, 581–584. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, A.; Kora, R. A comprehensive review on ensemble deep learning: Opportunities and challenges. J. King Saud. Univ.—Comput. Inf. Sci. 2023, 35, 757–774. [Google Scholar] [CrossRef]
- Žlahtič, B.; Završnik, J.; Kokol, P.; Blažun Vošner, H.; Sobotkiewicz, N.; Antolinc Schaubach, B.; Kirbiš, S. Trusting AI made decisions in healthcare by making them explainable. Sci. Prog. 2024, 107, 368504241266573. [Google Scholar] [CrossRef]
- Morgan, T.M.; Boorjian, S.A.; Buyyounouski, M.K.; Chapin, B.F.; Chen, D.Y.T.; Cheng, H.H.; Chou, R.; Jacene, H.A.; Kamran, S.C.; Kim, S.K.; et al. Salvage Therapy for Prostate Cancer: AUA/ASTRO/SUO Guideline Part II: Treatment Delivery for Non-metastatic Biochemical Recurrence After Primary Radical Prostatectomy. J. Urol. 2024, 211, 518–525. [Google Scholar] [CrossRef]
- Schaeffer, E.M.; Srinivas, S.; Adra, N.; An, Y.; Barocas, D.; Bitting, R.; Bryce, A.; Chapin, B.; Cheng, H.H.; D’Amico, A.V.; et al. Prostate Cancer, Version 4.2023, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2023, 21, 1067–1096. [Google Scholar] [CrossRef]
- Mottet, N.; van den Bergh, R.C.N.; Briers, E.; Van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer-2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2021, 79, 243–262. [Google Scholar] [CrossRef]
| Author and Year | Data Input | AI Models and/or Traditional Methods of BCR Prediction Used | Findings |
|---|---|---|---|
| Kim 2023 [20] | Clinicopathological variables | PCNN vs. SVM vs. RFC | Top three best-performing were PCNN, RF, and a tree-based algorithm, with the accuracy of all three models averaging 0.87. |
| Lee 2020 [23] | Clinicopathological variables | RFC vs. NN vs. LR vs. decision tree vs. gradient boosting classifier | Top three at predicting 5-year BCR were LR, NN, and RF (AUROCs of 0.81, 0.80, and 0.80, respectively). |
| Hu 2014 [24] | Clinicopathological variables | ANN vs. LR | The AUROCs of ANN (0.75) and LR (0.76) outperformed the Gleason score (0.71) and T-stage or PSA (0.62) in predicting 10-year BCR. |
| Han 2000 [26] | Clinicopathological variables | ANN vs. LR | The ANN outperformed LR in predicting 3-year BCR with an AUROC of 0.81 versus 0.68. |
| Park 2020 [31] | Clinicopathological variables and MRI | KNN vs. MLP vs. DT vs. auto-encoder | Auto-encoder showed the highest prediction ability in 1-year BCR after RP (AUC = 0.638), followed by MLP (AUC = 0.61), KNN (AUC = 0.60), and DT (AUC = 0.53). |
| Zhang 2016 [32] | Clinicopathological variables and MRI | SVM vs. LR | When compared to LR, SVM had significantly higher AUROC (0.96 vs. 0.89; p =0.007), sensitivity (93.3% vs. 83.3%; p = 0.025), specificity (91.7% vs. 77.2%; p =0.009), and accuracy (92.2% vs. 79.0%; p = 0.006) in predicting 3-year BCR. |
| Wong 2019 [38] | Clinicopathological variables, prostate ultrasound size, and operative variables | KNN vs. RFC vs. LR vs. conventional statistical regression model | KNN, RFC, and LR outperformed the conventional statistical regression model in predicting 1-year BCR. Respectively, the AUCs were 0.90, 0.92, and 0.94, and the accuracy values were 0.98, 0.95, and 0.98. |
| Ekşi 2021 [30] | Clinicopathological variables and mpMRI | RFC vs. KNN vs. LR vs. conventional statistical regression model | All ML models outperformed the conventional statistical regression model in the prediction of BCR. The AUROCs for RFC, KNN, and LR were 0.95, 0.93, and 0.93, respectively. |
| Tan 2021 [36] | Clinicopathological variables | Naive Bayes vs. RFC vs. SVM vs. traditional regression analyses vs. nomograms | AUCs for the prediction of BCR at 1, 3, and 5 years for Naive Bayes were 0.894, 0.876, and 0.894, for RFC were 0.846, 0.875, and 0.888, and for SVM were 0.835, 0.850, and 0.855, respectively. Although all three ML models were equivocal to traditional regression analyses, they outperformed existing nomograms (Kattan, John Hopkins [JHH], CAPSURE). |
| Sargos 2021 [21] | Clinicopathological variables | KNN vs. RFC vs. DNN vs. CAPRA score | The DNN model showed the highest AUC, 0.84, in predicting 3-year BCR when compared to LR, KNN, RF, and Cox regression, with AUC values of 0.77, 0.58, 0.74, and 0.75, respectively. The DNN developed based on CAPRA variables (AUROC of 0.7) outperformed the CAPRA score itself (AUROC of 0.63). |
| Hou 2023 [28] | Clinicopathological variables and mpMRI radiomics | Deep survival network vs. CAPRA score | The deep survival network could match a histopathological model (Concordance index 0.81 to 0.83 vs. 0.79 to 0.81, p > 0.05) and has a maximally 5.16-fold, 12.8-fold, and 2.09-fold (p < 0.05) benefit compared to the conventional D’Amico score, the CAPRA score, and the CAPRA Postsurgical score. |
| Shiradkar 2023 [29] | Biparametric MRI | RFC and ML vs. CAPRA score | Integration of RFC and ML performed the best at predicting BCR, with an AUC of 0.75 as compared to random forest classifier (0.70, p = 0.04) or ML (0.69, p = 0.01) alone. |
| Yan 2021 [35] | Quantitative features of MRI | DL vs. CAPRA score vs. NCCN model vs. Gleason grade group systems | The DL model (C-index of 0.80) developed outperformed Gleason grade group systems (C-index of 0.58), NCCN model (C-index of 0.59), and the CAPRA-S score (C-index of 0.68). |
| Poulakis 2004 [34] | clinicopathological variables, ultrasound, and MRI | ANN vs. Cox regression analysis vs. Kattan nomogram | ANN was comparable to Cox regression analysis and Kattan nomogram in terms of predicting 5-year BCR (AUROCs of 0.77, 0.74, and 0.73, respectively). With the addition of MRI findings, ANN outperformed Cox regression and Kattan nomogram, with an AUC of 0.897, in predicting 5-year BCR. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Zhang, H.; Woon, D.T.S.; Perera, M.; Lawrentschuk, N. Predicting Biochemical Recurrence of Prostate Cancer Post-Prostatectomy Using Artificial Intelligence: A Systematic Review. Cancers 2024, 16, 3596. https://doi.org/10.3390/cancers16213596
Liu J, Zhang H, Woon DTS, Perera M, Lawrentschuk N. Predicting Biochemical Recurrence of Prostate Cancer Post-Prostatectomy Using Artificial Intelligence: A Systematic Review. Cancers. 2024; 16(21):3596. https://doi.org/10.3390/cancers16213596
Chicago/Turabian StyleLiu, Jianliang, Haoyue Zhang, Dixon T. S. Woon, Marlon Perera, and Nathan Lawrentschuk. 2024. "Predicting Biochemical Recurrence of Prostate Cancer Post-Prostatectomy Using Artificial Intelligence: A Systematic Review" Cancers 16, no. 21: 3596. https://doi.org/10.3390/cancers16213596
APA StyleLiu, J., Zhang, H., Woon, D. T. S., Perera, M., & Lawrentschuk, N. (2024). Predicting Biochemical Recurrence of Prostate Cancer Post-Prostatectomy Using Artificial Intelligence: A Systematic Review. Cancers, 16(21), 3596. https://doi.org/10.3390/cancers16213596






