Deciphering the Role of ASPM in Breast Cancer: A Comprehensive Multicohort Study
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Discovery Cohort
2.2. Validation Cohorts
2.2.1. The METABRIC Cohort
2.2.2. The Uppsala Cohort
2.2.3. Combined Multicentric Cohorts
2.3. Proteomic Study
2.4. Selection of ASPM
2.5. Aberrations of Gene Copy Number
2.6. Western Blotting
2.7. Tissue Microarrays and Immunohistochemistry
2.8. Assessment of ASPM Expression
2.9. Ki67 Staining and Scoring
2.10. Statistical Analysis
3. Results
3.1. Differential Gene Expression Analysis and the Selection of ASPM
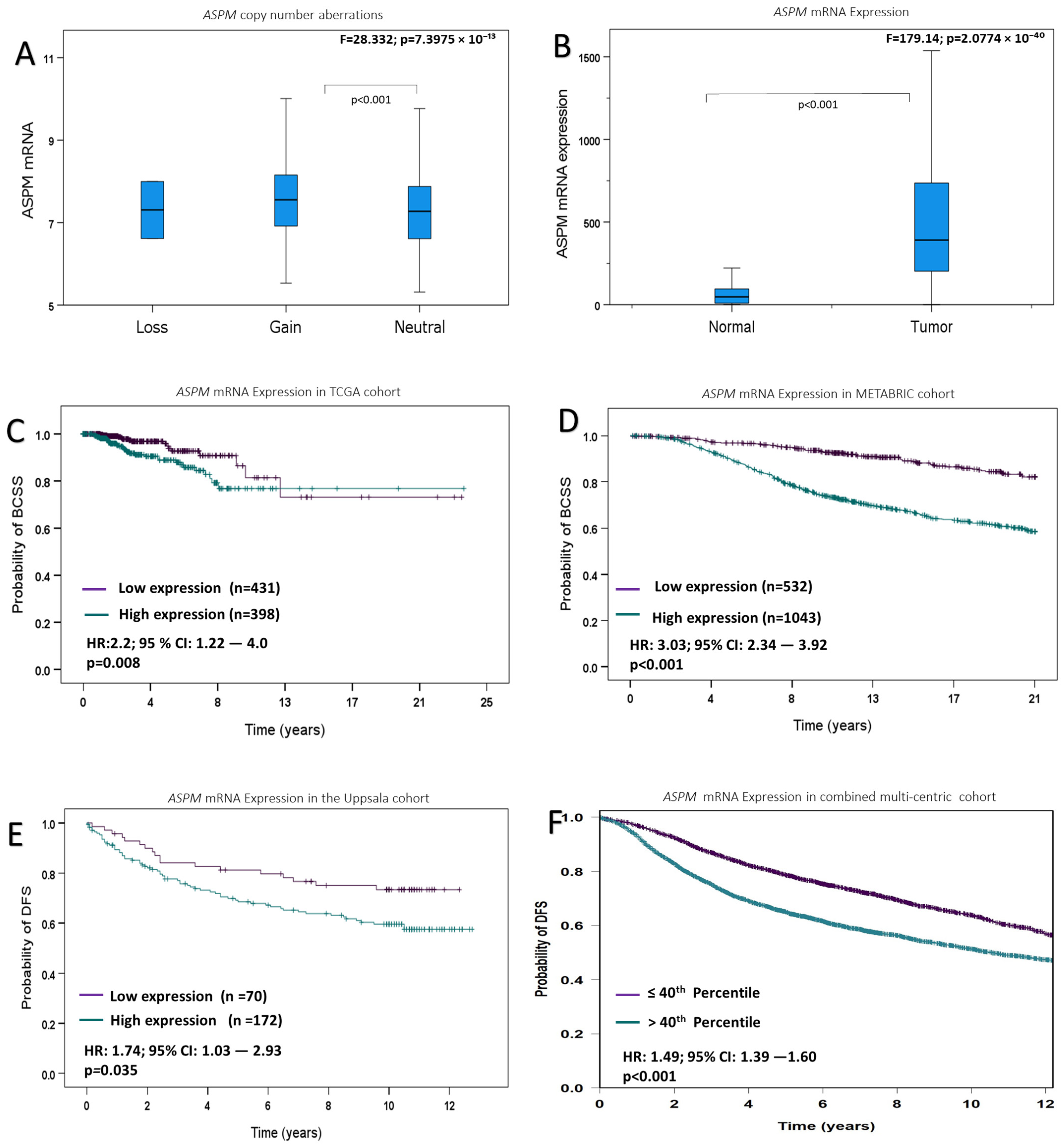
3.2. ASPM Gene Copy Number Aberrations and mRNA Levels
3.3. ASPM mRNA Expression
3.4. Association of ASPM mRNA Expression with Clinicopathological Parameters
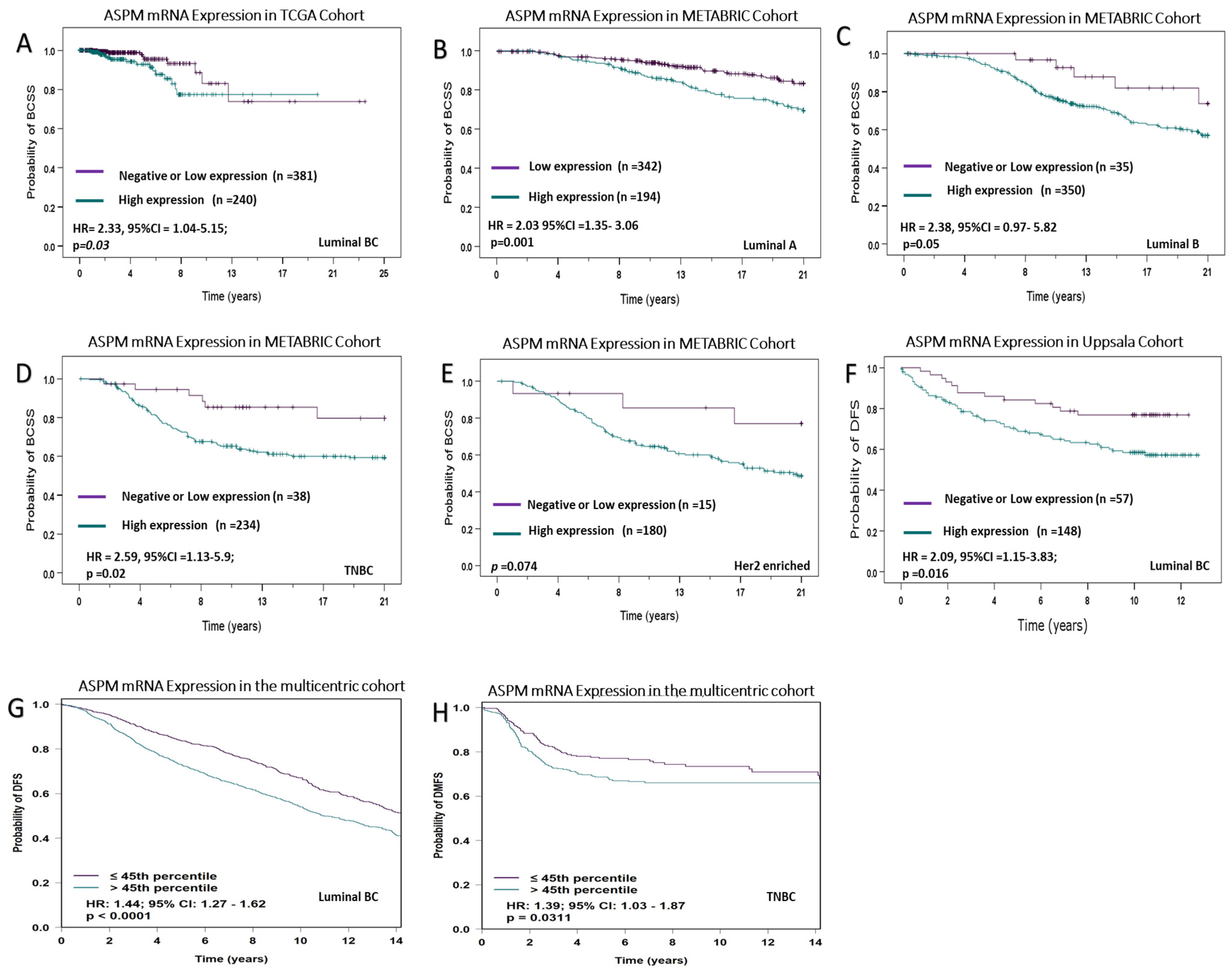
3.5. Association of ASPM mRNA Expression with the Patient Outcome
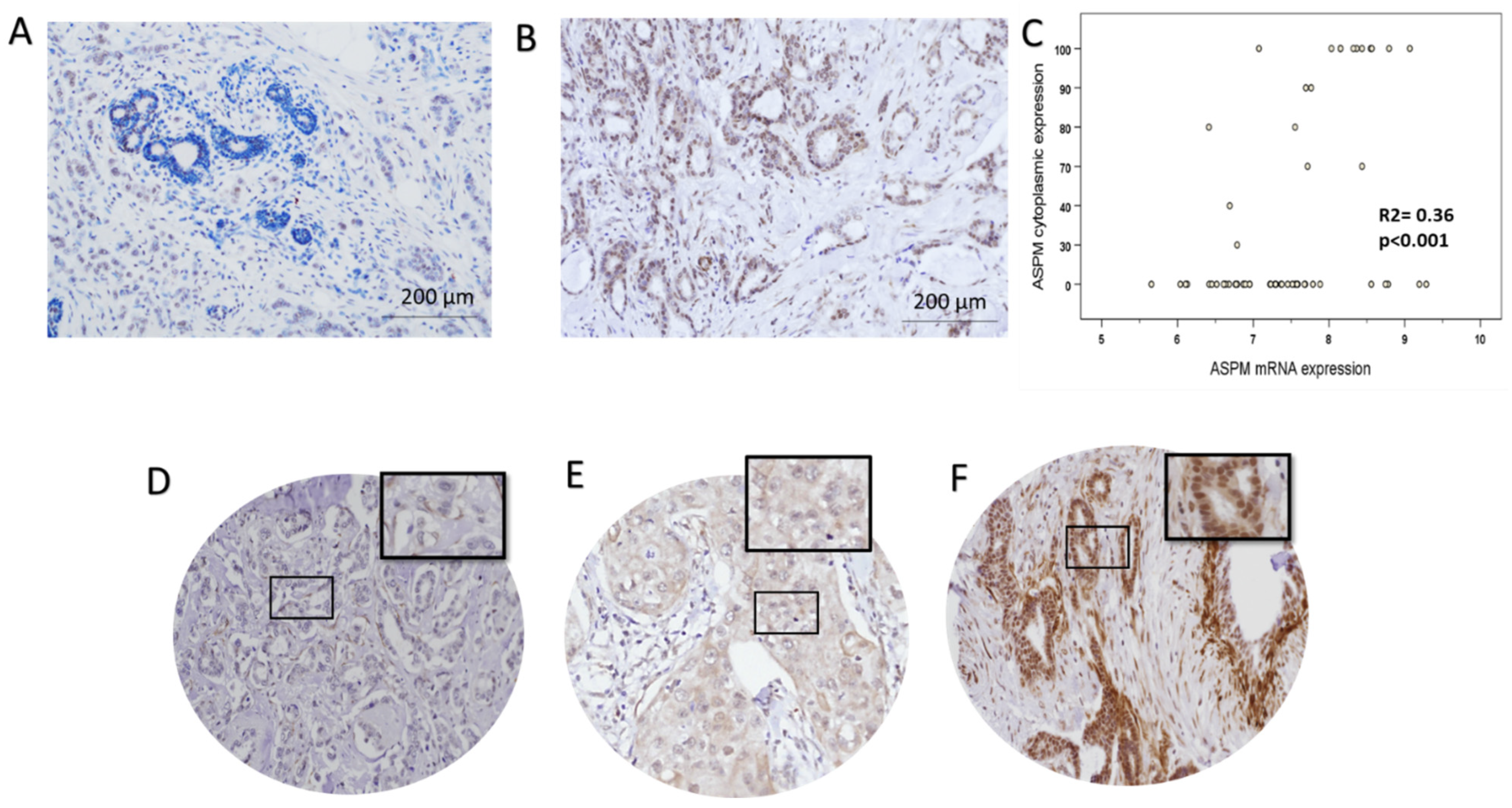
3.6. ASPM Protein Expression
3.7. Association of ASPM Protein Expression with Clinicopathological Parameters
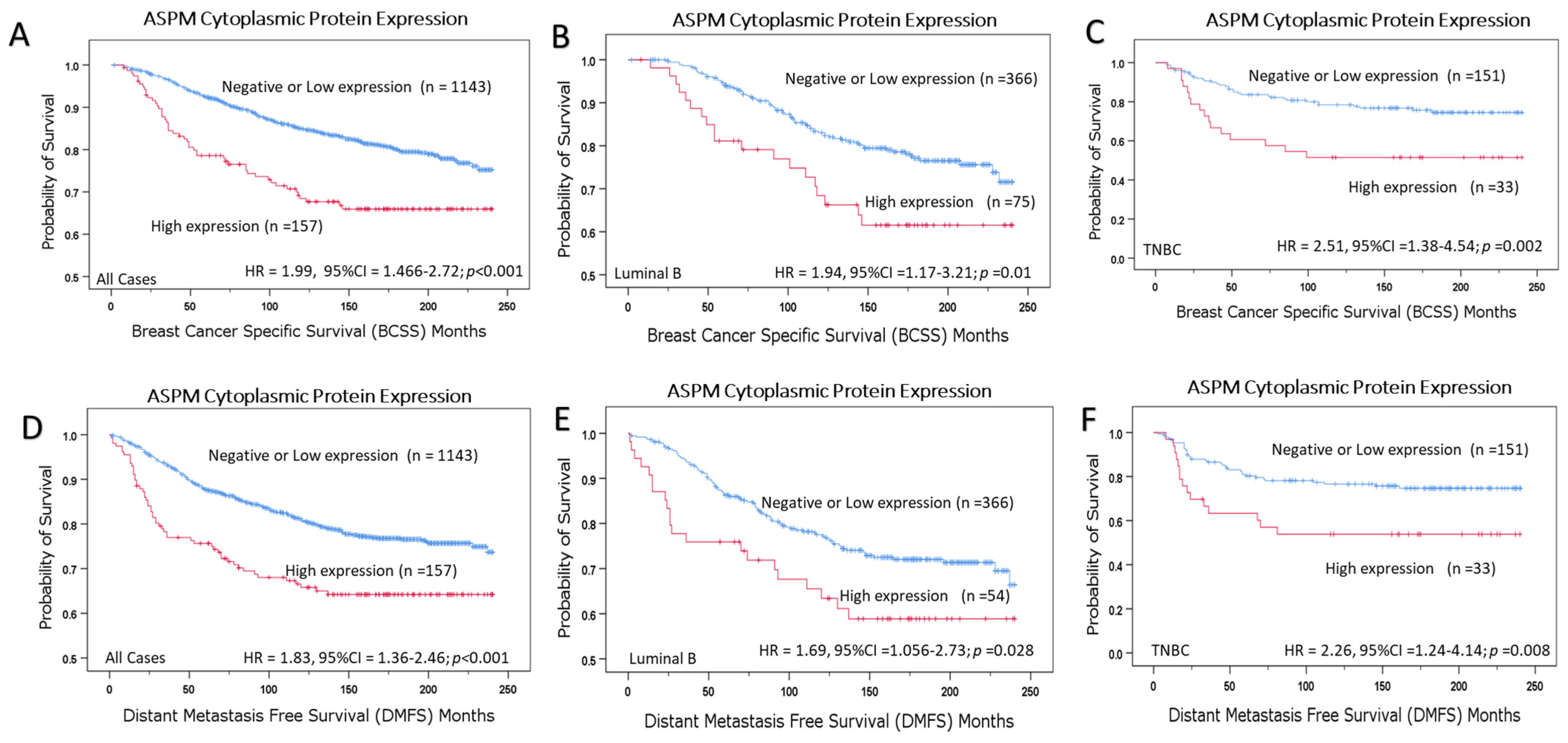
3.8. Association of ASPM Protein Expression with the Patient Outcome
3.9. ASPM mRNA and Protein Levels
3.10. Correlation Between ASPM and Ki67 Expression
3.11. Correlation Between ASPM Expression and Drug Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gonzalez-Angulo, A.M.; Morales-Vasquez, F.; Hortobagyi, G.N. Overview of resistance to systemic therapy in patients with breast cancer. Adv. Exp. Med. Biol. 2007, 608, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Mc Gee, M.M. Targeting the Mitotic Catastrophe Signaling Pathway in Cancer. Mediat. Inflamm. 2015, 2015, 146282. [Google Scholar] [CrossRef]
- Shahriyari, L.; Abdel-Rahman, M.; Cebulla, C. BAP1 expression is prognostic in breast and uveal melanoma but not colon cancer and is highly positively correlated with RBM15B and USP19. PLoS ONE 2019, 14, e0211507. [Google Scholar] [CrossRef]
- Lou, Y.; Fallah, Y.; Yamane, K.; Berg, P.E. BP1, a potential biomarker for breast cancer prognosis. Biomark. Med. 2018, 12, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Majidinia, M.; Yousefi, B. DNA repair and damage pathways in breast cancer development and therapy. DNA Repair 2017, 54, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.S.; Koh, C.G.; Li, H.Y. Mitosis-targeted anti-cancer therapies: Where they stand. Cell Death Dis. 2012, 3, e411. [Google Scholar] [CrossRef]
- Serrano-del Valle, A.; Reina-Ortiz, C.; Benedi, A.; Anel, A.; Naval, J.; Marzo, I. Future prospects for mitosis-targeted antitumor therapies. Biochem. Pharmacol. 2021, 190, 114655. [Google Scholar] [CrossRef]
- Mahalmani, V.; Sinha, S.; Prakash, A.; Medhi, B. Translational research: Bridging the gap between preclinical and clinical research. Indian J. Pharmacol. 2022, 54, 393–396. [Google Scholar] [CrossRef]
- Arai, T.; Okato, A.; Kojima, S.; Idichi, T.; Koshizuka, K.; Kurozumi, A.; Kato, M.; Yamazaki, K.; Ishida, Y.; Naya, Y. Regulation of spindle and kinetochore-associated protein 1 by antitumor miR-10a-5p in renal cell carcinoma. Cancer Sci. 2017, 108, 2088–2101. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Wei, Y.; Wu, J.; Zhang, P.; Shen, S.; Saiyin, H.; Wumaier, R.; Yang, X.; Wang, C. Molecular chaperone CCT3 supports proper mitotic progression and cell proliferation in hepatocellular carcinoma cells. Cancer Lett. 2016, 372, 101–109. [Google Scholar] [CrossRef]
- Ibrahim, A.; Toss, M.S.; Alsaleem, M.; Makhlouf, S.; Atallah, N.; Green, A.R.; Rakha, E.A. Novel 2 Gene Signatures Associated With Breast Cancer Proliferation: Insights From Predictive Differential Gene Expression Analysis. Mod. Pathol. 2023, 37, 100403. [Google Scholar] [CrossRef]
- Bikeye, S.-N.N.; Colin, C.; Marie, Y.; Vampouille, R.; Ravassard, P.; Rousseau, A.; Boisselier, B.; Idbaih, A.; Calvo, C.F.; Leuraud, P.; et al. ASPM-associated stem cell proliferation is involved in malignant progression of gliomas and constitutes an attractive therapeutic target. Cancer Cell Int. 2010, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Buchman, J.J.; Durak, O.; Tsai, L.-H. ASPM regulates Wnt signaling pathway activity in the developing brain. Genes Dev. 2011, 25, 1909–1914. [Google Scholar] [CrossRef] [PubMed]
- Major, M.B.; Roberts, B.S.; Berndt, J.D.; Marine, S.; Anastas, J.; Chung, N.; Ferrer, M.; Yi, X.; Stoick-Cooper, C.L.; Von Haller, P.D. New regulators of Wnt/β-catenin signaling revealed by integrative molecular screening. Sci. Signal. 2008, 1, ra12. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-Y.; Pan, H.-W.; Liu, S.-H.; Jeng, Y.-M.; Hu, F.-C.; Peng, S.-Y.; Lai, P.-L.; Hsu, H.-C. ASPM is a novel marker for vascular invasion, early recurrence, and poor prognosis of hepatocellular carcinoma. Clin. Cancer Res. 2008, 14, 4814–4820. [Google Scholar] [CrossRef]
- Drozdov, I.; Bornschein, J.; Wex, T.; Valeyev, N.V.; Tsoka, S.; Malfertheiner, P. Functional and topological properties in hepatocellular carcinoma transcriptome. PLoS ONE 2012, 7, e35510. [Google Scholar] [CrossRef]
- Jung, H.M.; Choi, S.J.; Kim, J.K. Expression profiles of SV40-immortalization-associated genes upregulated in various human cancers. J. Cell. Biochem. 2009, 106, 703–713. [Google Scholar] [CrossRef]
- Hagemann, C.; Anacker, J.; Gerngras, S.; Kühnel, S.; Said, H.M.; Patel, R.; Kämmerer, U.; Vordermark, D.; Roosen, K.; Vince, G.H. Expression analysis of the autosomal recessive primary microcephaly genes MCPH1 (microcephalin) and MCPH5 (ASPM, abnormal spindle-like, microcephaly associated) in human malignant gliomas. Oncol. Rep. 2008, 20, 301–308. [Google Scholar] [CrossRef]
- Horvath, S.; Zhang, B.; Carlson, M.; Lu, K.; Zhu, S.; Felciano, R.; Laurance, M.; Zhao, W.; Qi, S.; Chen, Z. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc. Natl. Acad. Sci. USA 2006, 103, 17402–17407. [Google Scholar] [CrossRef]
- Kouprina, N.; Pavlicek, A.; Collins, N.K.; Nakano, M.; Noskov, V.N.; Ohzeki, J.; Mochida, G.H.; Risinger, J.I.; Goldsmith, P.; Gunsior, M.; et al. The microcephaly ASPM gene is expressed in proliferating tissues and encodes for a mitotic spindle protein. Hum. Mol. Genet. 2005, 14, 2155–2165. [Google Scholar] [CrossRef]
- Peyre, M.; Commo, F.; Dantas-Barbosa, C.; Andreiuolo, F.; Puget, S.; Lacroix, L.; Drusch, F.; Scott, V.; Varlet, P.; Mauguen, A. Portrait of ependymoma recurrence in children: Biomarkers of tumor progression identified by dual-color microarray-based gene expression analysis. PLoS ONE 2010, 5, e12932. [Google Scholar] [CrossRef]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 19, A68–A77. [Google Scholar] [CrossRef]
- Ibrahim, A.; Lashen, A.G.; Katayama, A.; Mihai, R.; Ball, G.; Toss, M.S.; Rakha, E.A. Defining the area of mitoses counting in invasive breast cancer using whole slide image. Mod. Pathol. 2022, 35, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Hoadley, K.A.; Yau, C.; Wolf, D.M.; Cherniack, A.D.; Tamborero, D.; Ng, S.; Leiserson, M.D.M.; Niu, B.; McLellan, M.D.; Uzunangelov, V.; et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 2014, 158, 929–944. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Singh, D.; Zeng, Z.; Coleman, S.J.; Huang, Y.; Savich, G.L.; He, X.; Mieczkowski, P.; Grimm, S.A.; Perou, C.M.; et al. MapSplice: Accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010, 38, e178. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.-F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Ivshina, A.V.; George, J.; Senko, O.; Mow, B.; Putti, T.C.; Smeds, J.; Lindahl, T.; Pawitan, Y.; Hall, P.; Nordgren, H.; et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006, 66, 10292–10301. [Google Scholar] [CrossRef]
- Pawitan, Y.; Bjöhle, J.; Amler, L.; Borg, A.-L.; Egyhazi, S.; Hall, P.; Han, X.; Holmberg, L.; Huang, F.; Klaar, S.; et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: Derived and validated in two population-based cohorts. Breast Cancer Res. 2005, 7, R953. [Google Scholar] [CrossRef]
- Koboldt, D.; Fulton, R.; McLellan, M.; Schmidt, H.; Kalicki-Veizer, J.; McMichael, J.; Fulton, L.; Dooling, D.; Ding, L.; Mardis, E. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar]
- Rakha, E.A.; Martin, S.; Lee, A.H.S.; Morgan, D.; Pharoah, P.D.P.; Hodi, Z.; MacMillan, D.; Ellis, I.O. The prognostic significance of lymphovascular invasion in invasive breast carcinoma. Cancer 2012, 118, 3670–3680. [Google Scholar] [CrossRef] [PubMed]
- Aleskandarany, M.A.; Abduljabbar, R.; Ashankyty, I.; Elmouna, A.; Jerjees, D.; Ali, S.; Buluwela, L.; Diez-Rodriguez, M.; Caldas, C.; Green, A.R.; et al. Prognostic significance of androgen receptor expression in invasive breast cancer: Transcriptomic and protein expression analysis. Breast Cancer Res. Treat. 2016, 159, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Rakha, E.A.; Agarwal, D.; Green, A.R.; Ashankyty, I.; Ellis, I.O.; Ball, G.; Alaskandarany, M.A. Prognostic stratification of oestrogen receptor-positive HER2-negative lymph node-negative class of breast cancer. Histopathology 2017, 70, 622–631. [Google Scholar] [CrossRef]
- Rakha, E.A.; Elsheikh, S.E.; Aleskandarany, M.A.; Habashi, H.O.; Green, A.R.; Powe, D.G.; El-Sayed, M.E.; Benhasouna, A.; Brunet, J.S.; Akslen, L.A.; et al. Triple-negative breast cancer: Distinguishing between basal and nonbasal subtypes. Clin. Cancer Res. 2009, 15, 2302–2310. [Google Scholar] [CrossRef]
- Muftah, A.A.; Aleskandarany, M.A.; Al-Kaabi, M.M.; Sonbul, S.N.; Diez-Rodriguez, M.; Nolan, C.C.; Caldas, C.; Ellis, I.O.; Rakha, E.A.; Green, A.R. Ki67 expression in invasive breast cancer: The use of tissue microarrays compared with whole tissue sections. Breast Cancer Res. Treat. 2017, 164, 341–348. [Google Scholar] [CrossRef]
- Rakha, E.A.; Pinder, S.E.; Bartlett, J.M.; Ibrahim, M.; Starczynski, J.; Carder, P.J.; Provenzano, E.; Hanby, A.; Hales, S.; Lee, A.H.; et al. Updated UK Recommendations for HER2 assessment in breast cancer. J. Clin. Pathol. 2015, 68, 93–99. [Google Scholar] [CrossRef]
- Hammond, M.E.; Hayes, D.F.; Dowsett, M.; Allred, D.C.; Hagerty, K.L.; Badve, S.; Fitzgibbons, P.L.; Francis, G.; Goldstein, N.S.; Hayes, M.; et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Clin. Oncol. 2010, 28, 2784–2795. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thürlimann, B.; Senn, H.J. Strategies for subtypes--dealing with the diversity of breast cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Abd El-Rehim, D.M.; Ball, G.; Pinder, S.E.; Rakha, E.; Paish, C.; Robertson, J.F.; Macmillan, D.; Blamey, R.W.; Ellis, I.O. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int. J. Cancer 2005, 116, 340–350. [Google Scholar] [CrossRef]
- McCarty, K.S., Jr.; McCarty, K.S., Sr. Histochemical approaches to steroid receptor analyses. Semin. Diagn. Pathol. 1984, 1, 297–308. [Google Scholar] [PubMed]
- Camp, R.L.; Dolled-Filhart, M.; Rimm, D.L. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin. Cancer Res. 2004, 10, 7252–7259. [Google Scholar] [CrossRef] [PubMed]
- Polley, M.Y.; Leung, S.C.; McShane, L.M.; Gao, D.; Hugh, J.C.; Mastropasqua, M.G.; Viale, G.; Zabaglo, L.A.; Penault-Llorca, F.; Bartlett, J.M.; et al. An international Ki67 reproducibility study. J. Natl. Cancer Inst. 2013, 105, 1897–1906. [Google Scholar] [CrossRef] [PubMed]
- Ahlin, C.; Aaltonen, K.; Amini, R.-M.; Nevanlinna, H.; Fjällskog, M.-L.; Blomqvist, C. Ki67 and cyclin A as prognostic factors in early breast cancer. What are the optimal cut-off values? Histopathology 2007, 51, 491–498. [Google Scholar] [CrossRef] [PubMed]
- McShane, L.M.; Altman, D.G.; Sauerbrei, W.; Taube, S.E.; Gion, M.; Clark, G.M. REporting recommendations for tumour MARKer prognostic studies (REMARK). Br. J. Cancer 2005, 93, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Otto, T.; Sicinski, P. Cell cycle proteins as promising targets in cancer therapy. Nat. Rev. Cancer 2017, 17, 93–115. [Google Scholar] [CrossRef] [PubMed]
- Sauer, G.; Körner, R.; Hanisch, A.; Ries, A.; Nigg, E.A.; Silljé, H.H. Proteome analysis of the human mitotic spindle. Mol. Cell Proteom. 2005, 4, 35–43. [Google Scholar] [CrossRef]
- Higgins, J.; Midgley, C.; Bergh, A.M.; Bell, S.M.; Askham, J.M.; Roberts, E.; Binns, R.K.; Sharif, S.M.; Bennett, C.; Glover, D.M.; et al. Human ASPM participates in spindle organisation, spindle orientation and cytokinesis. BMC Cell Biol. 2010, 11, 85. [Google Scholar] [CrossRef]
- do Carmo Avides, M.; Glover, D.M. Abnormal spindle protein, Asp, and the integrity of mitotic centrosomal microtubule organizing centers. Science 1999, 283, 1733–1735. [Google Scholar] [CrossRef]
- Wakefield, J.G.; Bonaccorsi, S.; Gatti, M. The drosophila protein asp is involved in microtubule organization during spindle formation and cytokinesis. J. Cell Biol. 2001, 153, 637–648. [Google Scholar] [CrossRef]
- Capecchi, M.R.; Pozner, A. ASPM regulates symmetric stem cell division by tuning Cyclin E ubiquitination. Nat. Commun. 2015, 6, 8763. [Google Scholar] [CrossRef] [PubMed]
- Chenn, A.; Walsh, C.A. Increased neuronal production, enlarged forebrains and cytoarchitectural distortions in beta-catenin overexpressing transgenic mice. Cereb. Cortex 2003, 13, 599–606. [Google Scholar] [CrossRef]
- Pai, V.C.; Hsu, C.C.; Chan, T.S.; Liao, W.Y.; Chuu, C.P.; Chen, W.Y.; Li, C.R.; Lin, C.Y.; Huang, S.P.; Chen, L.T.; et al. ASPM promotes prostate cancer stemness and progression by augmenting Wnt-Dvl-3-β-catenin signaling. Oncogene 2019, 38, 1340–1353. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.Y.; Hsu, C.C.; Wang, T.Y.; Li, C.R.; Hou, Y.C.; Chu, J.M.; Lee, C.T.; Liu, M.S.; Su, J.J.; Jian, K.Y.; et al. A gene expression signature of epithelial tubulogenesis and a role for ASPM in pancreatic tumor progression. Gastroenterology 2013, 145, 1110–1120. [Google Scholar] [CrossRef]
- Xie, J.J.; Zhuo, Y.J.; Zheng, Y.; Mo, R.J.; Liu, Z.Z.; Li, B.W.; Cai, Z.D.; Zhu, X.J.; Liang, Y.X.; He, H.C.; et al. High expression of ASPM correlates with tumor progression and predicts poor outcome in patients with prostate cancer. Int. Urol. Nephrol. 2017, 49, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Liu, L.; Zhao, A.; Pfeifer, G.P.; Xu, X. The Abnormal Spindle-like, Microcephaly-associated (ASPM) Gene Encodes a Centrosomal Protein. Cell Cycle 2005, 4, 1227–1229. [Google Scholar] [CrossRef]
- Hsu, C.C.; Liao, W.Y.; Chan, T.S.; Chen, W.Y.; Lee, C.T.; Shan, Y.S.; Huang, P.J.; Hou, Y.C.; Li, C.R.; Tsai, K.K. The differential distributions of ASPM isoforms and their roles in Wnt signaling, cell cycle progression, and pancreatic cancer prognosis. J. Pathol. 2019, 249, 498–508. [Google Scholar] [CrossRef]
- Tang, J.; Lu, M.; Cui, Q.; Zhang, D.; Kong, D.; Liao, X.; Ren, J.; Gong, Y.; Wu, G. Overexpression of ASPM, CDC20, and TTK Confer a Poorer Prognosis in Breast Cancer Identified by Gene Co-expression Network Analysis. Front. Oncol. 2019, 9, 310. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, X.; Zhu, L.; Li, Z.; Zuo, P.; Wang, P.; Feng, J.; Mi, Y.; Zhang, C.; Xu, Y.; et al. ASPM promotes hepatocellular carcinoma progression by activating Wnt/β-catenin signaling through antagonizing autophagy-mediated Dvl2 degradation. FEBS Open Bio. 2021, 11, 2784–2799. [Google Scholar] [CrossRef]
- Zhou, J.W.; Wang, H.; Sun, W.; Han, N.N.; Chen, L. ASPM is a predictor of overall survival and has therapeutic potential in endometrial cancer. Am. J. Transl. Res. 2020, 12, 1942–1953. [Google Scholar]
- Brüning-Richardson, A.; Bond, J.; Alsiary, R.; Richardson, J.; Cairns, D.; McCormack, L.; Hutson, R.; Burns, P.; Wilkinson, N.; Hall, G. ASPM and microcephalin expression in epithelial ovarian cancer correlates with tumour grade and survival. Br. J. Cancer 2011, 104, 1602–1610. [Google Scholar] [CrossRef] [PubMed]
- Vulcani-Freitas, T.M.; Saba-Silva, N.; Cappellano, A.; Cavalheiro, S.; Marie, S.K.; Oba-Shinjo, S.M.; Malheiros, S.M.; de Toledo, S.R. ASPM gene expression in medulloblastoma. Childs Nerv. Syst. 2011, 27, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Zhang, J.; Zheng, Y.; Liu, J.; Duan, T.; Tian, T. Overexpression of abnormal spindle-like microcephaly-associated (ASPM) increases tumor aggressiveness and predicts poor outcome in patients with lung adenocarcinoma. Transl. Cancer Res. 2021, 10, 983–997. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.A.; Okayasu, R.; Jeggo, P.A.; Fujimori, A. ASPM influences DNA double-strand break repair and represents a potential target for radiotherapy. Int. J. Radiat. Biol. 2011, 87, 1189–1195. [Google Scholar] [CrossRef]
- Xu, S.; Wu, X.; Wang, P.; Cao, S.L.; Peng, B.; Xu, X. ASPM promotes homologous recombination-mediated DNA repair by safeguarding BRCA1 stability. iScience 2021, 24, 102534. [Google Scholar] [CrossRef]
- Yu, J.; Zhong, T.; Wang, J.; Xie, S.; Liu, L.; Wang, M.; Wu, F.; Xiao, C.; Chen, X.; Yan, W.; et al. ASPM induces radiotherapy resistance by disrupting microtubule stability leading to chromosome malsegregation in non-small cell lung cancer. 2022; preprint. [Google Scholar] [CrossRef]

| Parameter | METABRIC ASPM mRNA | TCGA ASPM mRNA | ||||
|---|---|---|---|---|---|---|
| Low No (%) | High No (%) | x2 p-Value | Low No (%) | High No (%) | x2 p-Value | |
| Patient age (years) <50 ≥50 | 112 (16.5) 568 (83.5) | 271 (21.5) 988 (78.5) | 7.116 0.008 | 108 (24.5) 333 (75.5) | 123 (29.8) 290 (70.2) | 3.027 0.082 |
| Tumor size <2 ≥2 | 264 (38.4) 424 (61.6) | 358 (28.3) 907 (71.7) | 20.827 <0.001 | 91 (30.0) 212 (70.0) | 60 (19.5) 247 (80.5) | 9.007 0.003 |
| Tumor grade 1 2 3 | 120 (18.7) 364 (56.6) 159 (24.7) | 50 (4.0) 406 (32.5) 793 (63.5) | 288.873 <0.001 | 67 (38.3) 89 (50.9) 19 (10.9) | 12 (5.9) 56 (27.5) 136 (66.7) | 132.675 <0.001 |
| Molecular subtypes Luminal A Luminal B Basal-like HER2 enriched. Normal | 459 (66.1) 49 (7.1) 19 (2.7) 20 (2.9) 147 (21.2) | 259 (20.1) 439 (34.1) 310 (24.1) 220 (17.1) 52 (4.0) | 726.897 <0.001 | 313 (71.0) 25 (5.7) 7 (1.6) 15 (3.4) 26 (5.9) | 92 (22.3) 116 (28.1) 126 (30.5) 41 (9.9) 4 (1.0) | 318.383 <0.001 |
| Mitotic score 1 2 3 | N/A | N/A | N/A | 199 (81.9) 29 (11.9) 15 (6.2) | 74 (33.6) 39 (17.7) 107 (48.6) | 127.254 <0.001 |
| Histological subtypes Invasive duct carcinoma (NST) Invasive lobular carcinoma Mixed NST and special type Other special types * | 465 (69.9) 75 (11.3) 47 (7.1) 76 (11.5) 2 (0.3) | 1079 (85.6) 72 (5.7) 43 (3.4) 37 (2.9) 30 (2.4) | 103.317 <0.001 | 274 (62.1) 128 (29.0) 9 (2.0) 30 (6.9) | 339 (82.0) 49 (11.9) 7 (1.7) 18 (4.4) | 52.294 <0.001 |
| Lymph nodal stage 1 2 3 | 394 (57.0) 209 (30.2) 88 (12.7) | 641 (50.0) 413 (32.2) 228 (17.8) | 11.917 0.003 | 101 (22.9) 237 (53.7) 103 (23.4) | 61 (14.8) 262 (63.4) 90 (21.8) | 11.099 0.005 |
| Nottingham Prognostic Index Good Moderate Poor | 362 (52.2) 299 (43.1) 33 (4.8) | 318 (24.7) 802 (62.4) 166 (12.9) | 158.723 <0.001 | N/A | N/A | N/A |
| Lympho-vascular invasion Negative Positive | 158 (23.2) 522 (76.8) | 578 (45.8) 685 (54.2) | 95.430 <0.001 | 312 (70.7) 129 (29.3) | 247 (59.8) 166 (40.2) | 11.293 <0.001 |
| Oestrogen receptor Negative Positive | 64 (9.2) 630 (90.8) | 410 (31.9) 876 (68.1) | 127.110 <0.001 | 38 (8.9) 390 (91.1) | 147 (37.1) 249 (62.9) | 94.234 <0.001 |
| Progesterone receptor Negative Positive | 218 (31.4) 476 (68.6) | 722 (56.1) 564 (43.9) | 110.557 <0.001 | 81 (19.0) 346 (81.0) | 191 (48.2) 205 (51.8) | 79.512 <0.001 |
| HER2 status Negative Positive | 670 (96.5) 24 (3.5) | 1063 (82.7) 223 (17.3) | 79.561 <0.001 | 387 (88.0) 53 (12.0) | 337 (81.6 76 (18.4) | 6.919 0.031 |
| Triple-negative status Non-triple negative Triple-negative | 644 (92.8) 50 (7.2) | 1016 (79.0) 270 (21.0) | 63.268 <0.001 | 430 (97.5) 11 (2.5) | 329 (79.6) 84 (20.3) | 68.715 <0.001 |
| P53 mutation Status Mutation Wild type | 11 (1.6) 275 (39.6) | 88 (6.8) 446 (34.7) | 27.956 <0.001 | N/A | N/A | N/A |
| Ki67 index Low High | 604 (98.1) 12 (1.9) | 359 (38.1) 583 (61.9) | 566.907 <0.001 | 356 (80.7) 85 (19.3) | 237 (57.4) 176 (42.6) | 54.749 <0.001 |
| Model Parameters | Breast Cancer Specific Survival (BCSS) | |||
|---|---|---|---|---|
| HR | 95% (CI) | p-Value | ||
| (A) | ASPM mRNA | 2.031 | 1.398–2.950 | <0.001 |
| Nodal stage | 1.96 | 1.694–2.268 | <0.001 | |
| Tumor grade | 1.304 | 1.050–1.620 | 0.016 | |
| Ki67 score | 1.538 | 1.166–2.029 | 0.002 | |
| (B) | ASPM mRNA | 1.856 | 1.005–3.428 | 0.04 |
| Nodal stage | 2.661 | 1.688–4.196 | <0.001 | |
| Ki67 score | 2.131 | 1.201–3.782 | 0.01 | |
| (C) | ASPM mRNA | 1.776 | 1.053–2.996 | 0.031 |
| ER status. | 0.816 | 0.442–1.505 | 0.514 | |
| Lymph node status | 1.91 | 1.234–2.957 | 0.004 | |
| Parameter | ASPM Cytoplasmic Expression | ||
|---|---|---|---|
| Low No (%) | High No (%) | x2 p-Value | |
| Patient age (years) <50 ≥50 | 346 (30.3) 797 (69.7) | 61 (38.9) 96 (61.1) | 4.728 0.030 |
| Tumor size (cm) <2 ≥2 | 712 (62.3) 431 (37.7) | 84 (53.5) 73 (46.5) | 4.492 0.034 |
| Tumor grade 1 2 3 | 181 (15.8) 461 (40.3) 501 (43.8) | 20 (12.7) 41 (26.1) 96 (61.1) | 17.094 <0.001 |
| Tubule formation 1 2 3 | 89 (7.8) 320 (28.0) 734 (64.2) | 10 (6.4) 49 (31.2) 98 (62.4) | 0.936 0.626 |
| Mitotic score 1 2 3 | 565 (49.4) 226 (19.8) 352 (30.8) | 50 (31.8) 38 (24.2) 69 (43.9) | 17.731 <0.001 |
| Nuclear pleomorphism 1 2 3 | 16 (1.4) 348 (30.4) 779 (68.2) | 2 (1.3) 22 (14.0) 133 (84.7) | 18.509 <0.001 |
| Molecular subtypes Luminal A Luminal B HER2 Triple negative breast cancer | 428 (43.1) 366 (36.9) 47 (4.7) 151 (15.2) | 39 (27.3) 54 (37.8) 17 (11.9) 33 (23.1) | 23.767 <0.001 |
| Histological subtypes Non-specific type (NST) Lobular Mixed NST and special type Other special types * | 718 (62.8) 105 (9.2) 267 (23.4) 53 (4.6) | 122 (77.7) 4 (2.5) 25 (15.9) 6 (3.8) | 16.089 0.003 |
| Axillary nodal stage 1 2 3 | 718 (62.8) 310 (27.1) 115 (10.1) | 81 (51.6) 53 (33.8) 23 (14.6) | 7.745 0.021 |
| Nottingham Prognostic Index Good Moderate Poor | 403 (35.3) 558 (48.8) 182 (15.9) | 37 (23.6) 82 (52.2) 38 (24.2) | 11.494 0.003 |
| Lympho-vascular invasion Negative Positive | 820 (71.7) 323 (28.3) | 108 (68.8) 49 (31.2) | 0.589 0.443 |
| Oestrogen receptor Negative Positive | 209 (18.3) 934 (81.7) | 51 (32.7) 105 (67.3) | 17.797 <0.001 |
| Progesterone receptor Negative Positive | 453 (39.8) 684 (60.2) | 82 (52.9) 73 (47.1) | 9.592 0.002 |
| HER2 status Negative Positive | 1000 (87.5) 143 (12.5) | 118 (75.6) 38 (24.4) | 16.068 <0.001 |
| Ki67 index Low High | 469 (54.5) 392 (45.5) | 42 (38.5) 67 (61.5) | 9.861 0.002 |
| Model Parameters | Breast Cancer Specific Survival (BCSS) | Distant Metastasis Free Survival (DMFS) | ||||
|---|---|---|---|---|---|---|
| HR | 95% (CI) | p-Value | HR | 95% (CI) | p-Value | |
| ASPM cytoplasmic expression | 1.702 | 1.154–2.510 | 0.007 | 1.545 | 1.063–2.246 | 0.022 |
| Nodal stage | 2.285 | 1.812–2.882 | <0.001 | 2.147 | 1.722–2.677 | <0.001 |
| Tumor grade | 2.413 | 1.506–3.865 | <0.001 | 1.925 | 1.267–2.926 | 0.002 |
| Mitosis score | 0.937 | 0.678–1.295 | 0.694 | 0.938 | 0.695–1.264 | 0.673 |
| Ki67 score | 1.122 | 0.774–1.628 | 0.543 | 1.333 | 0.939–1.894 | 0.108 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibrahim, A.; Atallah, N.M.; Makhlouf, S.; Toss, M.S.; Green, A.; Rakha, E. Deciphering the Role of ASPM in Breast Cancer: A Comprehensive Multicohort Study. Cancers 2024, 16, 3814. https://doi.org/10.3390/cancers16223814
Ibrahim A, Atallah NM, Makhlouf S, Toss MS, Green A, Rakha E. Deciphering the Role of ASPM in Breast Cancer: A Comprehensive Multicohort Study. Cancers. 2024; 16(22):3814. https://doi.org/10.3390/cancers16223814
Chicago/Turabian StyleIbrahim, Asmaa, Nehal M. Atallah, Shorouk Makhlouf, Michael S. Toss, Andrew Green, and Emad Rakha. 2024. "Deciphering the Role of ASPM in Breast Cancer: A Comprehensive Multicohort Study" Cancers 16, no. 22: 3814. https://doi.org/10.3390/cancers16223814
APA StyleIbrahim, A., Atallah, N. M., Makhlouf, S., Toss, M. S., Green, A., & Rakha, E. (2024). Deciphering the Role of ASPM in Breast Cancer: A Comprehensive Multicohort Study. Cancers, 16(22), 3814. https://doi.org/10.3390/cancers16223814







