Free Methylglyoxal as a Metabolic New Biomarker of Tumor Cell Proliferation in Cancers
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. MG Measurement
2.2. Statistical Analysis
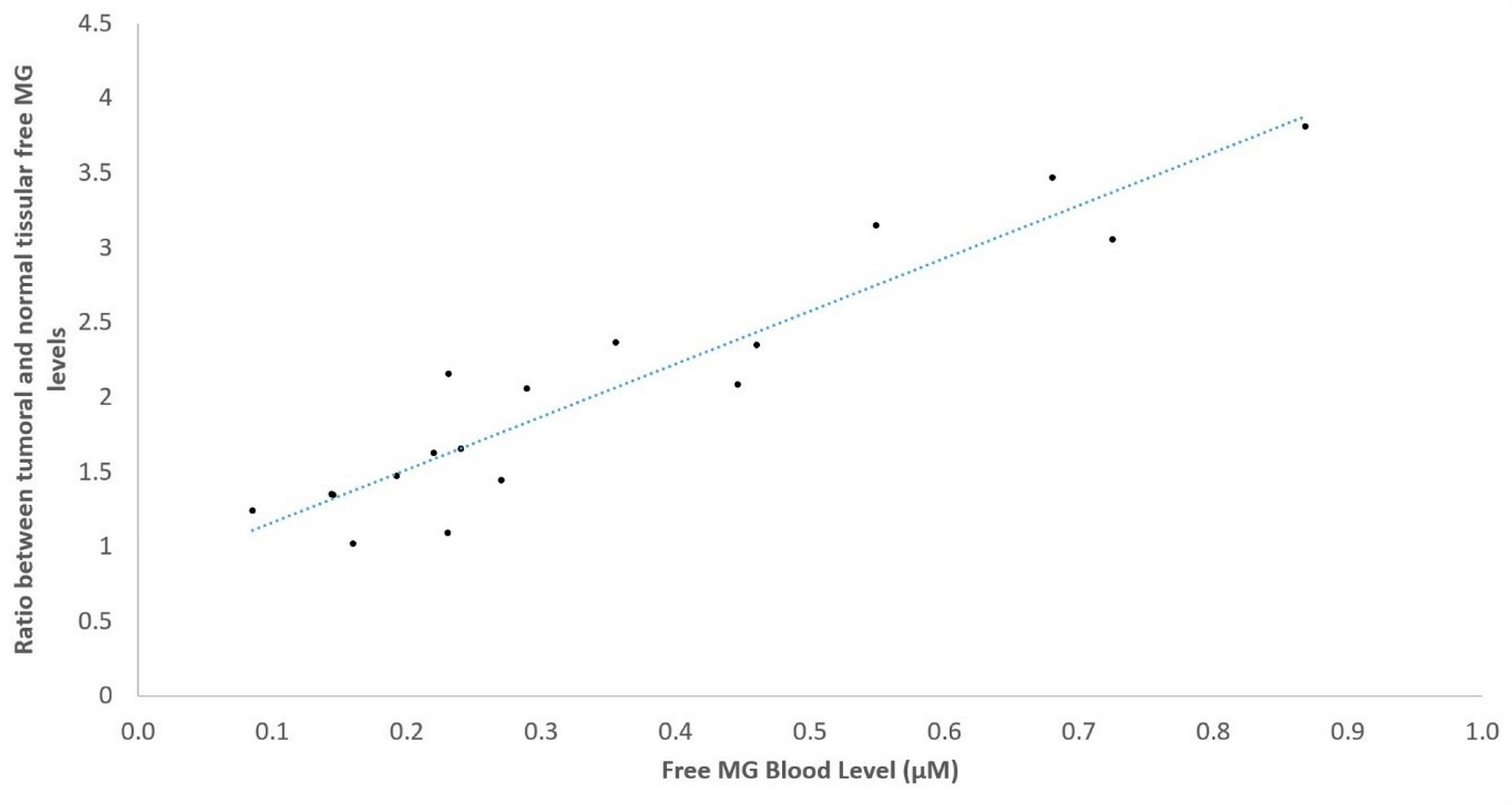
3. Results
3.1. Clinical Data
3.2. Experimental Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- WHO. Cancer—Key Facts; World Health Organization: Geneva, Switzerland, 3 February 2022; Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 14 February 2024).
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2020; Available online: https://gco.iarc.fr/today (accessed on 14 February 2024).
- Strimbu, K.; Tavel, J.A. What are biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466. [Google Scholar] [CrossRef] [PubMed]
- FDA-NIH Biomarker Working Group. BEST (Biomarkers, EndpointS, and Other Tools) Resource. Silver Spring (MD). Available online: https://www.ncbi.nlm.nih.gov/books/NBK326791/pdf/Bookshelf_NBK326791.pdf (accessed on 10 February 2024).
- Kamińska, K.; Nalejska, E.; Kubiak, M.; Wojtysiak, J.; Żołna, Ł.; Kowalewski, J.; Lewandowska, M.A. Prognostic and Predictive Epigenetic Biomarkers in Oncology. Mol. Diagn. Ther. 2019, 23, 83–95. [Google Scholar] [CrossRef]
- Nam, A.S.; Chaligne, R.; Landau, D.A. Integrating genetic and non-genetic determinants of cancer evolution by single-cell multi-omics. Nat. Rev. Genet. 2021, 22, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Irigaray, P.; Newby, J.A.; Clapp, R.; Hardell, L.; Howard, V.; Montagnier, L.; Epstein, S.; Belpomme, D. Lifestyle-related factors and environmental agents causing cancer: An overview. Biomed. Pharmacother. 2007, 61, 640–658. [Google Scholar] [CrossRef] [PubMed]
- Belpomme, D.; Irigaray, P.; Hardell, L.; Clapp, R.; Montagnier, L.; Epstein, S.; Sasco, A.J. The multitude and diversity of environmental carcinogens. Environ. Res. 2007, 105, 414–429. [Google Scholar] [CrossRef]
- US Department of Health and Human Services (NIH and NCI). 2008–2009 Annual President’s Cancer Panel: Environmental Factors in Cancer. NIH. April 2018. Available online: https://deainfo.nci.nih.gov/advisory/pcp/annualReports/pcp08-09rpt/PCP_Report_08-09_508.pdf (accessed on 14 February 2024).
- Irigaray, P.; Belpomme, D. Basic properties and molecular mechanisms of exogenous chemical carcinogens. Carcinogenesis 2010, 31, 135–148. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Lum, J.J.; Hatzivassiliou, G.; Thompson, C.B. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008, 7, 11–20. [Google Scholar] [CrossRef]
- Ward, P.S.; Thompson, C.B. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell 2012, 21, 297–308. [Google Scholar] [CrossRef]
- Caignard, A.; Pelletier, H.; Martin, F. Specificity of the immune response leading to protection or enhancement by regressive and progressive variants of a rat colon carcinoma. Int. J. Cancer 1988, 42, 883–886. [Google Scholar] [CrossRef]
- Irigaray, P.; Belpomme, D. Circulating free methylglyoxal as a metabolic tumor biomarker in a rat colon adenocarcinoma model. Mol. Clin. Oncol. 2020, 12, 311–316. [Google Scholar] [CrossRef]
- WHO. Handbook for Reporting Results of Cancer Treatment; World Health Organization Offset Publication: Geneva, Switzerland, 1979; Volume 48, Available online: https://iris.who.int/bitstream/handle/10665/37200/WHO_OFFSET_48.pdf?sequence=1&isAllowed=y (accessed on 14 February 2024).
- Rabbani, N.; Thornalley, P.J. Measurement of methylglyoxal by stable isotopic dilution analysis LC-MS/MS with corroborative prediction in physiological samples. Nat. Protoc. 2014, 9, 1969–1979. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.; Lu, M.; Jung, K.H.; Park, J.H.; Yu, L.; Onuchic, J.N.; Kaipparettu, B.A.; Levine, H. Elucidating cancer metabolic plasticity by coupling gene regulation with metabolic pathways. Proc. Natl. Acad. Sci. USA 2019, 116, 3909–3918. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.P.; Sabatini, D.M. Cancer cell metabolism: Warburg and beyond. Cell 2008, 134, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Vazquez, A.; Liu, J.; Zhou, Y.; Oltvai, Z.N. Catabolic efficiency of aerobic glycolysis: The Warburg effect revisited. BMC Syst. Biol. 2010, 4, 58. [Google Scholar] [CrossRef]
- Warburg, O.H. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Bensinger, S.J.; Christofk, H.R. New aspects of the Warburg effect in cancer cell biology. Semin. Cell Dev. Biol. 2012, 23, 352–361. [Google Scholar] [CrossRef]
- Jadvar, H.; Alavi, A.; Gambhir, S.S. 18F-FDG uptake in lung, breast, and colon cancers: Molecular biology correlates and disease characterization. J. Nucl. Med. 2009, 50, 1820–1827. [Google Scholar] [CrossRef]
- Phillips, S.A.; Thornalley, P.J. The formation of methylglyoxal from triose phosphates: Investigation using a specific assay for methylglyoxal. Eur. J. Biochem. 1993, 212, 101–105. [Google Scholar] [CrossRef]
- Lai, S.W.T.; Lopez Gonzalez, E.D.J.; Zoukari, T.; Ki, P.; Shuck, S.C. Methylglyoxal and Its Adducts: Induction, Repair, and Association with Disease. Chem. Res. Toxicol. 2022, 35, 1720–1746. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J. Pharmacology of methylglyoxal: Formation, modification of proteins and nucleic acids, and enzymatic detoxification—A role in pathogenesis and antiproliferative chemotherapy. Gen. Pharmacol. 1996, 27, 565–573. [Google Scholar] [CrossRef]
- Riboulet-Chavey, A.; Pierron, A.; Durand, I.; Murdaca, J.; Giudicelli, J.; Van Obberghen, E. Methylglyoxal impairs the insulin signaling pathways independently of the formation of intracellular reactive oxygen species. Diabetes 2006, 55, 1289–1299. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, H.; Mashima, T.; Yamamoto, K.; Tsuruo, T. Modulation of heat-shock protein 27 (Hsp27) anti-apoptotic activity by methylglyoxal modification. J. Biol. Chem. 2002, 277, 45770–45775. [Google Scholar] [CrossRef]
- Chaplen, F.W.; Fahl, W.E.; Cameron, D.C. Method for determination of free intracellular and extracellular methylglyoxal in animal cells grown in culture. Anal. Biochem. 1996, 238, 171–178. [Google Scholar] [CrossRef]
- Wallace, D.C. Mitochondria and cancer. Nat. Rev. Cancer 2012, 12, 685–698. [Google Scholar] [CrossRef]
- Pavlides, S.; Vera, I.; Gandara, R.; Sneddon, S.; Pestell, R.G.; Mercier, I.; Martinez-Outschoorn, U.E.; Whitaker-Menezes, D.; Howell, A.; Sotgia, F.; et al. Warburg meets autophagy: Cancer-associated fibroblasts accelerate tumor growth and metastasis via oxidative stress, mitophagy, and aerobic glycolysis. Antioxid. Redox Signal. 2012, 16, 1264–1284. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.C.; Chen, K.C.; Chang, G.C.; Lin, H.; Wu, C.C.; Kao, W.H.; Teng, C.J.; Hsu, S.L.; Yang, T.Y. RAGE acts as an oncogenic role and promotes the metastasis of human lung cancer. Cell Death Dis. 2020, 11, 265. [Google Scholar] [CrossRef]
- Kang, R.; Loux, T.; Tang, D.; Schapiro, N.E.; Vernon, P.; Livesey, K.M.; Krasinskas, A.; Lotze, M.T.; Zeh, H.J., 3rd. The expression of the receptor for advanced glycation endproducts (RAGE) is permissive for early pancreatic neoplasia. Proc. Natl. Acad. Sci. USA 2012, 109, 7031–7036. [Google Scholar] [CrossRef]
- Miki, S.; Kasayama, S.; Miki, Y.; Nakamura, Y.; Yamamoto, M.; Sato, B.; Kishimoto, T. Expression of receptors for advanced glycosylation end products on renal cell carcinoma cells in vitro. Biochem. Biophys. Res. Commun. 1993, 196, 984–989. [Google Scholar] [CrossRef]
- Leone, A.; Nigro, C.; Nicolò, A.; Prevenzano, I.; Formisano, P.; Beguinot, F.; Miele, C. The Dual-Role of Methylglyoxal in Tumor Progression—Novel Therapeutic Approaches. Front. Oncol. 2021, 11, 645686. [Google Scholar] [CrossRef]
- Kang, R.; Tang, D.; Schapiro, N.E.; Livesey, K.M.; Farkas, A.; Loughran, P.; Bierhaus, A.; Lotze, M.T.; Zeh, H.J. The receptor for advanced glycation end products (RAGE) sustains autophagy and limits apoptosis, promoting pancreatic tumor cell survival. Cell Death Differ. 2010, 17, 666–676. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J. The glyoxalase system in health and disease. Mol. Asp. Med. 1993, 14, 287–371. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kuramitsu, Y.; Ueno, T.; Suzuki, N.; Yoshino, S.; Iizuka, N.; Akada, J.; Kitagawa, T.; Oka, M.; Nakamura, K. Glyoxalase I (GLO1) is up-regulated in pancreatic cancerous tissues compared with related non-cancerous tissues. Anticancer. Res. 2012, 32, 3219–3222. [Google Scholar]
- Kreycy, N.; Gotzian, C.; Fleming, T.; Flechtenmacher, C.; Grabe, N.; Plinkert, P.; Hess, J.; Zaoui, K. Glyoxalase 1 expression is associated with an unfavorable prognosis of oropharyngeal squamous cell carcinoma. BMC Cancer 2017, 17, 382. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, N.; Thornalley, P.J. Dicarbonyl stress in cell and tissue dysfunction contributing to ageing and disease. Biochem. Biophys. Res. Commun. 2015, 458, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Jerzykowski, T.; Winter, R.; Matuszewski, W.; Piskorska, D. A re-evaluation of studies on the distribution of glyoxalases in animal and tumour tissues. Int. J. Biochem. 1978, 9, 853–860. [Google Scholar] [CrossRef]
- Di Ilio, C.; Angelucci, S.; Pennelli, A.; Zezza, A.; Tenaglia, R.; Sacchetta, P. Glyoxalase activities in tumor and non-tumor human urogenital tissues. Cancer Lett. 1995, 96, 189–193. [Google Scholar] [CrossRef]
- Dragani, B.; Cocco, R.; Ridderström, M.; Stenberg, G.; Mannervik, B.; Aceto, A. Unfolding and refolding of human glyoxalase II and its single-tryptophan mutants. J. Mol. Biol. 1999, 291, 481–490. [Google Scholar] [CrossRef]
- Thornalley, P.J.; Rabbani, N. Glyoxalase in tumourigenesis and multidrug resistance. Semin. Cell Dev. Biol. 2011, 22, 318–325. [Google Scholar] [CrossRef]
- Chiavarina, B.; Nokin, M.J.; Durieux, F.; Bianchi, E.; Turtoi, A.; Peulen, O.; Peixoto, P.; Irigaray, P.; Uchida, K.; Belpomme, D.; et al. Triple negative tumors accumulate significantly less methylglyoxal specific adducts than other human breast cancer subtypes. Oncotarget 2014, 5, 5472–5482. [Google Scholar] [CrossRef] [PubMed]
- Chiavarina, B.; Nokin, M.J.; Bellier, J.; Durieux, F.; Bletard, N.; Sherer, F.; Lovinfosse, P.; Peulen, O.; Verset, L.; Dehon, R.; et al. Methylglyoxal-Mediated Stress Correlates with High Metabolic Activity and Promotes Tumor Growth in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 213. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.I.; Ahmad, S. Preferential recognition of methylglyoxal-modified calf thymus DNA by circulating antibodies in cancer patients. Indian. J. Biochem. Biophys. 2011, 48, 290–296. [Google Scholar] [PubMed]
- Alamil, H.; Lechevrel, M.; Lagadu, S.; Galanti, L.; Dagher, Z.; Delépée, R. A validated UHPLC-MS/MS method for simultaneous quantification of 9 exocyclic DNA adducts induced by 8 aldehydes. J. Pharm. Biomed. Anal. 2020, 179, 113007. [Google Scholar] [CrossRef]
- Coluccio, M.L.; Presta, I.; Greco, M.; Gervasi, R.; La Torre, D.; Renne, M.; Voci, C.P.; Lunelli, L.; Donato, G.; Malara, N. Microenvironment Molecular Profile Combining Glycation Adducts and Cytokines Patterns on Secretome of Short-term Blood-derived Cultures during Tumour Progression. Int. J. Mol. Sci. 2020, 21, 4711. [Google Scholar] [CrossRef]
- Brownlee, M. Biochemistry and molecular cell biology of diabetic complications. Nature 2001, 414, 813–820. [Google Scholar] [CrossRef]
- Li, J.; Liu, D.; Sun, L.; Lu, Y.; Zhang, Z. Advanced glycation end products and neurodegenerative diseases: Mechanisms and perspective. J. Neurol. Sci. 2012, 317, 1–5. [Google Scholar] [CrossRef]


| Demographic Data | Normal Healthy Controls | Normo-Glycemic Treated Type 2 Diabetes | Overall Cancers Patients | Hyperglycemic Type 2 Diabetes |
|---|---|---|---|---|
| n | 68 | 12 | 139 | 10 |
| Age (mean +/− SD) | 45.32 +/− 15.61 | 62.33 +/− 10.79 | 62.01 +/− 10.76 | 55.50 +/− 15.61 |
| Age (median [range]) | 43 [23–83] | 60 [49–81] | 62.5 [28–90] | 67 [37–81] |
| Sex ratio (women/men) | 41/27 | 7/5 | 68/71 | 3/7 |
| Female (%) | 60.3 | 58.3 | 48.92 | 30 |
| Type | n | Mean Free MG (µM) | p * | p ** |
|---|---|---|---|---|
| Normal healthy Controls | 68 | 0.0647 +/− 0.0031 [0.021–0.121] | _ | _ |
| Normo-glycemic treated type 2 Diabetes | 12 | 0.0676 +/− 0.0281 [0.064–0.089] | 0.046 | _ |
| Overall cancers patients | 139 | 0.1758 +/− 0.0117 [0.035–0.868] | <0.0001 | _ |
| Hyperglycemic type 2 diabetes | 10 | 0.1624 +/− 0.0074 [0.138–0.199] | <0.0001 | 0.965 |
| Cancer Type | n | Mean Free MG (µM) | p * |
|---|---|---|---|
| Bronchus | 52 | 0.2354 +/− 0.0235 [0.085–0.868] | <0.0001 |
| Glioblastoma | 28 | 0.2201 +/− 0.0256 [0.068–0.814] | <0.0001 |
| Other digestive tumors | 8 | 0.1758 +/− 0.0371 [0.053–0.352] | 0.0002 |
| Colorectal | 9 | 0.1666 +/− 0.0347 [0.038–0.352] | <0.0001 |
| Pancreas | 7 | 0.1496 +/− 0.0210 [0.099–0.248] | <0.0001 |
| Gynecologic ** | 10 | 0.1479 +/− 0.0224 [0.053–0.223] | 0.0004 |
| Prostate | 8 | 0.1127 +/− 0.0157 [0.081–0.217] | 0.0003 |
| Breast | 12 | 0.0985 +/− 0.0114 [0.035–0.191] | 0.002 |
| Head and Neck | 5 | 0.0980 +/− 0.0330 [0.040–0.210] | 0.654 |
| Total | 139 | 0. 1758 +/− 0.0117 [0.035–0.868] | <0.0001 |
| Therapeutic Response | n | Mean Free MG (µM) +/− Standard Error |
|---|---|---|
| Complete response | 38 | 0.0610 +/− 0.0252 |
| Partial response | 31 | 0.1305 +/− 0.0721 |
| Stable/progressive | 29 | 0.1405 +/− 0.0617 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belpomme, D.; Lacomme, S.; Poletti, C.; Bonesso, L.; Hinault-Boyer, C.; Barbier, S.; Irigaray, P. Free Methylglyoxal as a Metabolic New Biomarker of Tumor Cell Proliferation in Cancers. Cancers 2024, 16, 3922. https://doi.org/10.3390/cancers16233922
Belpomme D, Lacomme S, Poletti C, Bonesso L, Hinault-Boyer C, Barbier S, Irigaray P. Free Methylglyoxal as a Metabolic New Biomarker of Tumor Cell Proliferation in Cancers. Cancers. 2024; 16(23):3922. https://doi.org/10.3390/cancers16233922
Chicago/Turabian StyleBelpomme, Dominique, Stéphanie Lacomme, Clément Poletti, Laurent Bonesso, Charlotte Hinault-Boyer, Sylvie Barbier, and Philippe Irigaray. 2024. "Free Methylglyoxal as a Metabolic New Biomarker of Tumor Cell Proliferation in Cancers" Cancers 16, no. 23: 3922. https://doi.org/10.3390/cancers16233922
APA StyleBelpomme, D., Lacomme, S., Poletti, C., Bonesso, L., Hinault-Boyer, C., Barbier, S., & Irigaray, P. (2024). Free Methylglyoxal as a Metabolic New Biomarker of Tumor Cell Proliferation in Cancers. Cancers, 16(23), 3922. https://doi.org/10.3390/cancers16233922







