Gut Microbiome Changes After Neoadjuvant Chemotherapy and Surgery in Patients with Gastric Cancer
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Ethics
2.2. Study Participants
2.3. Blood and Stool Sample Collection
2.4. 16S rRNA Gene Sequencing
2.5. Processing of the Sequencing Data
2.6. Measurement of Inflammation and Gut Permeability Biomarkers
2.7. Statistical Analysis
3. Results
3.1. Patient Characteristics
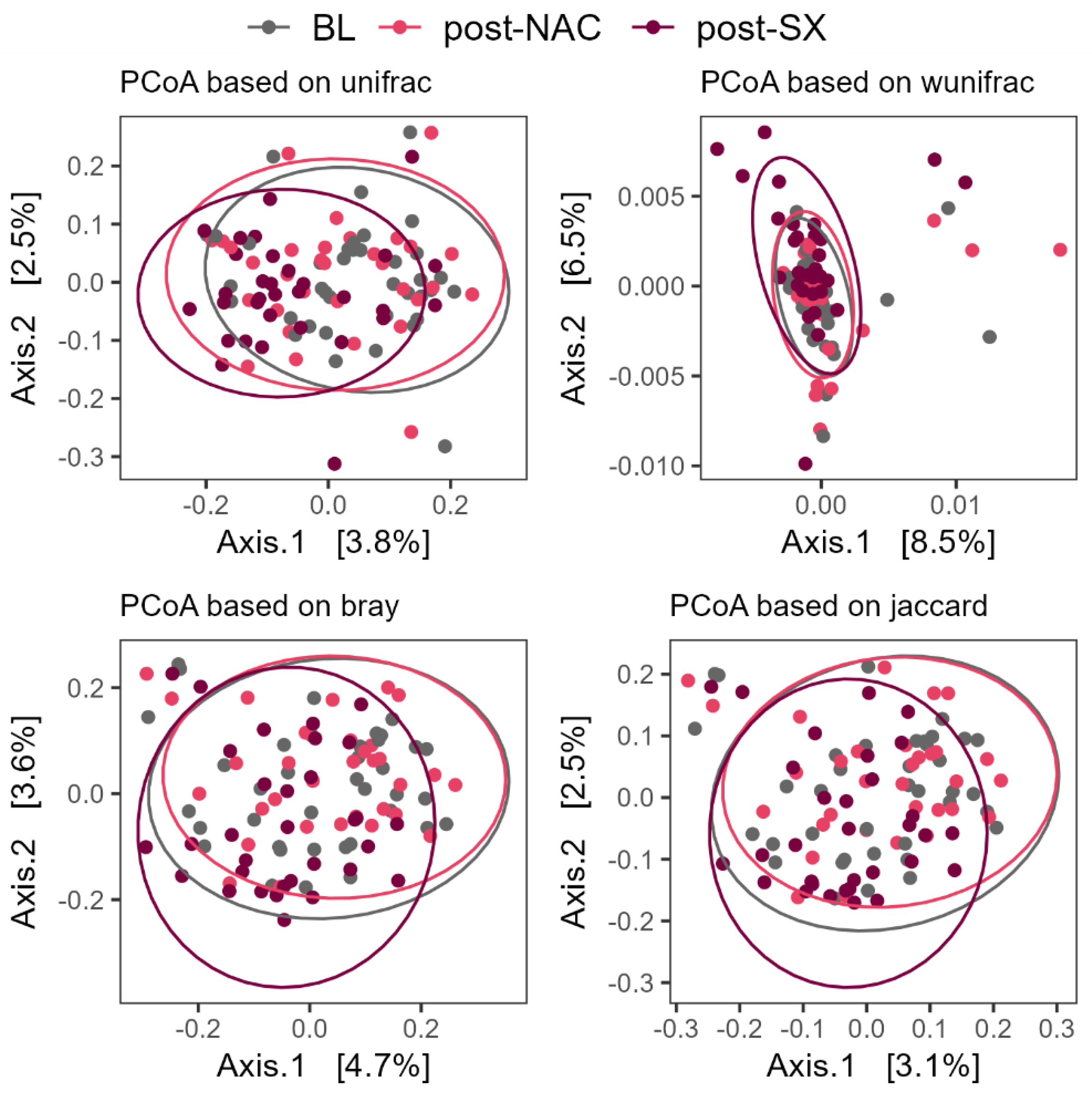
3.2. NAC and Gastrectomy Impact on Alpha and Beta Diversity
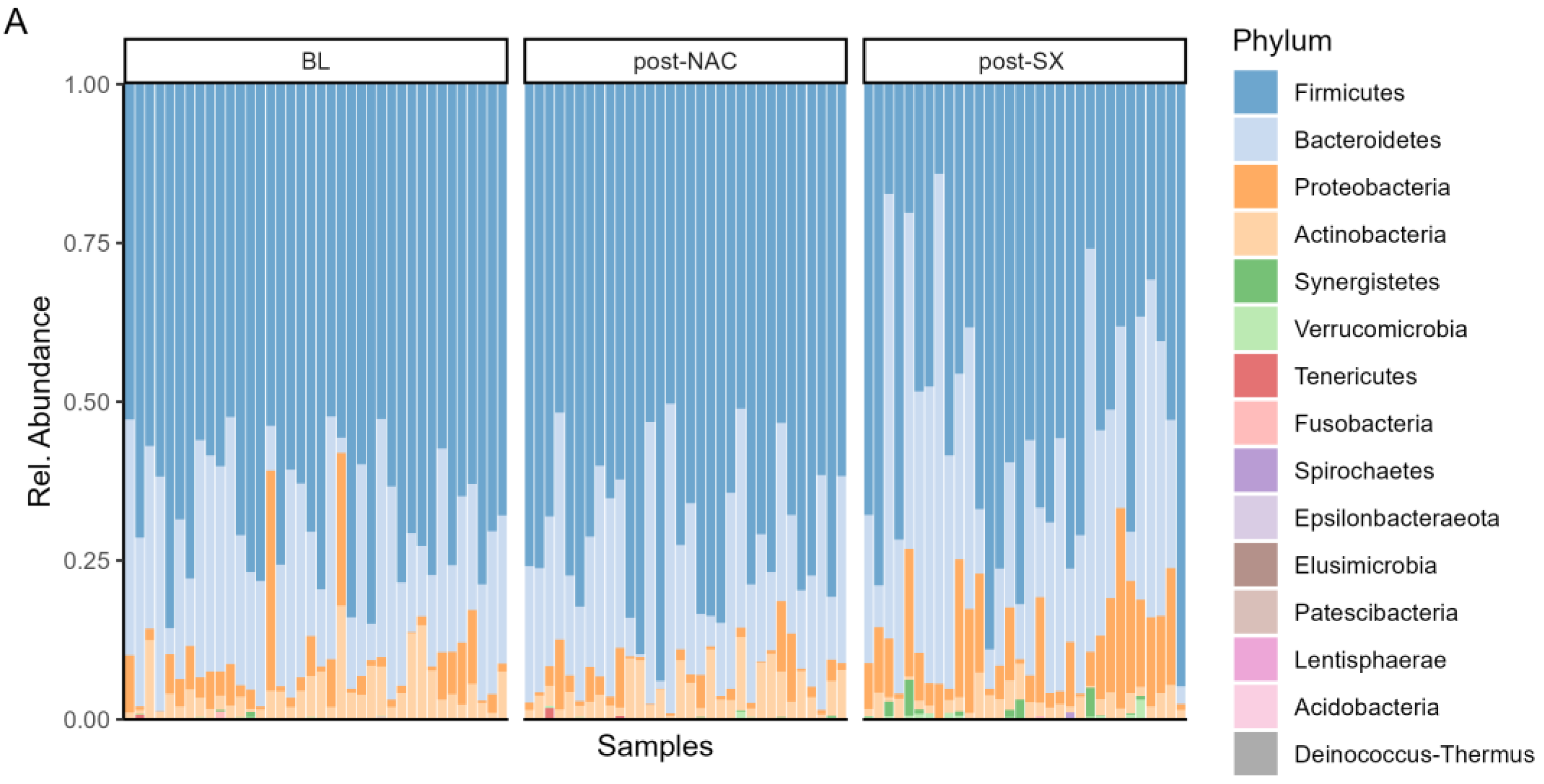
3.3. Microbiome Composition Changes Throughout the Treatment of GC
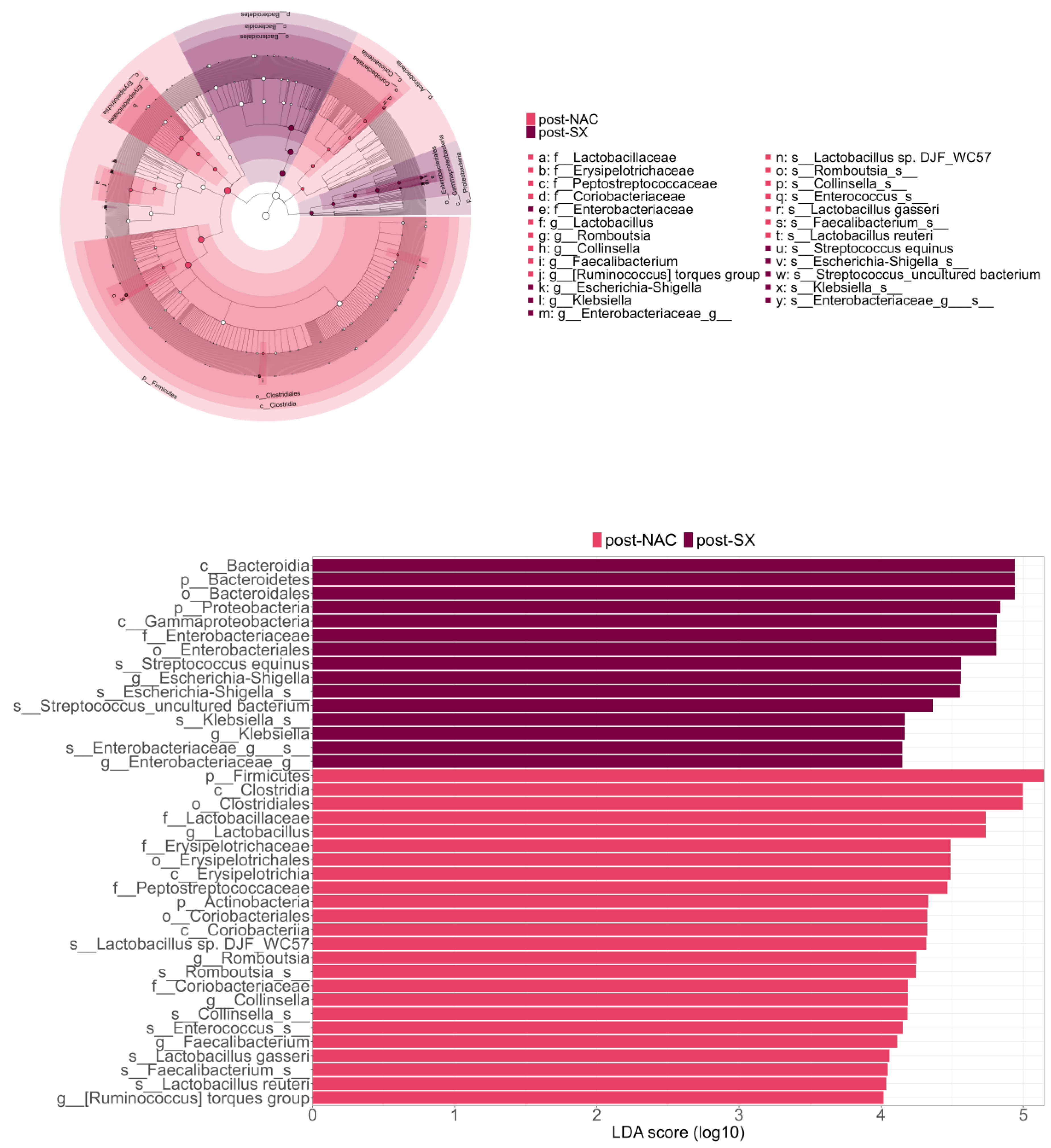
3.4. The Microbiome Composition After Surgery Is Enriched with Oralization-Associated Bacteria
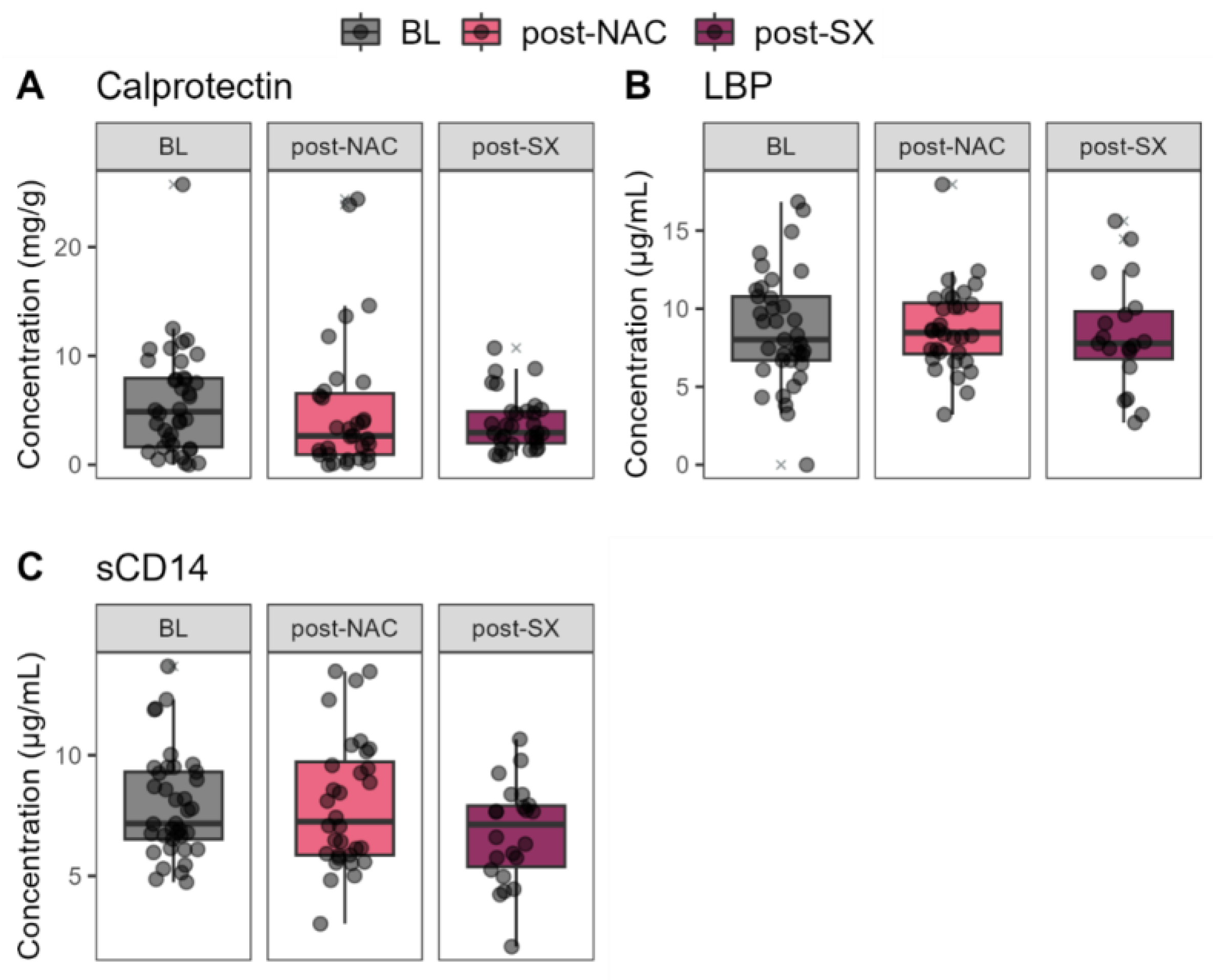
3.5. Fecal and Blood Biomarkers Throughout the Treatment of GC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Feng, A.; Zheng, S.; Chen, C.; Lyu, J. Recent Estimates and Predictions of 5-Year Survival in Patients with Gastric Cancer: A Model-Based Period Analysis. Cancer Control J. Moffitt Cancer Cent. 2022, 29, 10732748221099228. [Google Scholar] [CrossRef] [PubMed]
- Bausys, A.; Luksta, M.; Anglickiene, G.; Maneikiene, V.V.; Kryzauskas, M.; Rybakovas, A.; Dulskas, A.; Kuliavas, J.; Stratilatovas, E.; Macijauskiene, L.; et al. Effect of Home-Based Prehabilitation on Postoperative Complications after Surgery for Gastric Cancer: Randomized Clinical Trial. Br. J. Surg. 2023, 110, 1800–1807. [Google Scholar] [CrossRef] [PubMed]
- Karanicolas, P.J.; Graham, D.; Gönen, M.; Strong, V.E.; Brennan, M.F.; Coit, D.G. Quality of Life after Gastrectomy for Adenocarcinoma: A Prospective Cohort Study. Ann. Surg. 2013, 257, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.S.; Chung, H.Y.; Kwon, O.K.; Yu, W. Long-Term Quality of Life After Distal Subtotal and Total Gastrectomy: Symptom- and Behavior-Oriented Consequences. Ann. Surg. 2016, 263, 738–744. [Google Scholar] [CrossRef]
- Horvath, A.; Bausys, A.; Sabaliauskaite, R.; Stratilatovas, E.; Jarmalaite, S.; Schuetz, B.; Stiegler, P.; Bausys, R.; Stadlbauer, V.; Strupas, K. Distal Gastrectomy with Billroth II Reconstruction is Associated with Oralization of Gut Microbiome and Intestinal Inflammation: A Proof-of-Concept Study. Ann. Surg. Oncol. 2020, 28, 1198–1208. [Google Scholar] [CrossRef]
- Huang, L.; Xu, A.-M.; Li, T.-J.; Han, W.-X.; Xu, J. Should Peri-Gastrectomy Gastric Acidity Be Our Focus among Gastric Cancer Patients? World J. Gastroenterol. WJG 2014, 20, 6981. [Google Scholar] [CrossRef]
- Imhann, F.; Bonder, M.J.; Vich Vila, A.; Fu, J.; Mujagic, Z.; Vork, L.; Tigchelaar, E.F.; Jankipersadsing, S.A.; Cenit, M.C.; Harmsen, H.J.M.; et al. Proton Pump Inhibitors Affect the Gut Microbiome. Gut 2016, 65, 740–748. [Google Scholar] [CrossRef]
- Jackson, M.A.; Goodrich, J.K.; Maxan, M.-E.; Freedberg, D.E.; Abrams, J.A.; Poole, A.C.; Sutter, J.L.; Welter, D.; Ley, R.E.; Bell, J.T.; et al. Proton Pump Inhibitors Alter the Composition of the Gut Microbiota. Gut 2016, 65, 749–756. [Google Scholar] [CrossRef]
- Seto, C.T.; Jeraldo, P.; Orenstein, R.; Chia, N.; DiBaise, J.K. Prolonged Use of a Proton Pump Inhibitor Reduces Microbial Diversity: Implications for Clostridium Difficile Susceptibility. Microbiome 2014, 2, 42. [Google Scholar] [CrossRef]
- Shuwen, H.; Xi, Y.; Yuefen, P.; Jiamin, X.; Quan, Q.; Haihong, L.; Yizhen, J.; Wei, W. Effects of Postoperative Adjuvant Chemotherapy and Palliative Chemotherapy on the Gut Microbiome in Colorectal Cancer. Microb. Pathog. 2020, 149, 104343. [Google Scholar] [CrossRef] [PubMed]
- Kong, C.; Gao, R.; Yan, X.; Huang, L.; He, J.; Li, H.; You, J.; Qin, H. Alterations in Intestinal Microbiota of Colorectal Cancer Patients Receiving Radical Surgery Combined with Adjuvant CapeOx Therapy. Sci. China Life Sci. 2019, 62, 1178–1193. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Zhang, X.; Fan, Y.; Chen, L.; Ma, X.; Yu, H.; Li, J.; Guan, X.; Zhao, P.; Yang, J. Changes of Intestinal Microbiota in Ovarian Cancer Patients Treated with Surgery and Chemotherapy. Cancer Manag. Res. 2020, 12, 8125–8135. [Google Scholar] [CrossRef] [PubMed]
- Stringer, A.M.; Al-Dasooqi, N.; Bowen, J.M.; Tan, T.H.; Radzuan, M.; Logan, R.M.; Mayo, B.; Keefe, D.M.K.; Gibson, R.J. Biomarkers of Chemotherapy-Induced Diarrhoea: A Clinical Study of Intestinal Microbiome Alterations, Inflammation and Circulating Matrix Metalloproteinases. Support. Care Cancer Off. J. Multinatl. Assoc. Support. Care Cancer 2013, 21, 1843–1852. [Google Scholar] [CrossRef]
- Fei, Z.; Lijuan, Y.; Xi, Y.; Wei, W.; Jing, Z.; Miao, D.; Shuwen, H. Gut Microbiome Associated with Chemotherapy-Induced Diarrhea from the CapeOX Regimen as Adjuvant Chemotherapy in Resected Stage III Colorectal Cancer. Gut Pathog. 2019, 11, 18. [Google Scholar] [CrossRef]
- Bausys, A.; Luksta, M.; Kuliavas, J.; Anglickiene, G.; Maneikiene, V.; Gedvilaite, L.; Celutkiene, J.; Jamontaite, I.; Cirtautas, A.; Lenickiene, S.; et al. Personalized Trimodal Prehabilitation for Gastrectomy. Medicine 2020, 99, e20687. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Bisanz, J.E. qiime2R. Importing QIIME2 Artifacts and Associated Data into R Sessions. Version 0.99. 2018, Volume 13. Available online: https://github.com/jbisanz/qiime2R (accessed on 14 June 2024).
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.; Szoecs, E.; et al. Vegan: Community Ecology Package 2022. Available online: https://github.com/vegandevs/vegan (accessed on 14 June 2024).
- Cao, Y.; Dong, Q.; Wang, D.; Zhang, P.; Liu, Y.; Niu, C. MicrobiomeMarker: An R/Bioconductor Package for Microbiome Marker Identification and Visualization. Bioinforma. Oxf. Engl. 2022, 38, 4027–4029. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using Lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Yu, G. Convert Plot to “grob” or “Ggplot” Object. 2021. Available online: https://github.com/GuangchuangYu/ggplotify (accessed on 14 June 2024).
- Montassier, E.; Gastinne, T.; Vangay, P.; Al-Ghalith, G.A.; Bruley des Varannes, S.; Massart, S.; Moreau, P.; Potel, G.; de La Cochetière, M.F.; Batard, E.; et al. Chemotherapy-Driven Dysbiosis in the Intestinal Microbiome. Aliment. Pharmacol. Ther. 2015, 42, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Zwielehner, J.; Lassl, C.; Hippe, B.; Pointner, A.; Switzeny, O.J.; Remely, M.; Kitzweger, E.; Ruckser, R.; Haslberger, A.G. Changes in Human Fecal Microbiota Due to Chemotherapy Analyzed by TaqMan-PCR, 454 Sequencing and PCR-DGGE Fingerprinting. PLoS ONE 2011, 6, e28654. [Google Scholar] [CrossRef] [PubMed]
- Dörffel, Y.; Swidsinski, A.; Loening-Baucke, V.; Wiedenmann, B.; Pavel, M. Common Biostructure of the Colonic Microbiota in Neuroendocrine Tumors and Crohn’s Disease and the Effect of Therapy. Inflamm. Bowel Dis. 2012, 18, 1663–1671. [Google Scholar] [CrossRef]
- Galloway-Peña, J.R.; Smith, D.P.; Sahasrabhojane, P.; Ajami, N.J.; Wadsworth, W.D.; Daver, N.G.; Chemaly, R.F.; Marsh, L.; Ghantoji, S.S.; Pemmaraju, N.; et al. The Role of the Gastrointestinal Microbiome in Infectious Complications during Induction Chemotherapy for Acute Myeloid Leukemia. Cancer 2016, 122, 2186–2196. [Google Scholar] [CrossRef]
- Li, N.; Bai, C.; Zhao, L.; Sun, Z.; Ge, Y.; Li, X. The Relationship Between Gut Microbiome Features and Chemotherapy Response in Gastrointestinal Cancer. Front. Oncol. 2021, 11, 781697. [Google Scholar] [CrossRef]
- Chen, C.; Shen, J.; Du, Y.; Shi, X.; Niu, Y.; Jin, G.; Liu, Y.; Shi, Y.; Lyu, J.; Lin, L. Characteristics of Gut Microbiota in Patients with Gastric Cancer by Surgery, Chemotherapy and Lymph Node Metastasis. Clin. Transl. Oncol. Off. Publ. Fed. Span. Oncol. Soc. Natl. Cancer Inst. Mex. 2022, 24, 2181–2190. [Google Scholar] [CrossRef]
- Anbazhagan, A.N.; Priyamvada, S.; Alrefai, W.A.; Dudeja, P.K. Pathophysiology of IBD associated diarrhea. Tissue Barriers 2018, 6, e1463897. [Google Scholar] [CrossRef]
- Yu, W.; Park, K.B.; Chung, H.Y.; Kwon, O.K.; Lee, S.S. Chronological Changes of Quality of Life in Long-Term Survivors after Gastrectomy for Gastric Cancer. Cancer Res. Treat. 2016, 48, 1030–1036. [Google Scholar] [CrossRef]
- Kim, A.R.; Cho, J.; Hsu, Y.-J.; Choi, M.G.; Noh, J.H.; Sohn, T.S.; Bae, J.M.; Yun, Y.H.; Kim, S. Changes of Quality of Life in Gastric Cancer Patients after Curative Resection: A Longitudinal Cohort Study in Korea. Ann. Surg. 2012, 256, 1008–1013. [Google Scholar] [CrossRef]
- Aoki, T.; Yamaji, I.; Hisamoto, T.; Sato, M.; Matsuda, T. Irregular Bowel Movement in Gastrectomized Subjects: Bowel Habits, Stool Characteristics, Fecal Flora, and Metabolites. Gastric Cancer 2012, 15, 396–404. [Google Scholar] [CrossRef]
- Tsuda, A.; Suda, W.; Morita, H.; Takanashi, K.; Takagi, A.; Koga, Y.; Hattori, M. Influence of Proton-Pump Inhibitors on the Luminal Microbiota in the Gastrointestinal Tract. Clin. Transl. Gastroenterol. 2015, 6, e89. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J.S.; Cox, I.J.; Betrapally, N.S.; Heuman, D.M.; Schubert, M.L.; Ratneswaran, M.; Hylemon, P.B.; White, M.B.; Daita, K.; Noble, N.A.; et al. Systems Biology Analysis of Omeprazole Therapy in Cirrhosis Demonstrates Significant Shifts in Gut Microbiota Composition and Function. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 307, G951–G957. [Google Scholar] [CrossRef] [PubMed]
- Clooney, A.G.; Bernstein, C.N.; Leslie, W.D.; Vagianos, K.; Sargent, M.; Laserna-Mendieta, E.J.; Claesson, M.J.; Targownik, L.E. A Comparison of the Gut Microbiome between Long-Term Users and Non-Users of Proton Pump Inhibitors. Aliment. Pharmacol. Ther. 2016, 43, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Sachdev, A.H.; Pimentel, M. Gastrointestinal Bacterial Overgrowth: Pathogenesis and Clinical Significance. Ther. Adv. Chronic Dis. 2013, 4, 223–231. [Google Scholar] [CrossRef]
- Paik, C.N.; Choi, M.-G.; Lim, C.H.; Park, J.M.; Chung, W.C.; Lee, K.-M.; Jun, K.-H.; Song, K.Y.; Jeon, H.M.; Chin, H.-M.; et al. The Role of Small Intestinal Bacterial Overgrowth in Postgastrectomy Patients. Neurogastroenterol. Motil. Off. J. Eur. Gastrointest. Motil. Soc. 2011, 23, e191–e196. [Google Scholar] [CrossRef]
- Small, P.; Blankenhorn, D.; Welty, D.; Zinser, E.; Slonczewski, J.L. Acid and Base Resistance in Escherichia Coli and Shigella Flexneri: Role of RpoS and Growth PH. J. Bacteriol. 1994, 176, 1729–1737. [Google Scholar] [CrossRef]
- Engelbrektson, A.; Korzenik, J.R.; Pittler, A.; Sanders, M.E.; Klaenhammer, T.R.; Leyer, G.; Kitts, C.L. Probiotics to Minimize the Disruption of Faecal Microbiota in Healthy Subjects Undergoing Antibiotic Therapy. J. Med. Microbiol. 2009, 58, 663–670. [Google Scholar] [CrossRef]
- Yang, Q.; Huang, X.; Wang, P.; Yan, Z.; Sun, W.; Zhao, S.; Gun, S. Longitudinal development of the gut microbiota in healthy and diarrheic piglets induced by age-related dietary changes. Microbiologyopen 2019, 8, e923. [Google Scholar] [CrossRef]
- Martinsen, T.C.; Bergh, K.; Waldum, H.L. Gastric Juice: A Barrier against Infectious Diseases. Basic Clin. Pharmacol. Toxicol. 2005, 96, 94–102. [Google Scholar] [CrossRef]
- Carboni, M.; Guadagni, S.; Pistoia, M.A.; Amicucci, G.; Tuscano, D.; Negro, P.; Smith, P.L.; Walters, C.L. The Microflora of the Gastric Juice after Billroth I and Billroth II Partial Gastrectomy. Scand. J. Gastroenterol. 1986, 21, 461–470. [Google Scholar] [CrossRef]
- Vincent, Z.; Hornby, S.; Ball, S.; Sanders, G.; Ayling, R.M. Faecal Calprotectin as a Marker for Oesophago-Gastric Cancer. Ann. Clin. Biochem. 2015, 52, 660–664. [Google Scholar] [CrossRef]



| Characteristic | x | |
|---|---|---|
| Age (years), mean (SD) | 60 (11) | |
| Male: Female | 24:14 | |
| Charlson comorbidity index score, mean (SD) | 4 (1.4) | |
| BMI (kg/m2), mean (SD) | 25 (4) | |
| cT, n (%) | 1–2 | 4 (10.5) |
| 3–4 | 34 (89.5) | |
| cN, n (%) | 0 | 10 (26.3) |
| 1–3 | 28 (73.7) | |
| cM, n (%) | 0 | 36 (94.7) |
| 1 | 2 (5.3) | |
| Tumor localization, n (%) | Upper third | 6 (15.8) |
| Middle third | 14 (36.8) | |
| Lower third | 16 (42.1) | |
| Total | 2 (5.3) | |
| Type of neoadjuvant chemotherapy, n (%) | FLOT | 36 (94.7) |
| Cisplatin+5-FU | 2 (5.3) | |
| Number of cycles, mean (SD) | 4 (1) | |
| Type of surgery, n (%) | Total gastrectomy | 11 (28.9) |
| Subtotal gastrectomy | 27 (71.1) | |
| R0, n (%) | 37 (97.4) | |
| D2 lymphadenectomy, n (%) | 38 (100) | |
| Laparoscopic surgery, n (%) | 10 (26.3) | |
| ypT, n (%) | 0 | 2 (5.3) |
| 1 | 8 (21.0) | |
| 2 | 4 (10.5) | |
| 3 | 17 (44.7) | |
| 4 | 7 (18.5) | |
| ypN, n (%) | 0 | 16 (42.1) |
| 1 | 10 (26.3) | |
| 2 | 6 (15.8) | |
| 3 | 6 (15.8) | |
| ypM, n (%) | 0 | 38 (100) |
| 1 | 0 (0) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Žukauskaitė, K.; Baušys, B.; Horvath, A.; Sabaliauskaitė, R.; Šeštokaitė, A.; Mlynska, A.; Jarmalaitė, S.; Stadlbauer, V.; Baušys, R.; Baušys, A. Gut Microbiome Changes After Neoadjuvant Chemotherapy and Surgery in Patients with Gastric Cancer. Cancers 2024, 16, 4074. https://doi.org/10.3390/cancers16234074
Žukauskaitė K, Baušys B, Horvath A, Sabaliauskaitė R, Šeštokaitė A, Mlynska A, Jarmalaitė S, Stadlbauer V, Baušys R, Baušys A. Gut Microbiome Changes After Neoadjuvant Chemotherapy and Surgery in Patients with Gastric Cancer. Cancers. 2024; 16(23):4074. https://doi.org/10.3390/cancers16234074
Chicago/Turabian StyleŽukauskaitė, Kristina, Bernardas Baušys, Angela Horvath, Rasa Sabaliauskaitė, Agnė Šeštokaitė, Agata Mlynska, Sonata Jarmalaitė, Vanessa Stadlbauer, Rimantas Baušys, and Augustinas Baušys. 2024. "Gut Microbiome Changes After Neoadjuvant Chemotherapy and Surgery in Patients with Gastric Cancer" Cancers 16, no. 23: 4074. https://doi.org/10.3390/cancers16234074
APA StyleŽukauskaitė, K., Baušys, B., Horvath, A., Sabaliauskaitė, R., Šeštokaitė, A., Mlynska, A., Jarmalaitė, S., Stadlbauer, V., Baušys, R., & Baušys, A. (2024). Gut Microbiome Changes After Neoadjuvant Chemotherapy and Surgery in Patients with Gastric Cancer. Cancers, 16(23), 4074. https://doi.org/10.3390/cancers16234074








