Non-Metastatic Clear Cell Renal Cell Carcinoma Immune Cell Infiltration Heterogeneity and Prognostic Ability in Patients Following Surgery
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Discovery Cohort Patient Selection
2.2. Discovery Cohort Histology and Immunostaining
2.3. Discovery Cohort Slide Image Acquisition and Analysis
2.4. Validation Tissue Microarray Construction
2.5. Validation of TMA-Multiplexed Immunohistochemistry
2.6. Tissue Microarray Automated Image Acquisition and Analysis
2.7. Tissue Microarray Staining and Image Acquisition Using the PhenoCycler Platform
2.8. Cell Phenotype Labeling with PhenoCycler-Generated Images
2.9. Statistical Analysis and Spatial Analysis
3. Results
3.1. Discovery Cohort to Evaluate the Prognostic Impact of CD8+ T Cells
3.2. Validation Cohort to Evaluate the Prognostic Impact of CD8+ T Cells
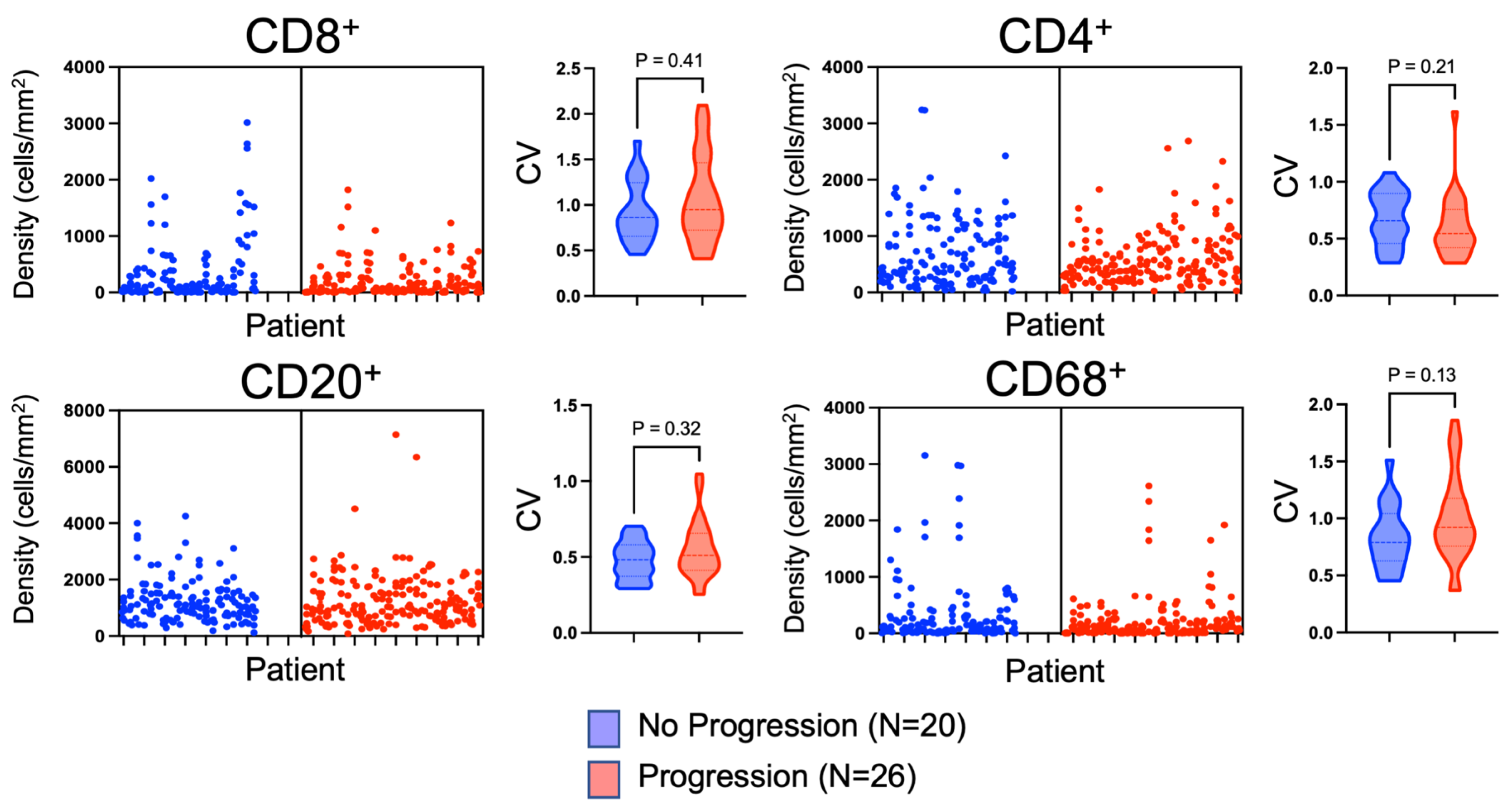
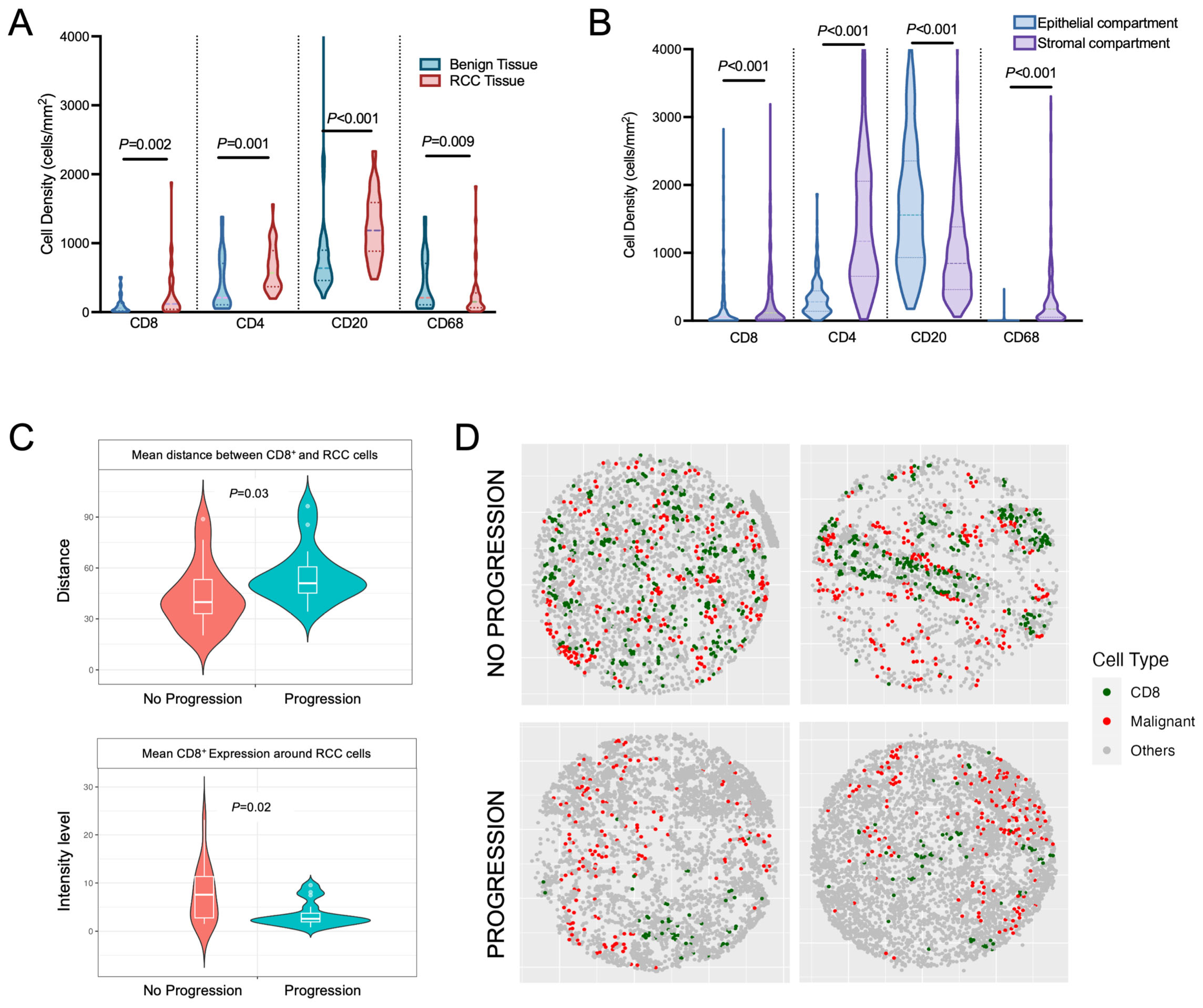
3.3. Evaluation of Tissue Heterogeneity and Spatial Variation of Immune Cell Infiltration within Non-Metastatic ccRCC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.C.; Clark, P.E.; Chang, S.S.; Karam, J.A.; Souter, L.; Uzzo, R.G. Renal mass and localized renal cancer: Evaluation, management, and follow-up: Aua guideline: Part I. J. Urol. 2021, 206, 199–208. [Google Scholar] [CrossRef]
- Campbell, S.C.; Uzzo, R.G.; Karam, J.A.; Chang, S.S.; Clark, P.E.; Souter, L. Renal mass and localized renal cancer: Evaluation, management, and follow-up: Aua guideline: Part II. J. Urol. 2021, 206, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Dabestani, S.; Beisland, C.; Stewart, G.D.; Bensalah, K.; Gudmundsson, E.; Lam, T.B.; Gietzmann, W.; Zakikhani, P.; Marconi, L.; Fernandéz-Pello, S.; et al. Long-term outcomes of follow-up for initially localised clear cell renal cell carcinoma: Recur database analysis. Eur. Urol. Focus 2019, 5, 857–866. [Google Scholar] [CrossRef]
- Abel, E.J.; Margulis, V.; Bauman, T.M.; Karam, J.A.; Christensen, W.P.; Krabbe, L.M.; Haddad, A.; Golla, V.; Wood, C.G. Risk factors for recurrence after surgery in non-metastatic rcc with thrombus: A contemporary multicentre analysis. BJU Int. 2016, 117, E87–E94. [Google Scholar] [CrossRef]
- Correa, A.F.; Jegede, O.; Haas, N.B.; Flaherty, K.T.; Pins, M.R.; Messing, E.M.; Manola, J.; Wood, C.G.; Kane, C.J.; Jewett, M.A.S.; et al. Predicting renal cancer recurrence: Defining limitations of existing prognostic models with prospective trial-based validation. J. Clin. Oncol. 2019, 37, 2062–2071. [Google Scholar] [CrossRef] [PubMed]
- Powles, T.; Tomczak, P.; Park, S.H.; Venugopal, B.; Ferguson, T.; Symeonides, S.N.; Hajek, J.; Gurney, H.; Chang, Y.H.; Lee, J.L.; et al. Pembrolizumab versus placebo as post-nephrectomy adjuvant therapy for clear cell renal cell carcinoma (keynote-564): 30-month follow-up analysis of a multicentre, randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2022, 23, 1133–1144. [Google Scholar] [CrossRef]
- Choueiri, T.K.; Tomczak, P.; Park, S.H.; Venugopal, B.; Ferguson, T.; Chang, Y.-H.; Hajek, J.; Symeonides, S.N.; Lee, J.L.; Sarwar, N.; et al. Adjuvant pembrolizumab after nephrectomy in renal-cell carcinoma. N. Engl. J. Med. 2021, 385, 683–694. [Google Scholar] [CrossRef]
- Şenbabaoğlu, Y.; Gejman, R.S.; Winer, A.G.; Liu, M.; Allen, E.M.V.; Velasco, G.d.; Miao, D.; Ostrovnaya, I.; Drill, E.; Luna, A.; et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger rna signatures. Genome Biol. 2016, 17, 231. [Google Scholar] [CrossRef]
- Lasorsa, F.; Rutigliano, M.; Milella, M.; Ferro, M.; Pandolfo, S.D.; Crocetto, F.; Tataru, O.S.; Autorino, R.; Battaglia, M.; Ditonno, P.; et al. Cellular and molecular players in the tumor microenvironment of renal cell carcinoma. J. Clin. Med. 2023, 12, 3888. [Google Scholar] [CrossRef]
- Motzer, R.J.; Tannir, N.M.; McDermott, D.F.; Frontera, O.A.; Melichar, B.; Choueiri, T.K.; Plimack, E.R.; Barthélémy, P.; Porta, C.; George, S.; et al. Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N. Engl. J. Med. 2018, 378, 1277–1290. [Google Scholar] [CrossRef]
- Motzer, R.J.; Penkov, K.; Haanen, J.; Rini, B.; Albiges, L.; Campbell, M.T.; Venugopal, B.; Kollmannsberger, C.; Negrier, S.; Uemura, M.; et al. Avelumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N. Engl. J. Med. 2019, 380, 1103–1115. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.; Alekseev, B.; Rha, S.Y.; Porta, C.; Eto, M.; Powles, T.; Grünwald, V.; Hutson, T.E.; Kopyltsov, E.; Méndez-Vidal, M.J.; et al. Lenvatinib plus pembrolizumab or everolimus for advanced renal cell carcinoma. N. Engl. J. Med. 2021, 384, 1289–1300. [Google Scholar] [CrossRef] [PubMed]
- Rini, B.I.; Plimack, E.R.; Stus, V.; Gafanov, R.; Hawkins, R.; Nosov, D.; Pouliot, F.; Alekseev, B.; Soulieres, D.; Melichar, B.; et al. Pembrolizumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N. Engl. J. Med. 2019, 380, 1116–1127. [Google Scholar] [CrossRef] [PubMed]
- Lasorsa, F.; di Meo, N.A.; Rutigliano, M.; Milella, M.; Ferro, M.; Pandolfo, S.D.; Crocetto, F.; Tataru, O.S.; Autorino, R.; Battaglia, M.; et al. Immune checkpoint inhibitors in renal cell carcinoma: Molecular basis and rationale for their use in clinical practice. Biomedicines 2023, 11, 1071. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, D.D.; Dolan, B.; Laklouk, I.A.; Rassi, S.; Lozar, T.; Emamekhoo, H.; Wentland, A.L.; Lubner, M.G.; Abel, E.J. Understanding the tumor immune microenvironment in renal cell carcinoma. Cancers 2023, 15, 2500. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, N.A.; Becht, E.; Pagès, F.; Skliris, G.; Verkarre, V.; Vano, Y.; Mejean, A.; Saint-Aubert, N.; Lacroix, L.; Natario, I.; et al. Orchestration and prognostic significance of immune checkpoints in the microenvironment of primary and metastatic renal cell cancer. Clin. Cancer Res. 2015, 21, 3031–3040. [Google Scholar] [CrossRef]
- Jansen, C.S.; Prokhnevska, N.; Master, V.A.; Sanda, M.G.; Carlisle, J.W.; Bilen, M.A.; Cardenas, M.; Wilkinson, S.; Lake, R.; Sowalsky, A.G.; et al. An intra-tumoral niche maintains and differentiates stem-like cd8 t cells. Nature 2019, 576, 465–470. [Google Scholar] [CrossRef]
- Bauman, T.M.; Huang, W.; Lee, M.H.; Abel, E.J. Neovascularity as a prognostic marker in renal cell carcinoma. Hum. Pathol. 2016, 57, 98–105. [Google Scholar] [CrossRef]
- Black, S.; Phillips, D.; Hickey, J.W.; Kennedy-Darling, J.; Venkataraaman, V.G.; Samusik, N.; Goltsev, Y.; Schürch, C.M.; Nolan, G.P. Codex multiplexed tissue imaging with dna-conjugated antibodies. Nat. Protoc. 2021, 16, 3802–3835. [Google Scholar] [CrossRef]
- Zhang, W.; Li, I.; Reticker-Flynn, N.E.; Good, Z.; Chang, S.; Samusik, N.; Saumyaa, S.; Li, Y.; Zhou, X.; Liang, R.; et al. Identification of cell types in multiplexed in situ images by combining protein expression and spatial information using celesta. Nat. Methods 2022, 19, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Baitsch, L.; Baumgaertner, P.; Devêvre, E.; Raghav, S.K.; Legat, A.; Barba, L.; Wieckowski, S.; Bouzourene, H.; Deplancke, B.; Romero, P.; et al. Exhaustion of tumor-specific cd8+ t cells in metastases from melanoma patients. J. Clin. Investig. 2011, 121, 2350–2360. [Google Scholar] [CrossRef] [PubMed]
- Ahmadzadeh, M.; Johnson, L.A.; Heemskerk, B.; Wunderlich, J.R.; Dudley, M.E.; White, D.E.; Rosenberg, S.A. Tumor antigen–specific cd8 t cells infiltrating the tumor express high levels of pd-1 and are functionally impaired. Blood 2009, 114, 1537–1544. [Google Scholar] [CrossRef] [PubMed]
- Giles, J.R.; Globig, A.-M.; Kaech, S.M.; Wherry, E.J. Cd8+ t cells in the cancer-immunity cycle. Immunity 2023, 56, 2231–2253. [Google Scholar] [CrossRef]
- Feng, Y.; Yang, T.; Zhu, J.; Li, M.; Doyle, M.; Ozcoban, V.; Bass, G.T.; Pizzolla, A.; Cain, L.; Weng, S.; et al. Spatial analysis with spiat and spasim to characterize and simulate tissue microenvironments. Nat. Commun. 2023, 14, 2697. [Google Scholar] [CrossRef]
- Giraldo, N.A.; Becht, E.; Vano, Y.; Petitprez, F.; Lacroix, L.; Validire, P.; Sanchez-Salas, R.; Ingels, A.; Oudard, S.; Moatti, A.; et al. Tumor-infiltrating and peripheral blood t-cell immunophenotypes predict early relapse in localized clear cell renal cell carcinoma. Clin. Cancer Res. 2017, 23, 4416–4428. [Google Scholar] [CrossRef]
- Braun, D.A.; Hou, Y.; Bakouny, Z.; Ficial, M.; Angelo, M.S.; Forman, J.; Ross-Macdonald, P.; Berger, A.C.; Jegede, O.A.; Elagina, L.; et al. Interplay of somatic alterations and immune infiltration modulates response to pd-1 blockade in advanced clear cell renal cell carcinoma. Nat. Med. 2020, 26, 909–918. [Google Scholar] [CrossRef]
- Varn, F.S.; Wang, Y.; Mullins, D.W.; Fiering, S.; Cheng, C. Systematic pan-cancer analysis reveals immune cell interactions in the tumor microenvironment. Cancer Res. 2017, 77, 1271–1282. [Google Scholar] [CrossRef]
- Braun, D.A.; Street, K.; Burke, K.P.; Cookmeyer, D.L.; Denize, T.; Pedersen, C.B.; Gohil, S.H.; Schindler, N.; Pomerance, L.; Hirsch, L.; et al. Progressive immune dysfunction with advancing disease stage in renal cell carcinoma. Cancer Cell 2021, 39, 632–648.e638. [Google Scholar] [CrossRef]
- Clark, D.J.; Dhanasekaran, S.M.; Petralia, F.; Pan, J.; Song, X.; Hu, Y.; Leprevost, F.d.V.; Reva, B.; Lih, T.-S.M.; Chang, H.-Y.; et al. Integrated proteogenomic characterization of clear cell renal cell carcinoma. Cell 2019, 179, 964–983.e931. [Google Scholar] [CrossRef]
- Hakimi, A.A.; Voss, M.H.; Kuo, F.; Sanchez, A.; Liu, M.; Nixon, B.G.; Vuong, L.; Ostrovnaya, I.; Chen, Y.-B.; Reuter, V.; et al. Transcriptomic profiling of the tumor microenvironment reveals distinct subgroups of clear cell renal cell cancer—Data from a randomized phase iii trial. Cancer Discov. 2019, 9, CD-18-0957. [Google Scholar] [CrossRef] [PubMed]
- Turajlic, S.; Xu, H.; Litchfield, K.; Rowan, A.; Horswell, S.; Chambers, T.; O’Brien, T.; Lopez, J.I.; Watkins, T.B.K.; Nicol, D.; et al. Deterministic evolutionary trajectories influence primary tumor growth: Tracerx renal. Cell 2018, 173, 595–610.e11. [Google Scholar] [CrossRef] [PubMed]
- Schürch, C.M.; Bhate, S.S.; Barlow, G.L.; Phillips, D.J.; Noti, L.; Zlobec, I.; Chu, P.; Black, S.; Demeter, J.; McIlwain, D.R.; et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell 2020, 182, 1341–1359.e19. [Google Scholar] [CrossRef] [PubMed]
- Bhate, S.S.; Barlow, G.L.; Schürch, C.M.; Nolan, G.P. Tissue schematics map the specialization of immune tissue motifs and their appropriation by tumors. Cell Syst. 2021, 13, 109–130.e6. [Google Scholar] [CrossRef]
- Sharma, V.; Wymer, K.M.; Joyce, D.D.; Moriarty, J.; Khanna, A.; Borah, B.J.; Thompson, R.H.; Costello, B.A.; Leibovich, B.C.; Boorjian, S.A. Cost-effectiveness of adjuvant pembrolizumab after nephrectomy for high-risk renal cell carcinoma: Insights for patient selection from a markov model. J. Urol. 2023, 209, 89–98. [Google Scholar] [CrossRef]

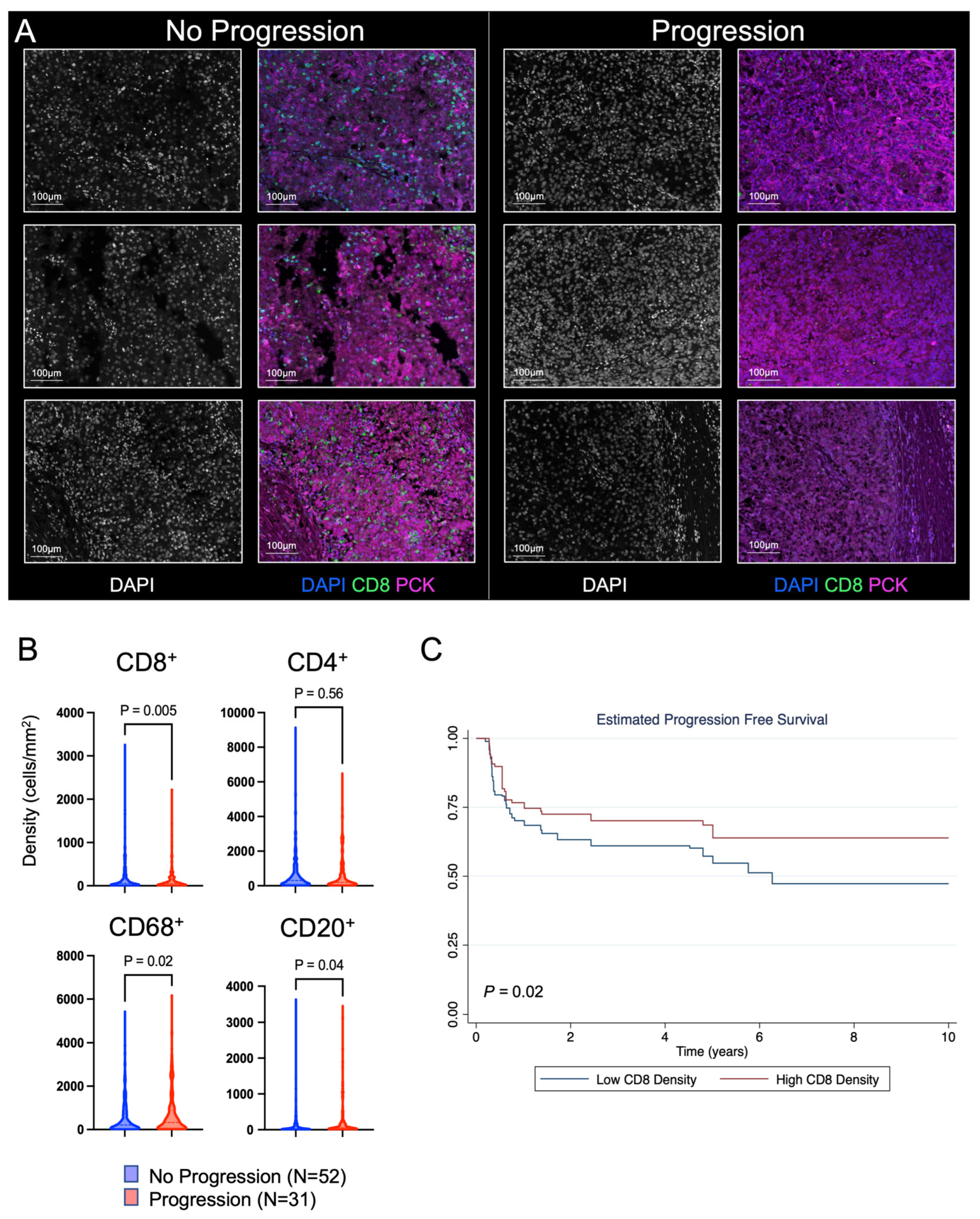
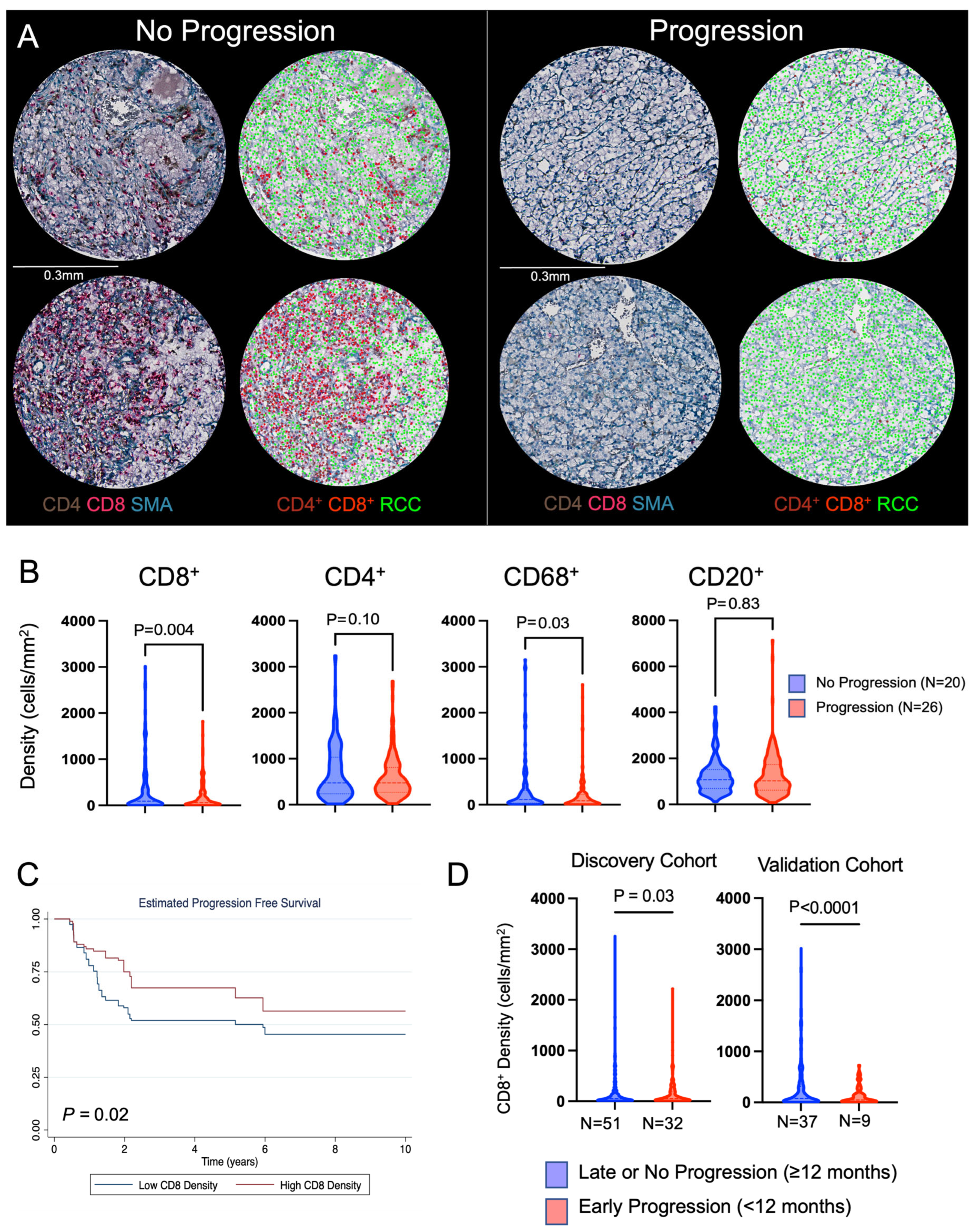
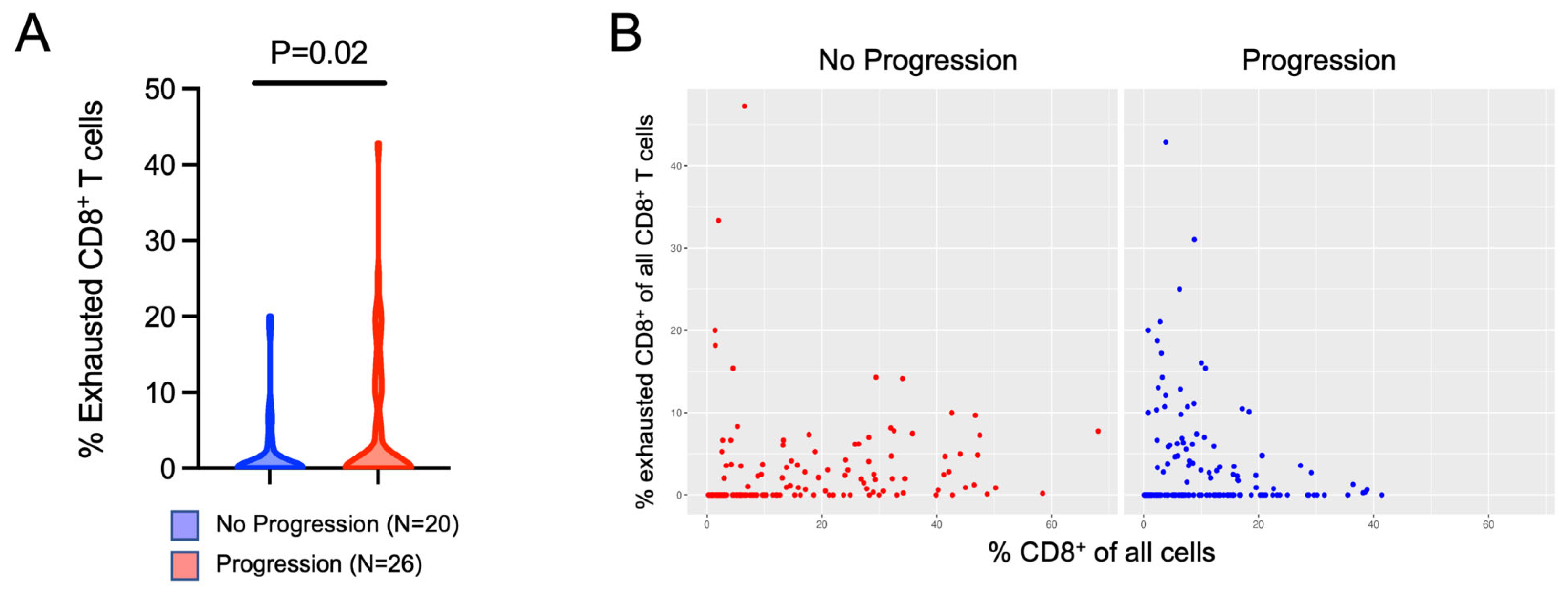
| Discovery Cohort | No Progression | Progression | p Value |
|---|---|---|---|
| N = 52 | N = 31 | ||
| Median age, years (IQR) | 58 (54–64) | 64 (55–74) | 0.03 |
| Gender, no. of females (%) | 15 (31) | 6 (19) | 0.3 |
| ECOG Performance Status, no. (%) | 1 | ||
| 0 | 43 (91) | 29 (93) | |
| 1 | 4 (9) | 2 (7) | |
| Pathologic T-stage | 1 | ||
| T2 | 2 (4) | 1 (3) | |
| T3–T4 | 50 (96) | 30 (97) | |
| Median maximum pathologic tumor diameter, cm (IQR) | 9.5 (8–11) | 9.8 (8–12.5) | 0.4 |
| Grade, no. (%) | 0.02 | ||
| 1–2 | 14 (27) | 2 (6) | |
| 3–4 | 38 (73) | 29 (94) | |
| Thrombus | 29 (56) | 22 (71) | 0.2 |
| Died, no. (%) | 11 (21) | 17 (55) | 0.004 |
| Median follow-up, months (IQR) | 37 (13–81) | 33 (14–77) | 0.9 |
| Validation Cohort | No Progression | Progression | p Value |
|---|---|---|---|
| N = 20 | N = 26 | ||
| Median age, years (IQR) | 61 (51–70) | 57 (50–66) | 0.3 |
| Gender, no. of females (%) | 8 (40) | 9 (35) | 0.8 |
| ECOG Performance Status, no. (%) | 0.6 | ||
| 0 | 19 (95) | 23 (88) | |
| 1 | 1 (5) | 3 (12) | |
| Median preop NLR, (IQR) | 4.4 (2.7–6.1) | 2.9 (2.3–4.2) | 0.2 |
| Median preop CRP, (IQR) | 2 (2–11) | 1 (1–3) | 0.2 |
| Pathologic T-stage | 0.2 | ||
| T2 | 10 (50) | 8 (31) | |
| T3–T4 | 10 (50) | 18 (69) | |
| Median maximum pathologic tumor diameter, cm (IQR) | 9 (7.2–9.4) | 9.1 (8–13) | 0.2 |
| Grade, no. (%) | 0.4 | ||
| 1–2 | 11 (55) | 10 (38) | |
| 3–4 | 9 (45) | 16 (62) | |
| Thrombus | 7 (35) | 8 (31) | 1 |
| Died, no. (%) | 5 (25) | 14 (54) | 0.07 |
| Median follow-up, years (IQR) | 11 (8–15) | 7 (5–11) | 0.08 |
| Median Coefficient of Variation | No Progression (N = 20) | Progression (N = 26) | p Value |
|---|---|---|---|
| CD8 CV | 0.86 | 0.94 | 0.4 |
| CD4 CV | 0.66 | 0.54 | 0.2 |
| CD20 CV | 0.48 | 0.51 | 0.3 |
| CD68 CV | 0.79 | 0.92 | 0.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shapiro, D.D.; Lozar, T.; Cheng, L.; Xie, E.; Laklouk, I.; Lee, M.H.; Huang, W.; Jarrard, D.F.; Allen, G.O.; Hu, R.; et al. Non-Metastatic Clear Cell Renal Cell Carcinoma Immune Cell Infiltration Heterogeneity and Prognostic Ability in Patients Following Surgery. Cancers 2024, 16, 478. https://doi.org/10.3390/cancers16030478
Shapiro DD, Lozar T, Cheng L, Xie E, Laklouk I, Lee MH, Huang W, Jarrard DF, Allen GO, Hu R, et al. Non-Metastatic Clear Cell Renal Cell Carcinoma Immune Cell Infiltration Heterogeneity and Prognostic Ability in Patients Following Surgery. Cancers. 2024; 16(3):478. https://doi.org/10.3390/cancers16030478
Chicago/Turabian StyleShapiro, Daniel D., Taja Lozar, Lingxin Cheng, Elliot Xie, Israa Laklouk, Moon Hee Lee, Wei Huang, David F. Jarrard, Glenn O. Allen, Rong Hu, and et al. 2024. "Non-Metastatic Clear Cell Renal Cell Carcinoma Immune Cell Infiltration Heterogeneity and Prognostic Ability in Patients Following Surgery" Cancers 16, no. 3: 478. https://doi.org/10.3390/cancers16030478
APA StyleShapiro, D. D., Lozar, T., Cheng, L., Xie, E., Laklouk, I., Lee, M. H., Huang, W., Jarrard, D. F., Allen, G. O., Hu, R., Kinoshita, T., Esbona, K., Lambert, P. F., Capitini, C. M., Kendziorski, C., & Abel, E. J. (2024). Non-Metastatic Clear Cell Renal Cell Carcinoma Immune Cell Infiltration Heterogeneity and Prognostic Ability in Patients Following Surgery. Cancers, 16(3), 478. https://doi.org/10.3390/cancers16030478







