In Silico and In Vitro Mapping of Receptor-Type Protein Tyrosine Phosphatase Receptor Type D in Health and Disease: Implications for Asprosin Signalling in Endometrial Cancer and Neuroblastoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Immunofluorescence (IF)
2.3. Immunohistochemistry (IHC) of Tissue Microarray
2.4. RNA Isolation, cDNA Synthesis, and Reverse Transcription Quantitative PCR (RT qPCR)
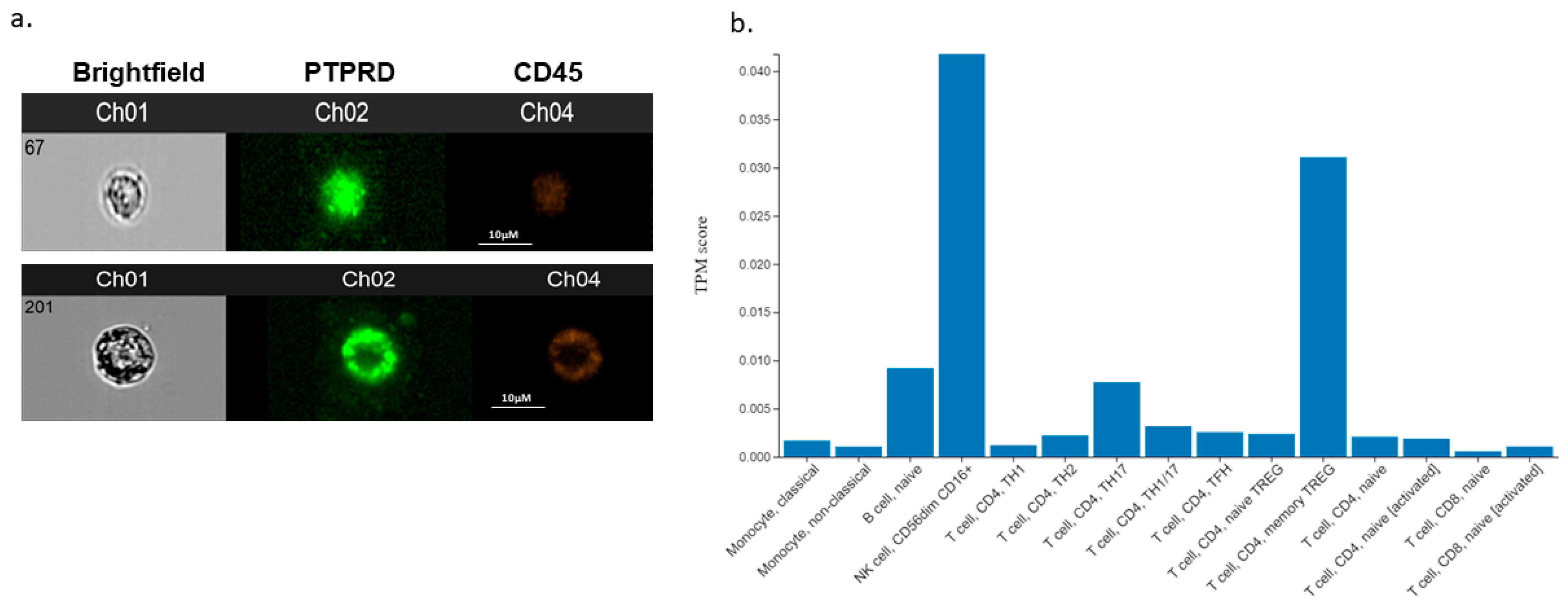
2.5. Imaging Flow Cytometry of Clinical Samples
2.6. Statistical Analysis
3. Results
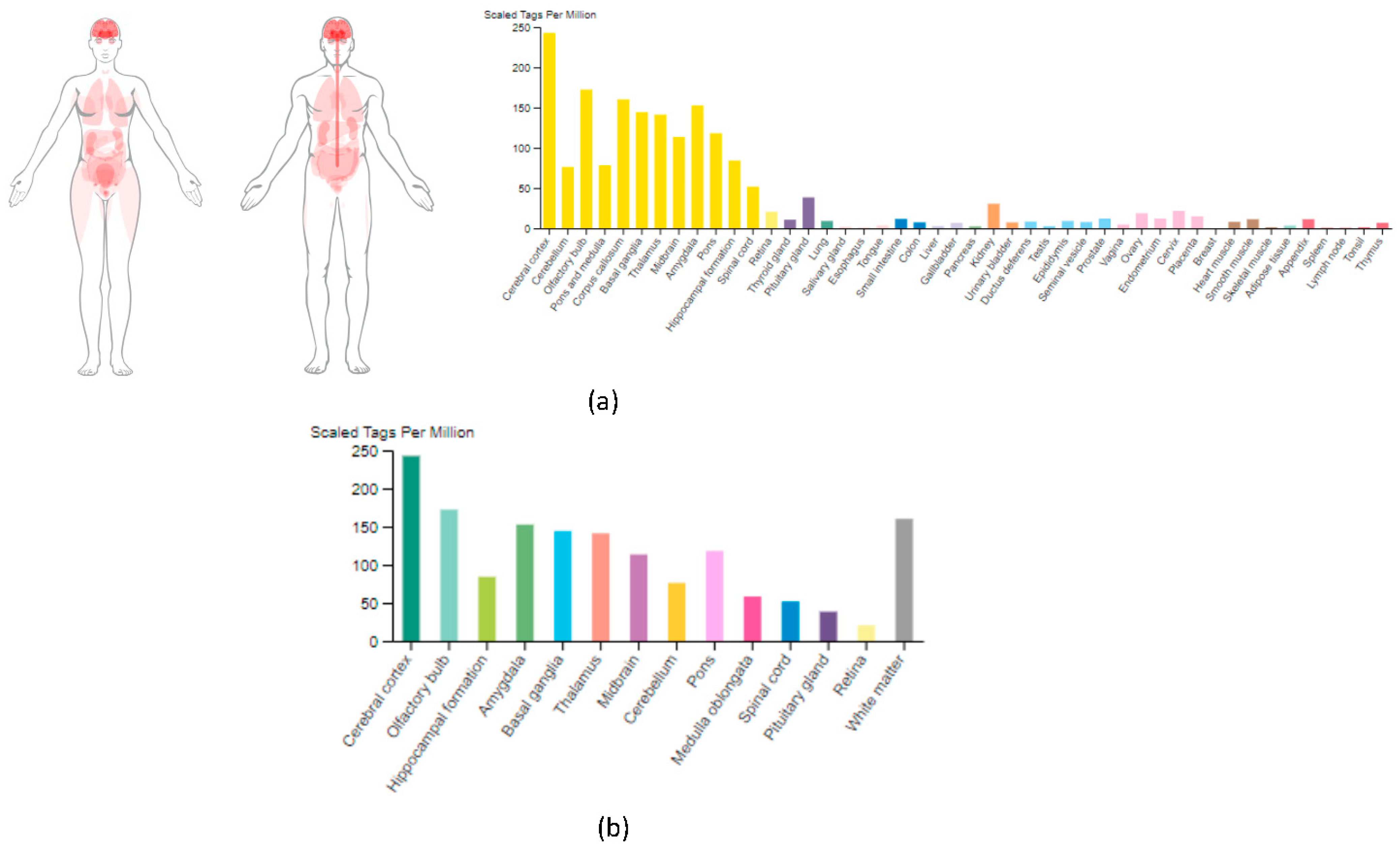
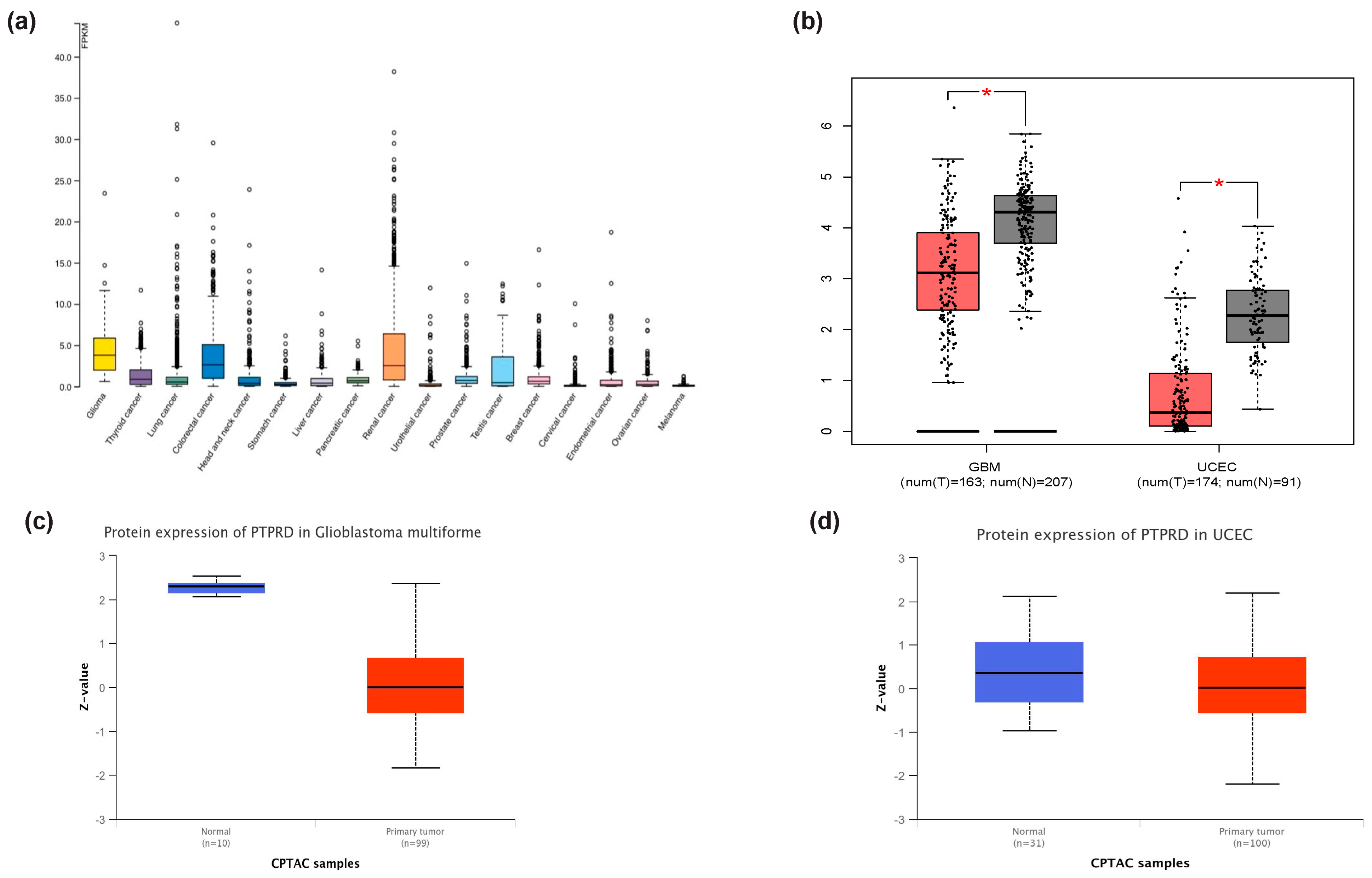
3.1. PTPRD mRNA Expression across Human Tissues
3.2. PTPRD Regional Distribution within the Brain
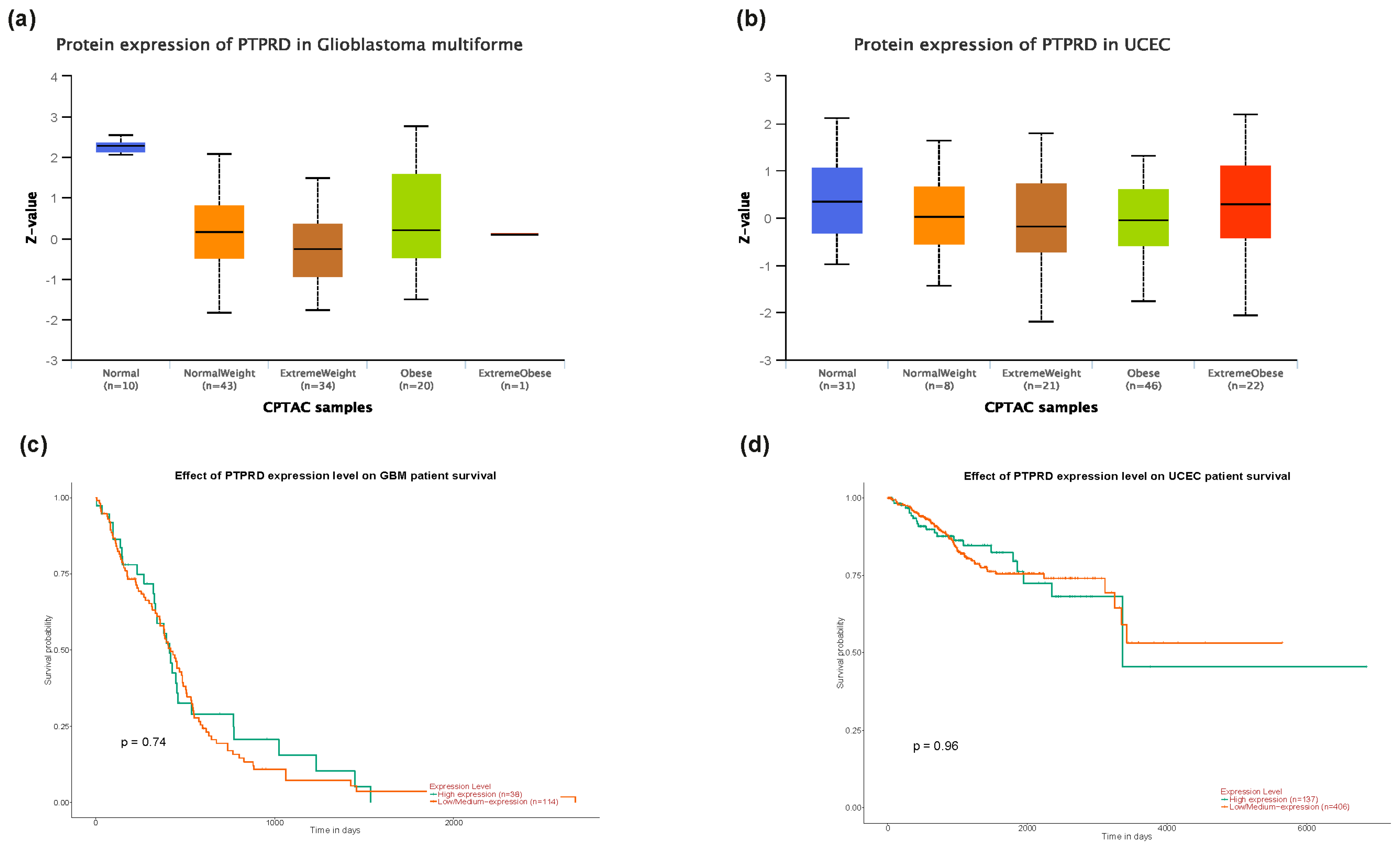
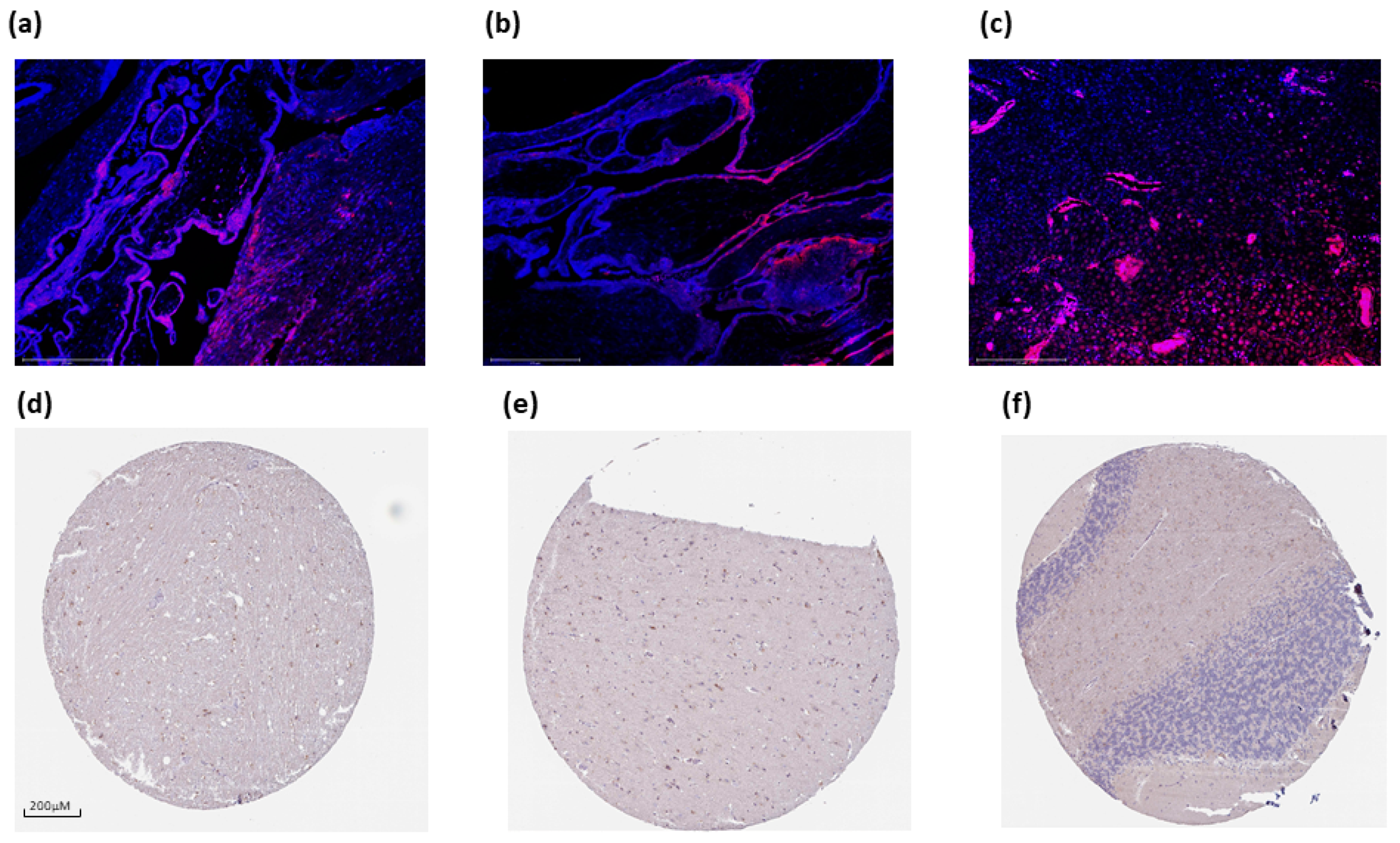
3.3. PTPRD Relative Expression in Endometrial Cancer and GBM
3.4. PTPRD Expression in Endometrial Cancer
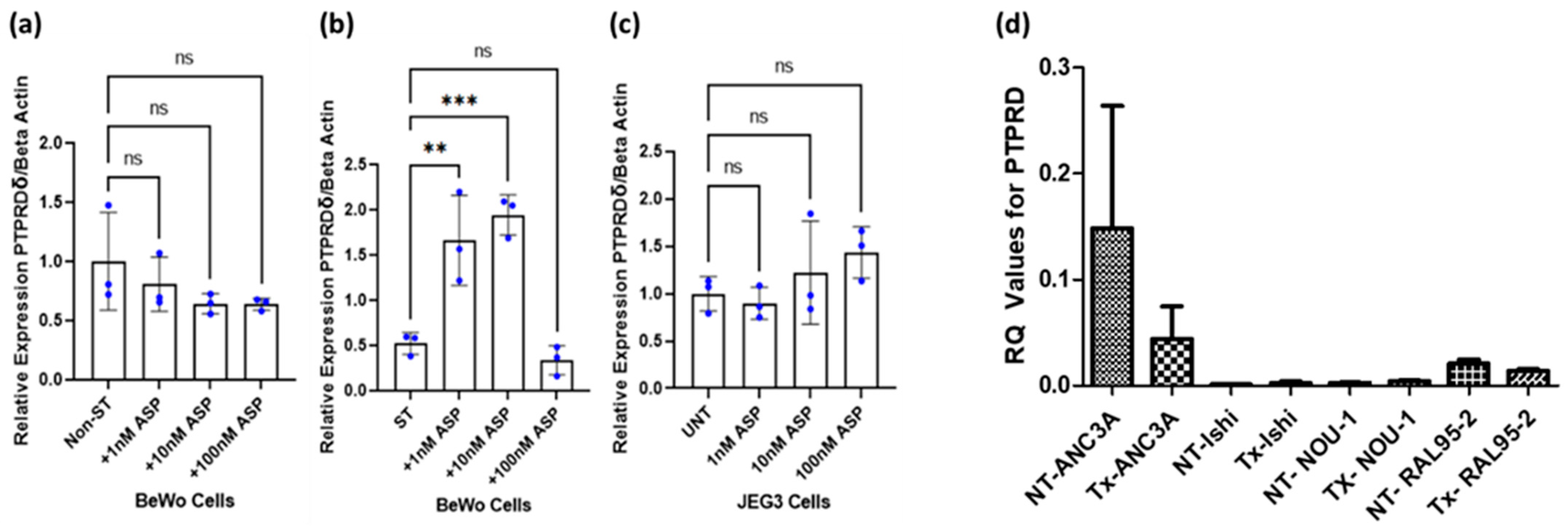
3.5. PTPRD Expression In Vitro
3.6. Effect of Asprosin on PTPRD Expression
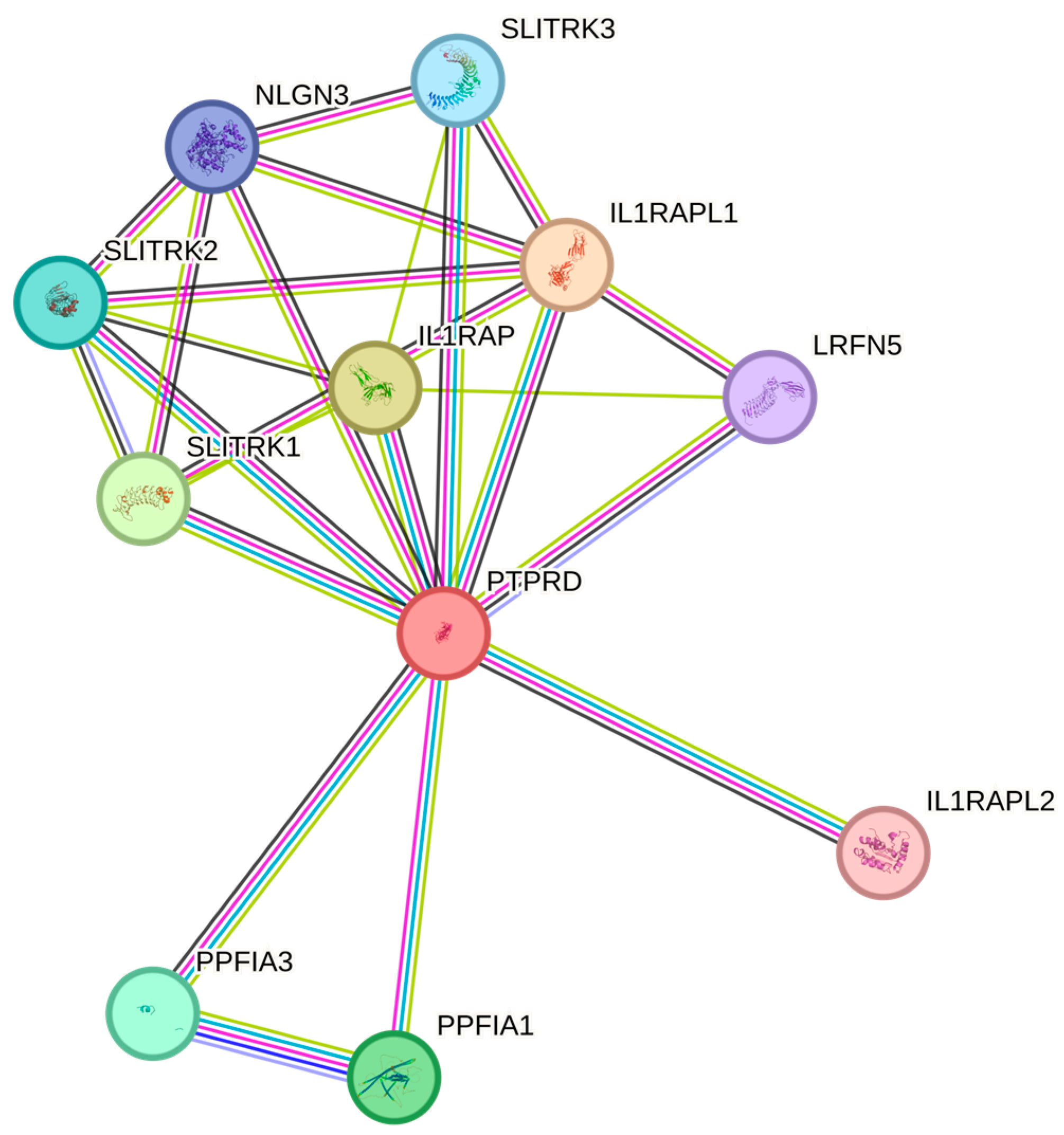
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Romere, C.; Duerrschmid, C.; Bournat, J.; Constable, P.; Jain, M.; Xia, F.; Saha, P.K.; Del Solar, M.; Zhu, B.; York, B.; et al. Asprosin, a Fasting-Induced Glucogenic Protein Hormone. Cell 2016, 165, 566–579. [Google Scholar] [CrossRef]
- Summers, K.M.; Bush, S.J.; Davis, M.R.; Hume, D.A.; Keshvari, S.; West, J.A. Fibrillin-1 and asprosin, novel players in metabolic syndrome. Mol. Genet. Metab. 2023, 138, 106979. [Google Scholar] [CrossRef] [PubMed]
- Alan, M.; Gurlek, B.; Yilmaz, A.; Aksit, M.; Aslanipour, B.; Gulhan, I.; Mehmet, C.; Taner, C.E. Asprosin: A novel peptide hormone related to insulin resistance in women with polycystic ovary syndrome. Gynecol. Endocrinol. 2019, 35, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.L.; Huang, S.Y.; Hsu, Y.C.; Chin, T.H.; Soong, Y.K. The serum level of irisin, but not asprosin, is abnormal in polycystic ovary syndrome patients. Sci. Rep. 2019, 9, 6447. [Google Scholar] [CrossRef] [PubMed]
- Baykus, Y.; Yavuzkir, S.; Ustebay, S.; Ugur, K.; Deniz, R.; Aydin, S. Asprosin in umbilical cord of newborns and maternal blood of gestational diabetes, preeclampsia, severe preeclampsia, intrauterine growth retardation and macrosemic fetus. Peptides 2019, 120, 170132. [Google Scholar] [CrossRef]
- Zhong, L.; Long, Y.; Wang, S.; Lian, R.; Deng, L.; Ye, Z.; Wang, Z.; Liu, B. Continuous elevation of plasma asprosin in pregnant women complicated with gestational diabetes mellitus: A nested case-control study. Placenta 2020, 93, 17–22. [Google Scholar] [CrossRef]
- Hutt, S.; Mihaies, D.; Karteris, E.; Michael, A.; Payne, A.M.; Chatterjee, J. Statistical Meta-Analysis of Risk Factors for Endometrial Cancer and Development of a Risk Prediction Model Using an Artificial Neural Network Algorithm. Cancers 2021, 13, 3689. [Google Scholar] [CrossRef]
- Kerslake, R.; Hall, M.; Vagnarelli, P.; Jeyaneethi, J.; Randeva, H.S.; Pados, G.; Kyrou, I.; Karteris, E. A pancancer overview of FBN1, asprosin and its cognate receptor OR4M1 with detailed expression profiling in ovarian cancer. Oncol. Lett. 2021, 22, 650. [Google Scholar] [CrossRef]
- Kerslake, R.; Sisu, C.; Panfilov, S.; Hall, M.; Khan, N.; Jeyaneethi, J.; Randeva, H.; Kyrou, I.; Karteris, E. Differential Regulation of Genes by the Glucogenic Hormone Asprosin in Ovarian Cancer. J. Clin. Med. 2022, 11, 5942. [Google Scholar] [CrossRef] [PubMed]
- Mirzaoglu, M.; Yavuzkir, S.; Mirzaoglu, C.; Yurt, N.; Dagli, A.F.; Yildirim, S.O.; Sahin, I.; Aydin, S. Use of asprosin and subfatin for differential diagnosis of serous ovarian tumors. Biotech. Histochem. 2023, 98, 140–146. [Google Scholar] [CrossRef]
- Nam, H.; Hong, S.-S.; Jung, K.H.; Kang, S.; Park, M.S.; Kang, S.; Kim, H.S.; Mai, V.-H.; Kim, J.; Lee, H.; et al. A Serum Marker for Early Pancreatic Cancer with a Possible Link to Diabetes. JNCI J. Natl. Cancer Inst. 2022, 114, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Uzun, M.; Ilhan, Y.S.; Bozdag, A.; Yilmaz, M.; Artas, G.; Kuloglu, T. Asprosin, irisin, and meteorin-like protein immunoreactivity in different stages of colorectal adenocarcinoma. Pathol. Res. Pract. 2023, 245, 154432. [Google Scholar] [CrossRef] [PubMed]
- Mishra, I.; Xie, W.R.; Bournat, J.C.; He, Y.; Wang, C.; Silva, E.S.; Liu, H.; Ku, Z.; Chen, Y.; Erokwu, B.O.; et al. Protein tyrosine phosphatase receptor δ serves as the orexigenic asprosin receptor. Cell Metab. 2022, 34, 549–563.e8. [Google Scholar] [CrossRef] [PubMed]
- Ou, C.; Peng, Q.; Zeng, C. An integrative prognostic and immune analysis of PTPRD in cancer. Math. Biosci. Eng. 2022, 19, 5361–5379. [Google Scholar] [CrossRef] [PubMed]
- Tomita, H.; Cornejo, F.; Aranda-Pino, B.; Woodard, C.L.; Rioseco, C.C.; Neel, B.G.; Alvarez, A.R.; Kaplan, D.R.; Miller, F.D.; Cancino, G.I. The Protein Tyrosine Phosphatase Receptor Delta Regulates Developmental Neurogenesis. Cell Rep. 2020, 30, 215–228.e5. [Google Scholar] [CrossRef]
- Nunes-Xavier, C.E.; Zaldumbide, L.; Mosteiro, L.; López-Almaraz, R.; de Andoin, N.G.; Aguirre, P.; Emaldi, M.; Torices, L.; López, J.I.; Pulido, R. Protein Tyrosine Phosphatases in Neuroblastoma: Emerging Roles as Biomarkers and Therapeutic Targets. Front. Cell Dev. Biol. 2021, 9, 811297. [Google Scholar] [CrossRef]
- Uetani, N.; Kato, K.; Ogura, H.; Mizuno, K.; Kawano, K.; Mikoshiba, K.; Yakura, H.; Asano, M.; Iwakura, Y. Impaired learning with enhanced hippocampal long-term potentiation in PTPδ-deficient mice. EMBO J. 2000, 19, 2775. [Google Scholar] [CrossRef]
- Painter, J.N.; O’Mara, T.A.; Morris, A.P.; Cheng, T.H.T.; Gorman, M.; Martin, L.; Hodson, S.; Jones, A.; Martin, N.G.; Gordon, S.; et al. Genetic overlap between endometriosis and endometrial cancer: Evidence from cross-disease genetic correlation and GWAS meta-analyses. Cancer Med. 2018, 7, 1978–1987. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Traglia, M.; Croen, L.A.; Lyall, K.; Windham, G.C.; Kharrazi, M.; DeLorenze, G.N.; Torres, A.R.; Weiss, L.A. Independent Maternal and Fetal Genetic Effects on Midgestational Circulating Levels of Environmental Pollutants. G3 Genes Genomes Genet. 2017, 7, 1287–1299. [Google Scholar] [CrossRef]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 6 December 2023).
- Brain Map—Brain-Map.org. Available online: https://portal.brain-map.org/ (accessed on 6 December 2023).
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- GEPIA 2. Available online: http://gepia2.cancer-pku.cn/#index (accessed on 6 December 2023).
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Single Cell Portal. Available online: https://singlecell.broadinstitute.org/single_cell (accessed on 6 December 2023).
- Saravi, S.; Katsuta, E.; Jeyaneethi, J.; Amin, H.A.; Kaspar, M.; Takabe, K.; Pados, G.; Drenos, F.; Hall, M.; Karteris, E. H2A Histone Family Member X (H2AX) Is Upregulated in Ovarian Cancer and Demonstrates Utility as a Prognostic Biomarker in Terms of Overall Survival. J. Clin. Med. 2020, 9, 2844. [Google Scholar] [CrossRef] [PubMed]
- Vestergaard, A.L.; Knudsen, U.B.; Munk, T.; Rosbach, H.; Martensen, P.M. Transcriptional expression of type-I interferon response genes and stability of housekeeping genes in the human endometrium and endometriosis. Mol. Hum. Reprod. 2011, 17, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Hawrylycz, M.J.; Lein, E.S.; Guillozet-Bongaarts, A.L.; Shen, E.H.; Ng, L.; Miller, J.A.; Van De Lagemaat, L.N.; Smith, K.A.; Ebbert, A.; Riley, Z.L.; et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 2012, 489, 391–399. [Google Scholar] [CrossRef]
- Shabir, K.; Gharanei, S.; Orton, S.; Patel, V.; Chauhan, P.; Karteris, E.; Randeva, H.S.; Brown, J.E.; Kyrou, I. Asprosin Exerts Pro-Inflammatory Effects in THP-1 Macrophages Mediated via the Toll-like Receptor 4 (TLR4) Pathway. Int. J. Mol. Sci. 2023, 24, 227. [Google Scholar] [CrossRef] [PubMed]
- Hossain, A.; Islam, S.M.S.; Quinn, J.M.; Huq, F.; Moni, M.A. Machine learning and bioinformatics models to identify gene expression patterns of ovarian cancer associated with disease progression and mortality. J. Biomed. Inform. 2019, 100, 103313. [Google Scholar] [CrossRef]
- Khanlarkhani, N.; Azizi, E.; Amidi, F.; Khodarahmian, M.; Salehi, E.; Pazhohan, A.; Farhood, B.; Mortezae, K.; Goradel, N.H.; Nashtaei, M.S. Metabolic risk factors of ovarian cancer: A review. Jbra Assist. Reprod. 2022, 26, 335–347. [Google Scholar] [CrossRef]
- Rahman, F.; Mahmud, P.; Karim, R.; Hossain, T.; Islam, F. Determination of novel biomarkers and pathways shared by colorectal cancer and endometrial cancer via comprehensive bioinformatics analysis. Inform. Med. Unlocked 2020, 20, 100376. [Google Scholar] [CrossRef]
- Daley-Brown, D.; Harbuzariu, A.; Kurian, A.A.; Oprea-Ilies, G.; Gonzalez-Perez, R.R. Leptin-induced Notch and IL-1 signaling crosstalk in endometrial adenocarcinoma is associated with invasiveness and chemoresistance. World J. Clin. Oncol. 2019, 10, 222–233. [Google Scholar] [CrossRef]
- Dong, X.; Liao, Z.; Gritsch, D.; Hadzhiev, Y.; Bai, Y.; Locascio, J.J.; Guennewig, B.; Liu, G.; Blauwendraat, C.; Wang, T.; et al. Enhancers active in dopamine neurons are a primary link between genetic variation and neuropsychiatric disease. Nat. Neurosci. 2018, 21, 1482–1492. [Google Scholar] [CrossRef]
- Grochans, S.; Cybulska, A.M.; Simińska, D.; Korbecki, J.; Kojder, K.; Chlubek, D.; Baranowska-Bosiacka, I. Epidemiology of Glioblastoma Multiforme—Literature Review. Cancers 2022, 14, 2412. [Google Scholar] [CrossRef]
- Ortiz, B.; Fabius, A.W.M.; Wu, W.H.; Pedraza, A.; Brennan, C.W.; Schultz, N.; Pitter, K.L.; Bromberg, J.F.; Huse, J.T.; Holland, E.C.; et al. Loss of the tyrosine phosphatase PTPRD leads to aberrant STAT3 activation and promotes gliomagenesis. Proc. Natl. Acad. Sci. USA 2014, 111, 8149–8154. [Google Scholar] [CrossRef] [PubMed]
- Bralten, L.B.C.; Gravendeel, A.M.; Kloosterhof, N.K.; Sacchetti, A.; Vrijenhoek, T.; A Veltman, J.; Bent, M.J.v.D.; Kros, J.M.; Hoogenraad, C.C.; Smitt, P.A.E.S.; et al. The CASPR2 cell adhesion molecule functions as a tumor suppressor gene in glioma. Oncogene 2010, 29, 6138–6148. [Google Scholar] [CrossRef] [PubMed]
- Bae, W.J.; Ahn, J.M.; Byeon, H.E.; Kim, S.; Lee, D. PTPRD-inactivation-induced CXCL8 promotes angiogenesis and metastasis in gastric cancer and is inhibited by metformin. J. Exp. Clin. Cancer Res. 2019, 38, 484. [Google Scholar] [CrossRef]
- Funato, K.; Yamazumi, Y.; Oda, T.; Akiyama, T. Tyrosine phosphatase PTPRD suppresses colon cancer cell migration in coordination with CD44. Exp. Ther. Med. 2011, 2, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Kohno, T.; Otsuka, A.; Girard, L.; Sato, M.; Iwakawa, R.; Ogiwara, H.; Sanchez-Cespedes, M.; Minna, J.D.; Yokota, J. A catalog of genes homozygously deleted in human lung cancer and the candidacy of PTPRD as a tumor suppressor gene. Genes, Chromosom. Cancer 2010, 49, 342–352. [Google Scholar] [CrossRef]
- Chen, C.-L.; Hsieh, F.-C.; Lieblein, J.C.; Brown, J.; Chan, C.; A Wallace, J.; Cheng, G.; Hall, B.M.; Lin, J. Stat3 activation in human endometrial and cervical cancers. Br. J. Cancer 2007, 96, 591–599. [Google Scholar] [CrossRef]
- PTPRD Gene—Somatic Mutations in Cancer. Available online: https://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=PTPRD#gene-view%20PTPRD%20Gene%20-%20Somatic%20Mutations%20in%20Cancer%20(sanger.ac.uk) (accessed on 6 December 2023).
- Parrillo, L.; Spinelli, R.; Longo, M.; Desiderio, A.; Mirra, P.; Nigro, C.; Fiory, F.; Hedjazifar, S.; Mutarelli, M.; Carissimo, A.; et al. Altered PTPRD DNA methylation associates with restricted adipogenesis in healthy first-degree relatives of Type 2 diabetes subjects. Epigenomics 2020, 12, 873–888. [Google Scholar] [CrossRef]
- Chen, T.; Xu, J.; Liu, G.; Liu, H.; Chen, M.; Qin, Y.; Wu, W.; Xia, Y.; Ji, C.; Guo, X.; et al. Genetic variants in PTPRD and risk of gestational diabetes mellitus. Oncotarget 2016, 7, 76101–76107. [Google Scholar] [CrossRef][Green Version]
- Kang, Y.; Huang, H.; Li, H.; Sun, W.; Zhang, C. Functional genetic variants in the 3’UTR of PTPRD associated with the risk of gestational diabetes mellitus. Exp. Ther. Med. 2021, 21, 562. [Google Scholar] [CrossRef]
- Hong, X.; Surkan, P.J.; Zhang, B.; Keiser, A.; Ji, Y.; Ji, H.; Burd, I.; Bustamante-Helfrich, B.; Ogunwole, S.M.; Tang, W.-Y.; et al. Genome-wide association study identifies a novel maternal gene × stress interaction associated with spontaneous preterm birth. Pediatr. Res. 2020, 89, 1549–1556. [Google Scholar] [CrossRef]
- Kodavanti, P.R.S.; Loganathan, B.G. Organohalogen Pollutants and Human Health. In International Encyclopedia of Public Health; Elsevier: Amsterdam, The Netherlands, 2017; pp. 359–366. [Google Scholar]
- NLS Mapper. Available online: https://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi (accessed on 6 December 2023).
- NLStradamus. Available online: http://www.moseslab.csb.utoronto.ca/NLStradamus/ (accessed on 6 December 2023).
- Ba, A.N.N.; Pogoutse, A.; Provart, N.; Moses, A.M. NLStradamus: A simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinform. 2009, 10, 202. [Google Scholar]
- Costa, M.A. The endocrine function of human placenta: An overview. Reprod. Biomed. Online 2016, 32, 14–43. [Google Scholar] [CrossRef]
- Malassiné, A.; Cronier, L. Hormones and human trophoblast differentiation: A review. Endocrine 2002, 19, 3–11. [Google Scholar] [CrossRef]
- Rebut-Bonneton, C.; Boutemy-Roulier, S.; Evain-Brion, D. Modulation of pp60c-src activity and cellular localization during differentiation of human trophoblast cells in culture. J. Cell Sci. 1993, 105, 629–636. [Google Scholar] [CrossRef]
- Greca, S.-C.d.A.; Kyrou, I.; Pink, R.; Randeva, H.; Grammatopoulos, D.; Silva, E.; Karteris, E. Involvement of the Endocrine-Disrupting Chemical Bisphenol A (BPA) in Human Placentation. J. Clin. Med. 2020, 9, 405. [Google Scholar] [CrossRef]
- Fogarty, N.; Burton, G.; Ferguson-Smith, A. Different epigenetic states define syncytiotrophoblast and cytotrophoblast nuclei in the trophoblast of the human placenta. Placenta 2015, 36, 796–802. [Google Scholar] [CrossRef]
- Xu, J.; Zhong, Y.; Yin, H.; Linneman, J.; Luo, Y.; Xia, S.; Xia, Q.; Yang, L.; Huang, X.; Kang, K.; et al. Methylation-mediated silencing of PTPRD induces pulmonary hypertension by promoting pulmonary arterial smooth muscle cell migration via the PDGFRB/PLCγ1 axis. J. Hypertens. 2022, 40, 1795–1807. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Spoelstra, N.S.; Jean, A.; Howe, E.; Torkko, K.C.; Clark, H.R.; Darling, D.S.; Shroyer, K.R.; Horwitz, K.B.; Broaddus, R.R.; et al. ZEB1 expression in type I vs type II endometrial cancers: A marker of aggressive disease. Mod. Pathol. 2008, 21, 912–923. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, L.; Zhou, J.; Pan, J.; Qian, W.; Fu, J.; Zhang, G.; Zhu, Y.; Liu, C.; Wang, C.; et al. Reduced Expression of PTPRD Correlates with Poor Prognosis in Gastric Adenocarcinoma. PLoS ONE 2014, 9, e113754. [Google Scholar] [CrossRef]
- Wu, L.; Gao, L.; Kong, D.; Xue, H. Loss of Tyrosine Phosphatase Delta Promotes Gastric Cancer Progression via Signal Transducer and Activator of Transcription 3 Pathways. Dig. Dis. Sci. 2019, 64, 3164–3172. [Google Scholar] [CrossRef]



| Gene | Forward Primer Sequence | Reverse Primer Sequence |
|---|---|---|
| YWHAZ | AGACGGAAGGTGCTGAGAAA | GAAGCATTGGGGATCAAGAA |
| PTPRD | CAGGCGGAAGCGTTAATATCA | TTGGCATATCATCTTCAGGTGTC |
| GO Term | Description |
|---|---|
| GO:0099545 | Trans-synaptic signalling by trans-synaptic complex |
| GO:0099537 | Trans-synaptic signalling |
| GO:0099560 | Synaptic membrane adhesion |
| GO:0050808 | Synapse organisation |
| GO:0007416 | Synapse assembly |
| GO:0051963 | Regulation of synapse assembly |
| GO:1905606 | Regulation of presynaptic assembly |
| GO:0099151 | Regulation of postsynaptic density assembly |
| GO:0099175 | Regulation of postsynaptic organisation |
| GO:0051128 | Regulation of cellular component organisation |
| GO:0044087 | Regulation of cellular component biogenesis |
| GO:0065008 | Regulation of biological quality |
| GO:0097105 | Presynaptic membrane assembly |
| GO:0099172 | Presynaptic organisation |
| GO:0051965 | Positive regulation of synapse assembly |
| GO:0030182 | Neuron differentiation |
| GO:0098742 | Cell–cell adhesion via plasma membrane adhesion molecules |
| GO:0098609 | Cell–cell adhesion |
| GO:0007155 | Cell adhesion |
| GO Term | Description |
|---|---|
| GO:0061809 | NAD + nucleotidase, cyclic ADP-ribose generation |
| GO:0050135 | NAD(P) + nucleosidase activity |
| GO:0004908 | Interleukin-1 receptor activity |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orton, S.; Karkia, R.; Mustafov, D.; Gharanei, S.; Braoudaki, M.; Filipe, A.; Panfilov, S.; Saravi, S.; Khan, N.; Kyrou, I.; et al. In Silico and In Vitro Mapping of Receptor-Type Protein Tyrosine Phosphatase Receptor Type D in Health and Disease: Implications for Asprosin Signalling in Endometrial Cancer and Neuroblastoma. Cancers 2024, 16, 582. https://doi.org/10.3390/cancers16030582
Orton S, Karkia R, Mustafov D, Gharanei S, Braoudaki M, Filipe A, Panfilov S, Saravi S, Khan N, Kyrou I, et al. In Silico and In Vitro Mapping of Receptor-Type Protein Tyrosine Phosphatase Receptor Type D in Health and Disease: Implications for Asprosin Signalling in Endometrial Cancer and Neuroblastoma. Cancers. 2024; 16(3):582. https://doi.org/10.3390/cancers16030582
Chicago/Turabian StyleOrton, Sophie, Rebecca Karkia, Denis Mustafov, Seley Gharanei, Maria Braoudaki, Alice Filipe, Suzana Panfilov, Sayeh Saravi, Nabeel Khan, Ioannis Kyrou, and et al. 2024. "In Silico and In Vitro Mapping of Receptor-Type Protein Tyrosine Phosphatase Receptor Type D in Health and Disease: Implications for Asprosin Signalling in Endometrial Cancer and Neuroblastoma" Cancers 16, no. 3: 582. https://doi.org/10.3390/cancers16030582
APA StyleOrton, S., Karkia, R., Mustafov, D., Gharanei, S., Braoudaki, M., Filipe, A., Panfilov, S., Saravi, S., Khan, N., Kyrou, I., Karteris, E., Chatterjee, J., & Randeva, H. S. (2024). In Silico and In Vitro Mapping of Receptor-Type Protein Tyrosine Phosphatase Receptor Type D in Health and Disease: Implications for Asprosin Signalling in Endometrial Cancer and Neuroblastoma. Cancers, 16(3), 582. https://doi.org/10.3390/cancers16030582










