Generation of a Realistic Synthetic Laryngeal Cancer Cohort for AI Applications
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Motivation for Synthetic Data Generation
- Privacy concerns: Real patient data often contains sensitive personal information that must be protected to ensure patient privacy;
- Limited availability: Real patient data is often difficult to obtain in large quantities, particularly for specific populations or conditions;
- High costs: Obtaining real patient data can be costly, particularly when it requires the use of special equipment or specialized personnel;
- Compliance of patients to approve of the utilization of their personal data.
1.2. Synthetic Patient Population Simulator
1.3. Clinical Use Case of Laryngeal Cancer
2. Materials and Methods
2.1. Generating Patient Datasets Using Synthea Engine
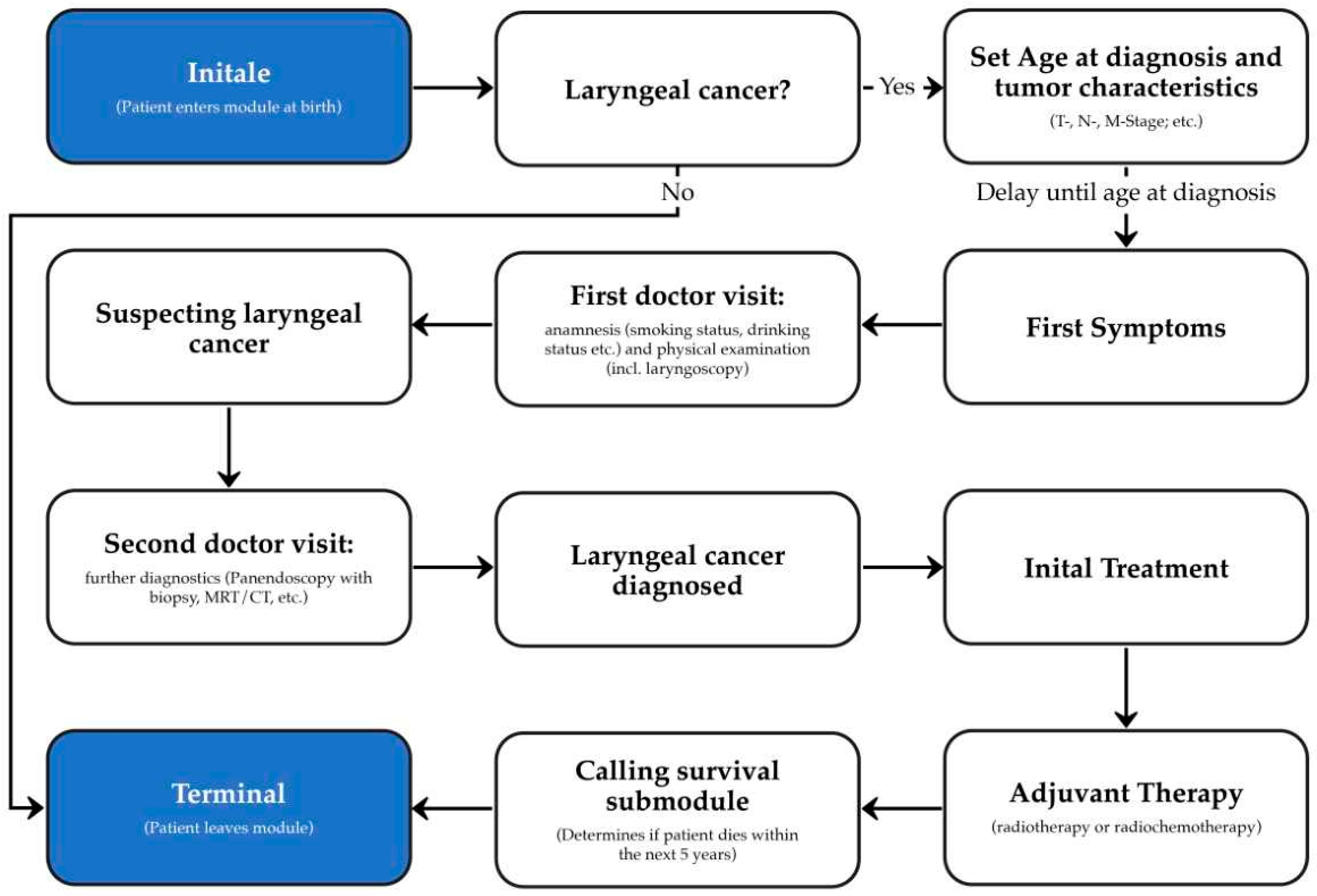
2.2. Development of the Laryngeal Cancer Synthea Module
2.3. Generation of Datasets
2.4. Validation of the Laryngeal Cancer Module
3. Results
3.1. Prevalence Approach: Dataset C32-1
3.2. Enriched Approach: Dataset C32-2
3.3. Cohort Approach: Dataset C32-3
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tucker, A.; Wang, Z.; Rotalinti, Y.; Myles, P. Generating High-Fidelity Synthetic Patient Data for Assessing Machine Learning Healthcare Software. NPJ Digit. Med. 2020, 3, 147. [Google Scholar] [CrossRef]
- Chen, A.; Chen, D.O. Simulation of a Machine Learning Enabled Learning Health System for Risk Prediction Using Synthetic Patient Data. Sci. Rep. 2022, 12, 17917. [Google Scholar] [CrossRef]
- Weldon, J.; Ward, T.; Brophy, E. Generation of Synthetic Electronic Health Records Using a Federated GAN. arXiv 2021, arXiv:2109.02543. [Google Scholar]
- Ive, J.; Viani, N.; Kam, J.; Yin, L.; Verma, S.; Puntis, S.; Cardinal, R.N.; Roberts, A.; Stewart, R.; Velupillai, S. Generation and Evaluation of Artificial Mental Health Records for Natural Language Processing. NPJ Digit. Med. 2020, 3, 69. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Chen, H.; Loew, M.; Ko, H. COVID-19 CT Image Synthesis With a Conditional Generative Adversarial Network. IEEE J. Biomed. Health Inform. 2021, 25, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Das, H.P.; Tran, R.; Singh, J.; Yue, X.; Tison, G.; Sangiovanni-Vincentelli, A.; Spanos, C.J. Conditional Synthetic Data Generation for Robust Machine Learning Applications with Limited Pandemic Data. arXiv 2021, arXiv:2109.06486. [Google Scholar]
- Levine, A.B.; Peng, J.; Farnell, D.; Nursey, M.; Wang, Y.; Naso, J.R.; Ren, H.; Farahani, H.; Chen, C.; Chiu, D.; et al. Synthesis of Diagnostic Quality Cancer Pathology Images by Generative Adversarial Networks. J. Pathol. 2020, 252, 178–188. [Google Scholar] [CrossRef]
- Synthea (TM). Synthetic Patient Population Simulator. Available online: https://github.com/synthetichealth/synthea (accessed on 26 November 2022).
- Walonoski, J.; Kramer, M.; Nichols, J.; Quina, A.; Moesel, C.; Hall, D.; Duffett, C.; Dube, K.; Gallagher, T.; McLachlan, S. Synthea: An Approach, Method, and Software Mechanism for Generating Synthetic Patients and the Synthetic Electronic Health Care Record. J. Am. Med. Inform. Assoc. JAMIA 2018, 25, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Bala, S.; Keniston, A.; Burden, M. Patient Perception of Plain-Language Medical Notes Generated Using Artificial Intelligence Software: Pilot Mixed-Methods Study. JMIR Form. Res. 2020, 4, e16670. [Google Scholar] [CrossRef] [PubMed]
- Scalfani, R.; Shamsnaz, V.B. Health Insurance and Its Impact on the Survival Rates of Breast Cancer Patients in Synthea. Bachelor’s Thesis, Worcester Polytechnic Institute, Worcester, MA, USA, 2020. [Google Scholar]
- Murtaza, H.; Ahmed, M.; Khan, N.F.; Murtaza, G.; Zafar, S.; Bano, A. Synthetic Data Generation: State of the Art in Health Care Domain. Comput. Sci. Rev. 2023, 48, 100546. [Google Scholar] [CrossRef]
- Hernandez, M.; Epelde, G.; Alberdi, A.; Cilla, R.; Rankin, D. Synthetic Data Generation for Tabular Health Records: A Systematic Review. Neurocomputing 2022, 493, 28–45. [Google Scholar] [CrossRef]
- Buczak, A.L.; Babin, S.; Moniz, L. Data-Driven Approach for Creating Synthetic Electronic Medical Records. BMC Med. Inform. Decis. Mak. 2010, 10, 59. [Google Scholar] [CrossRef]
- Choi, E.; Biswal, S.; Bradley, M.; Duke, J.; Stewart, W.F.; Sun, J. Generating Multi-Label Discrete Patient Records Using Generative Adversarial Networks. arXiv 2018, arXiv:1703.06490. [Google Scholar]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Burki, T.K. Symptoms Associated with Risk of Laryngeal Cancer. Lancet Oncol. 2019, 20, e135. [Google Scholar] [CrossRef] [PubMed]
- Robert Koch-Institute. Cancer in Germany 2017/2018, 13th ed.; Association of Population-Based Cancer Registries in Germany: Berlin, Germany, 2022; ISBN 978-3-89606-315-1. [Google Scholar]
- Altieri, A.; Bosetti, C.; Talamini, R.; Gallus, S.; Franceschi, S.; Levi, F.; Dal Maso, L.; Negri, E.; La Vecchia, C. Cessation of Smoking and Drinking and the Risk of Laryngeal Cancer. Br. J. Cancer 2002, 87, 1227–1229. [Google Scholar] [CrossRef] [PubMed]
- Maier, H.; Gewelke, U.; Dietz, A.; Heller, W.D. Risk Factors of Cancer of the Larynx: Results of the Heidelberg Case-Control Study. Otolaryngol. Head Neck Surg. Off. J. Am. Acad. Otolaryngol. Head Neck Surg. 1992, 107, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Leitlinienprogramm Onkologie (Deutsche Krebsgesellschaft). Deutsche Krebshilfe, AWMF: Diagnostik, Therapie und Nachsorge Des Larynxkarzinoms, Langversion 1.1; 2019; AWMF-Registernummer: 017/076OL. 2019. Available online: http://www.leitlinienprogramm-onkologie.de/leitlinien/larynxkarzinom%20cancer (accessed on 19 September 2023).
- Krishnatreya, M.; Rahman, T.; Kataki, A.C.; Sharma, J.D.; Nandy, P.; Baishya, N. Pre-Treatment Performance Status and Stage at Diagnosis in Patients with Head and Neck Cancers. Asian Pac. J. Cancer Prev. APJCP 2014, 15, 8479–8482. [Google Scholar] [CrossRef] [PubMed]
- Patrick, E. Die Behandlung von Larynx-/Hypopharynxkarzinomen Und Die Laryngektomie Im Wandel Der Zeit; University Ulm: Ulm, Germany, 2018. [Google Scholar]
- Castellsagué, X.; Alemany, L.; Quer, M.; Halec, G.; Quirós, B.; Tous, S.; Clavero, O.; Alòs, L.; Biegner, T.; Szafarowski, T.; et al. HPV Involvement in Head and Neck Cancers: Comprehensive Assessment of Biomarkers in 3680 Patients. J. Natl. Cancer Inst. 2016, 108, djv403. [Google Scholar] [CrossRef] [PubMed]
- Sannino, N.J.B.; Mehlum, C.S.; Grøntved, Å.M.; Kjaergaard, T.; Kiss, K.; Godballe, C.; Tvedskov, J.F. Incidence and Malignant Transformation of Glottic Precursor Lesions in Denmark. Acta Oncol. Stockh. Swed. 2020, 59, 596–602. [Google Scholar] [CrossRef]
- Shephard, E.A.; Parkinson, M.A.; Hamilton, W.T. Recognising Laryngeal Cancer in Primary Care: A Large Case-Control Study Using Electronic Records. Br. J. Gen. Pract. J. R. Coll. Gen. Pract. 2019, 69, e127–e133. [Google Scholar] [CrossRef]
- Brierley, J.D.; Gospodarowicz, M.K.; Wittekind, C. (Eds.) TNM Classification of Malignant Tumours, 8th ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2016; ISBN 978-1-119-26357-9. [Google Scholar]
- Ketterer, M.C.; Lemus Moraga, L.A.; Beitinger, U.; Pfeiffer, J.; Knopf, A.; Becker, C. Surgical Nodal Management in Hypopharyngeal and Laryngeal Cancer. Eur. Arch. Oto-Rhino-Laryngol. Off. J. Eur. Fed. Oto-Rhino-Laryngol. Soc. EUFOS Affil. Ger. Soc. Oto-Rhino-Laryngol.-Head Neck Surg. 2020, 277, 1481–1489. [Google Scholar] [CrossRef] [PubMed]
- Spector, G.J. Distant Metastases from Laryngeal and Hypopharyngeal Cancer. ORL J. Oto-Rhino-Laryngol. Its Relat. Spec. 2001, 63, 224–228. [Google Scholar] [CrossRef] [PubMed]
- Laryngeal Cancer Module Git Page. Available online: https://git.iccas.de/synthea/laryngeal-cancer (accessed on 29 January 2024).
- German Center for Cancer Registry Data (ZfKD). Interactive Database—Laryngeal Cancer. Available online: https://www.krebsdaten.de/Krebs/DE/Datenbankabfrage/datenbankabfrage_stufe1_node.html (accessed on 20 May 2023).
- Cancer Registry Baden-Wüttenberg. Interactive Database—Laryngeal Cancer. Available online: https://www.krebsregister-bw.de/CARESS/index.html#/diagnoses/overview (accessed on 27 July 2023).
- Cancer Registry Lower Saxony. Interactive Database—Laryngeal Cancer. Available online: https://www.krebsregister-niedersachsen.de/Online-Jahresbericht/#/diagnoses/overview (accessed on 19 September 2023).
- Cancer Registry North Rhine Westphalia. Interactive Database—Laryngeal Cancer. Available online: https://www.landeskrebsregister.nrw/online-jahresbericht (accessed on 19 September 2023).
- Cancer Registry Munich. Laryngeal Cancer—Survival. Available online: https://www.tumorregister-muenchen.de/facts/surv/sC32__G-ICD-10-C32-Larynxkarzinom-Survival.pdf (accessed on 19 September 2023).
- Markou, K.; Christoforidou, A.; Karasmanis, I.; Tsiropoulos, G.; Triaridis, S.; Constantinidis, I.; Vital, V.; Nikolaou, A. Laryngeal Cancer: Epidemiological Data from Νorthern Greece and Review of the Literature. Hippokratia 2013, 17, 313–318. [Google Scholar] [PubMed]
- Katalinic, A.; Halber, M.; Meyer, M.; Pflüger, M.; Eberle, A.; Nennecke, A.; Kim-Wanner, S.-Z.; Hartz, T.; Weitmann, K.; Stang, A.; et al. Population-Based Clinical Cancer Registration in Germany. Cancers 2023, 15, 3934. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer. Globocan 2020—Laryngeal Cancer. Available online: https://gco.iarc.fr/today/data/factsheets/cancers/14-Larynx-fact-sheet.pdf (accessed on 20 May 2023).

| Variable | Description | Source |
|---|---|---|
| Age, gender | Age distribution of laryngeal cancer patients in 5-year groups reflecting gender- and age-specific probability of disease, age <40 years was excluded | Cancer Registry SH |
| Performance status (ECOG) | Frequency of ECOG score depending on UICC (0–5) | Manigreeva Krishnatreya, 2014 [22] |
| Alcohol | Frequency of alcohol consumption in laryngeal cancer patients (no, occasionally, regular, excessive and former excessive alcohol consumption) | Engel, 2018 [23] |
| Smoking | Prevalence of smokers, non-smokers and former smokers in laryngeal cancer patients (non-smoker, former smoker, active smoker) | Engel, 2018 [23] |
| Human papilloma virus (HPV) | Frequency of HPV-associated laryngeal cancer (Yes/No) | Castellsagué, 2016 [24] |
| Glottic Precursor Lesion | Occurrence and differentiation of laryngeal precursor lesions prior to development of laryngeal cancer (carcinoma in situ, NOS, mild, moderat, severe) | Sanninoa, 2020 [25] |
| Symptoms | Frequency of symptoms (hoarseness, otalgia, dysphagia, insomnia, mouth symptoms, chest infection, sore throat and dysphonia) that onset up to one year before diagnosis of laryngeal cancer (Yes/No) | Shephard, 2019 [26] |
| T staging | Distribution of tumor size (T category of TNM) depending on gender (T1–T4, including T1a, T1b, T4a and T4b) | Cancer Registry SH |
| N staging | Distribution of regional lymph node involvement (N category of TNM) depending on gender and T staging (N0–N3) | Cancer Registry SH |
| M staging | Distribution of distant metastasis (M category, TNM) depending on gender, T and N stage (M0, M1) | Cancer Registry SH |
| UICC | Condensed tumor staging, compiled form T, N, M according UICC (I–IV) | Brierly, 2016 [27] |
| Grading | Frequency of levels of histopathological differentiation in laryngeal cancer cells (G1–G3) | Ketterer, 2020 [28] |
| Localisation of primary tumor | Distribution of primary cancer sites (glottis, subglottis, supraglottis) | German S3 guideline for laryngeal cancer [21] |
| Localisation of metastasis | distribution of the localisation of laryngeal cancer metastasis (lung, bone, skin, central nervous system, other organs) | Spector, 2001 [29] |
| Initial treatment | Frequency of initial treatments depending on TNM and UICC staging (open partial laryngectomy, transoral partial laryngectomy, laryngectomy, radiotherapy, chemotherapy, radio-chemotherapy) | Engel, 2018 [23], German S3 guideline for laryngeal cancer [21] |
| R-status | Frequency of resection status in surgically treated (R0–R2) | Engel, 2018 [23] |
| Adjuvant therapy | Frequency for a postoperative treatment (postoperative radiotherapy, postoperative radio-chemotherapy, no postoperative therapy) | German S3 guideline for laryngeal cancer [21] |
| Neck dissection | Operation including regional lymph node (Yes (curative or elective)/No) | German S3 guideline for laryngeal cancer [21] |
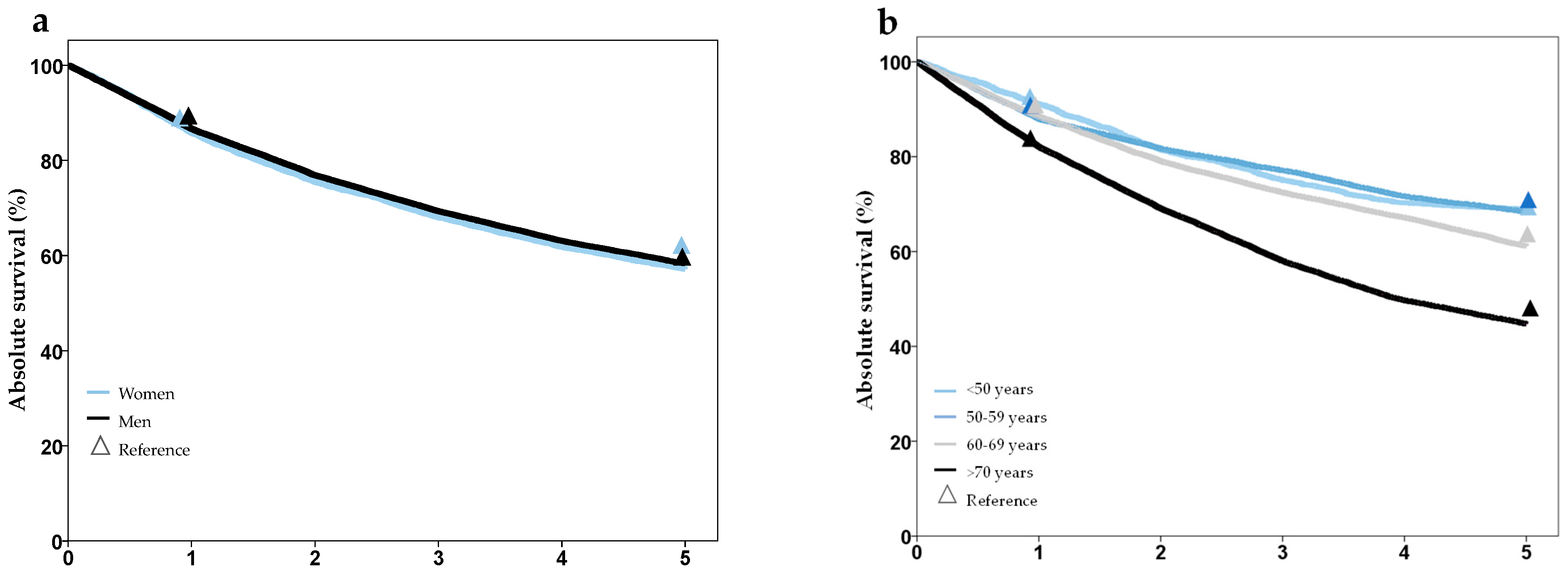
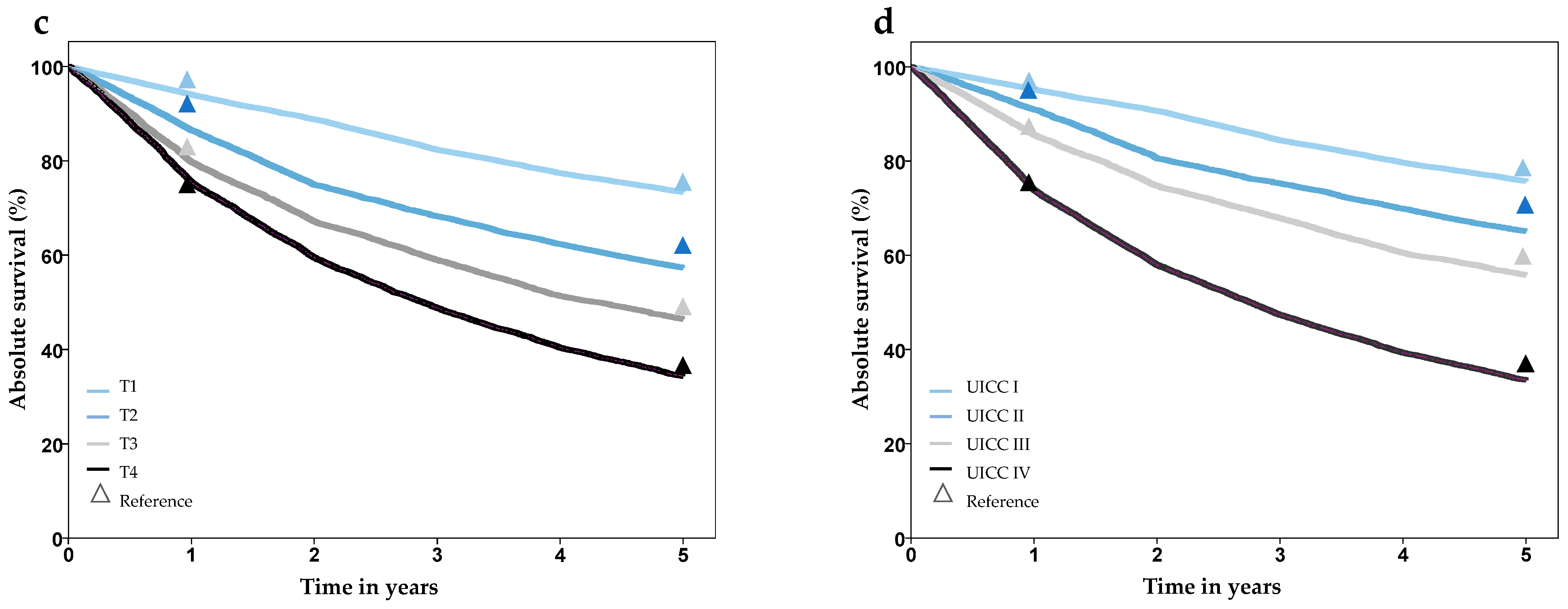
| Survival | 1-, 2-, 3-, 4-, 5- year overall survival (%) stratified by age, gender and T, N, M stage | Cancer Registry SH |
| Dataset C32-1 | Dataset C32-2 | Dataset C32-3 | |
|---|---|---|---|
| Description | Population-based prevalence | Enriched prevalence | Cohort approach (age and gender matched) |
| Patients synthetized | alive: 2,000,000 (total: 2,061,998) | alive: 249,999 (total: 366,429) | alive: 27,095 (total: 50,007) |
| … with laryngeal cancer (ICD10: C32) | alive: 879 (total: 1394) | alive: 98,084 (total: 227,643) | alive: 27,095 (total: 50,007) |
| Gender (in patients with laryngeal cancer) women/men | 173/1221 | 113,316/114,327 | 7354/42,653 |
| Age at diagnosis (Mean, SD) | 60.9 (±10.5) | 63.1 (±10.2) | 65.5 Years (±10.6) |
| Total | Women | Men | |
|---|---|---|---|
| Synthea data (Average of the last five simulated years) | 3.63 | 0.83 | 6.51 |
| ZfKD data (average from 1999–2019) | 4.68 | 1.2 | 8.3 |
| Deviation% | −22.3% | −30.8% | −21.4% |
| Synthea | CR SH | |||||
|---|---|---|---|---|---|---|
| Variable | Female | Male | Female | Male | p-Value | |
| Gender (%, row) | 14.7 | 85.3 | 15.8 | 84.2 | n. s. | |
| Age (%) | 40–49 | 8.7 | 6.1 | 9.4 | 6.6 | n. s. |
| 50–59 | 24.0 | 23.7 | 23.6 | 23.1 | ||
| 60–69 | 32.9 | 35.1 | 32.8 | 35.0 | ||
| 70–79 | 22.1 | 25.0 | 21.9 | 25.2 | ||
| 80+ | 12.3 | 10.1 | 12.3 | 10.1 | ||
| T-Staging (%) | T1 | 36.4 | 43.2 | 43.2 | 43.3 | n. s. |
| T2 | 23.5 | 21.0 | 21.0 | 21.0 | ||
| T3 | 23.9 | 18.9 | 18.9 | 18.5 | ||
| T4 | 16.2 | 16.9 | 16.9 | 17.1 | ||
| N-Staging (%) | N0 | 67.3 | 73.9 | 66.6 | 72.0 | n. s. |
| N1 | 7.9 | 7.0 | 9.2 | 7.6 | ||
| N2 | 22.0 | 16.5 | 22.4 | 17.7 | ||
| N3 | 2.8 | 2.6 | 2.9 | 2.7 | ||
| M-Staging (%) | M0 | 94.6 | 95.6 | 93.9 | 95.6 | n. s. |
| M1 | 5.4 | 4.4 | 6.1 | 4.4 | ||
| UICC Staging (%) | I | 34.9 | 40.5 | 35.1 | 41.4 | n. s. |
| II | 15.4 | 14.5 | 15.5 | 14.7 | ||
| II | 15.1 | 15.1 | 15.3 | 15.3 | ||
| IV | 34.6 | 29.9 | 34.1 | 34.1 | ||
| Grading (%) | 1 | 7.3 | 6.7 | 8.3 | 8.7 | n. s. |
| 2 | 68.9 | 69.6 | 62.5 | 67.8 | ||
| 3 | 23.0 | 23.7 | 29.2 | 23.5 | ||
| Survival (%) | 1-year | 85.3 | 86.5 | 85.7 | 86.9 | n. s. |
| 5-year | 57 | 58.1 | 63.6 | 58.9 | ||
| Survival 5-year (%) by UICC | I | 76.3 | 75.3 | 80.2 | 76.9 | n. s. |
| II | 67.4 | 64.0 | 76.2 | 67.9 | ||
| II | 58.0 | 56.0 | 67.4 | 58.9 | ||
| IV | 35.5 | 33.5 | 44.8 | 35.8 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Katalinic, M.; Schenk, M.; Franke, S.; Katalinic, A.; Neumuth, T.; Dietz, A.; Stoehr, M.; Gaebel, J. Generation of a Realistic Synthetic Laryngeal Cancer Cohort for AI Applications. Cancers 2024, 16, 639. https://doi.org/10.3390/cancers16030639
Katalinic M, Schenk M, Franke S, Katalinic A, Neumuth T, Dietz A, Stoehr M, Gaebel J. Generation of a Realistic Synthetic Laryngeal Cancer Cohort for AI Applications. Cancers. 2024; 16(3):639. https://doi.org/10.3390/cancers16030639
Chicago/Turabian StyleKatalinic, Mika, Martin Schenk, Stefan Franke, Alexander Katalinic, Thomas Neumuth, Andreas Dietz, Matthaeus Stoehr, and Jan Gaebel. 2024. "Generation of a Realistic Synthetic Laryngeal Cancer Cohort for AI Applications" Cancers 16, no. 3: 639. https://doi.org/10.3390/cancers16030639
APA StyleKatalinic, M., Schenk, M., Franke, S., Katalinic, A., Neumuth, T., Dietz, A., Stoehr, M., & Gaebel, J. (2024). Generation of a Realistic Synthetic Laryngeal Cancer Cohort for AI Applications. Cancers, 16(3), 639. https://doi.org/10.3390/cancers16030639






