The Influence of Telomere-Related Gene Variants, Serum Levels, and Relative Leukocyte Telomere Length in Pituitary Adenoma Occurrence and Recurrence
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Group
2.2. DNA Extraction
2.3. Genotyping
2.4. Relative Leukocyte Telomere Length Measurement
2.5. Serum Levels Measurement
2.6. Study Characteristics
2.7. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ajlan, A.M.; Harsh, G.R. Functional pituitary adenoma recurrence after surgical resection. World Neurosurg. 2013, 81, 494–496. [Google Scholar] [CrossRef] [PubMed]
- Gupta, T.; Chatterjee, A. Modern Radiation Therapy for Pituitary Adenoma: Review of Techniques and Outcomes. Neurol. India 2020, 68, S113–S122. [Google Scholar] [CrossRef] [PubMed]
- Møller, M.W.; Andersen, M.S.; Glintborg, D.; Pedersen, C.B.; Halle, B.; Kristensen, B.W.; Poulsen, F.R. Pituitary adenoma. Ugeskr. Laeger. 2019, 181, V05180331. [Google Scholar] [PubMed]
- Shammas, M.A. Telomeres, lifestyle, cancer, and aging. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Zhou, Z.; Wei, S.; Liu, Z.; Pooley, K.A.; Dunning, A.M.; Svenson, U.; Roos, G.; Hosgood, H.D.; Shen, M.; et al. Shortened Telomere Length Is Associated with Increased Risk of Cancer: A Meta-Analysis. PLoS ONE 2011, 6, e20466. [Google Scholar] [CrossRef] [PubMed]
- Mir, S.M.; Samavarchi Tehrani, S.; Goodarzi, G.; Jamalpoor, Z.; Asadi, J.; Khelghati, N.; Qujeq, D.; Maniati, M. Shelterin Complex at Telomeres: Implications in Ageing. Clin. Interv. Aging 2020, 15, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Bandaria, J.N.; Qin, P.; Berk, V.; Chu, S.; Yildiz, A. Shelterin protects chromosome ends by compacting telomeric chromatin. Cell 2016, 164, 735–746. [Google Scholar] [CrossRef]
- Ruis, P.; Boulton, S.J. The end protection problem—An unexpected twist in the tail. Genes Dev. 2021, 35, 1–21. [Google Scholar] [CrossRef]
- Rice, C.; Skordalakes, E. Structure and function of the telomeric CST complex. Comput. Struct. Biotechnol. J. 2016, 14, 161–167. [Google Scholar] [CrossRef]
- Martínez, P.; Blasco, M.A. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat. Rev. Cancer 2011, 11, 161–176. [Google Scholar] [CrossRef]
- Smogorzewska, A.; van Steensel, B.; Bianchi, A.; Oelmann, S.; Schaefer, M.R.; Schnapp, G.; de Lange, T. Control of Human Telomere Length by TRF1 and TRF2. Mol. Cell. Biol. 2000, 20, 1659–1668. [Google Scholar] [CrossRef]
- Kang, H.-W.; Wang, F.; Wei, Q.; Zhao, Y.-F.; Liu, M.; Li, X.; Tang, H. miR-20a promotes migration and invasion by regulating TNKS2 in human cervical cancer cells. FEBS Lett. 2012, 586, 897–904. [Google Scholar] [CrossRef]
- Mangino, M.; Hwang, S.-J.; Spector, T.D.; Hunt, S.C.; Kimura, M.; Fitzpatrick, A.L.; Christiansen, L.; Petersen, I.; Elbers, C.C.; Harris, T.; et al. Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Hum. Mol. Genet. 2012, 21, 5385–5394. [Google Scholar] [CrossRef]
- Amir, M.; Khan, P.; Queen, A.; Dohare, R.; Alajmi, M.F.; Hussain, A.; Islam, A.; Ahmad, F.; Hassan, I. Structural Features of Nucleoprotein CST/Shelterin Complex Involved in the Telomere Maintenance and Its Association with Disease Mutations. Cells 2020, 9, 359. [Google Scholar] [CrossRef] [PubMed]
- Rodier, F.; Kim, S.-H.; Nijjar, T.; Yaswen, P.; Campisi, J. Cancer and aging: The importance of telomeres in genome maintenance. Int. J. Biochem. Cell Biol. 2005, 37, 977–990. [Google Scholar] [CrossRef] [PubMed]
- de Araújo, L.J.T.; Lerario, A.M.; de Castro, M.; Martins, C.S.; Bronstein, M.D.; Machado, M.C.; Trarbach, E.B.; Fragoso, M.C.B.V. Transcriptome Analysis Showed a Differential Signature between Invasive and Non-invasive Corticotrophinomas. Front. Endocrinol. 2017, 8, 55, Erratum in: Front. Endocrinol. 2019, 10, 567. [Google Scholar] [CrossRef] [PubMed]
- Gedvilaite, G.; Vilkeviciute, A.; Glebauskiene, B.; Kriauciuniene, L.; Liutkeviciene, R. Molecular Markers of Telomerase Complex for Patients with Pituitary Adenoma. Brain Sci. 2022, 12, 980. [Google Scholar] [CrossRef] [PubMed]
- Cawthon, R.M. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002, 30, e47. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real time quantitative PCR and the method. Methods 2011, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Holesova, Z.; Krasnicanova, L.; Saade, R.; Pös, O.; Budis, J.; Gazdarica, J.; Repiska, V.; Szemes, T. Telomere Length Changes in Cancer: Insights on Carcinogenesis and Potential for Non-Invasive Diagnostic Strategies. Genes 2023, 14, 715. [Google Scholar] [CrossRef] [PubMed]
- Crocco, P.; Barale, R.; Rose, G.; Rizzato, C.; Santoro, A.; De Rango, F.; Carrai, M.; Fogar, P.; Monti, D.; Biondi, F.; et al. Population-specific association of genes for telomere-associated proteins with longevity in an Italian population. Biogerontology 2015, 16, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Varadi, V.; Brendle, A.; Brandt, A.; Johansson, R.; Enquist, K.; Henriksson, R.; Svenson, U.; Tavelin, B.; Roos, G.; Hemminki, K.; et al. Polymorphisms in telomere-associated genes, breast cancer susceptibility and prognosis. Eur. J. Cancer 2009, 45, 3008–3016. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, F.; Han, N. Recurrence Rate and Exploration of Clinical Factors after Pituitary Adenoma Surgery: A Systematic Review and Meta-Analysis based on Computer Artificial Intelligence System. Comput. Intell. Neurosci. 2022, 2022, 6002672. [Google Scholar] [CrossRef] [PubMed]
- Salhab, M.; Jiang, W.G.; Newbold, R.F.; Mokbel, K. The expression of gene transcripts of telomere-associated genes in human breast cancer: Correlation with clinico-pathological parameters and clinical outcome. Breast Cancer Res. Treat. 2007, 109, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Bhari, V.K.; Kumar, D.; Kumar, S.; Mishra, R. Shelterin complex gene: Prognosis and therapeutic vulnerability in cancer. Biochem. Biophys. Rep. 2021, 26, 100937. [Google Scholar] [CrossRef] [PubMed]
- Morcos, Y.A.T.; Najjar, G.; Meessen, S.; Witt, B.; Azoitei, A.; Kumar, M.; Wakileh, G.; Schwarz, K.; Schrezenmeier, H.; Zengerling, F.; et al. A Novel Tissue and Stem Cell Specific TERF1 Splice Variant Is Downregulated in Tumour Cells. Int. J. Mol. Sci. 2019, 21, 85. [Google Scholar] [CrossRef]
- Heaphy, C.M.; Bi, W.L.; Coy, S.; Davis, C.; Gallia, G.L.; Santagata, S.; Rodriguez, F.J. Telomere length alterations and ATRX/DAXX loss in pituitary adenomas. Mod. Pathol. 2020, 33, 1475–1481. [Google Scholar] [CrossRef]
- van Steensel, B.; de Lange, T. Control of telomere length by the human telomeric protein TRF1. Nature 1997, 385, 740–743. [Google Scholar] [CrossRef]
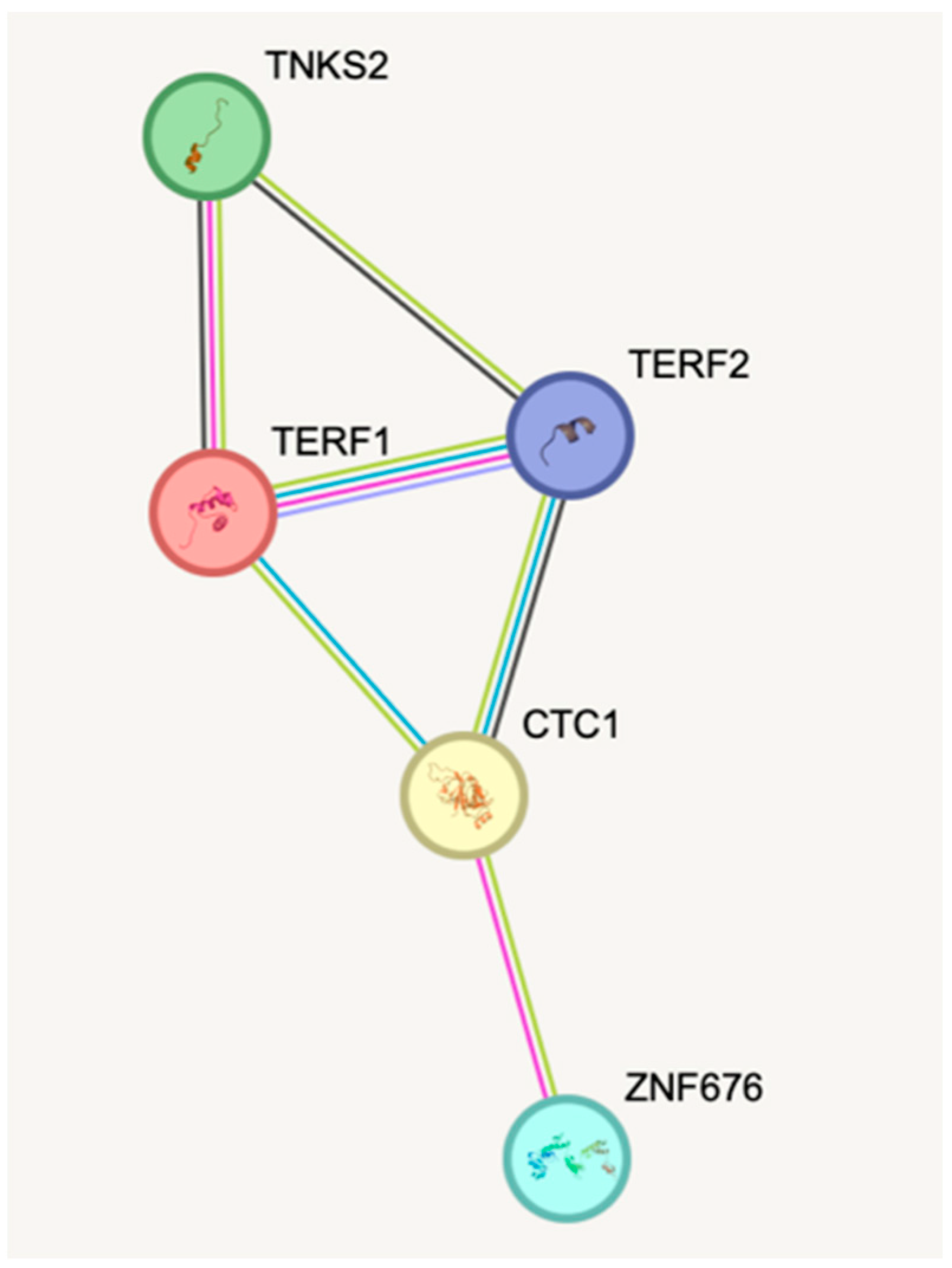
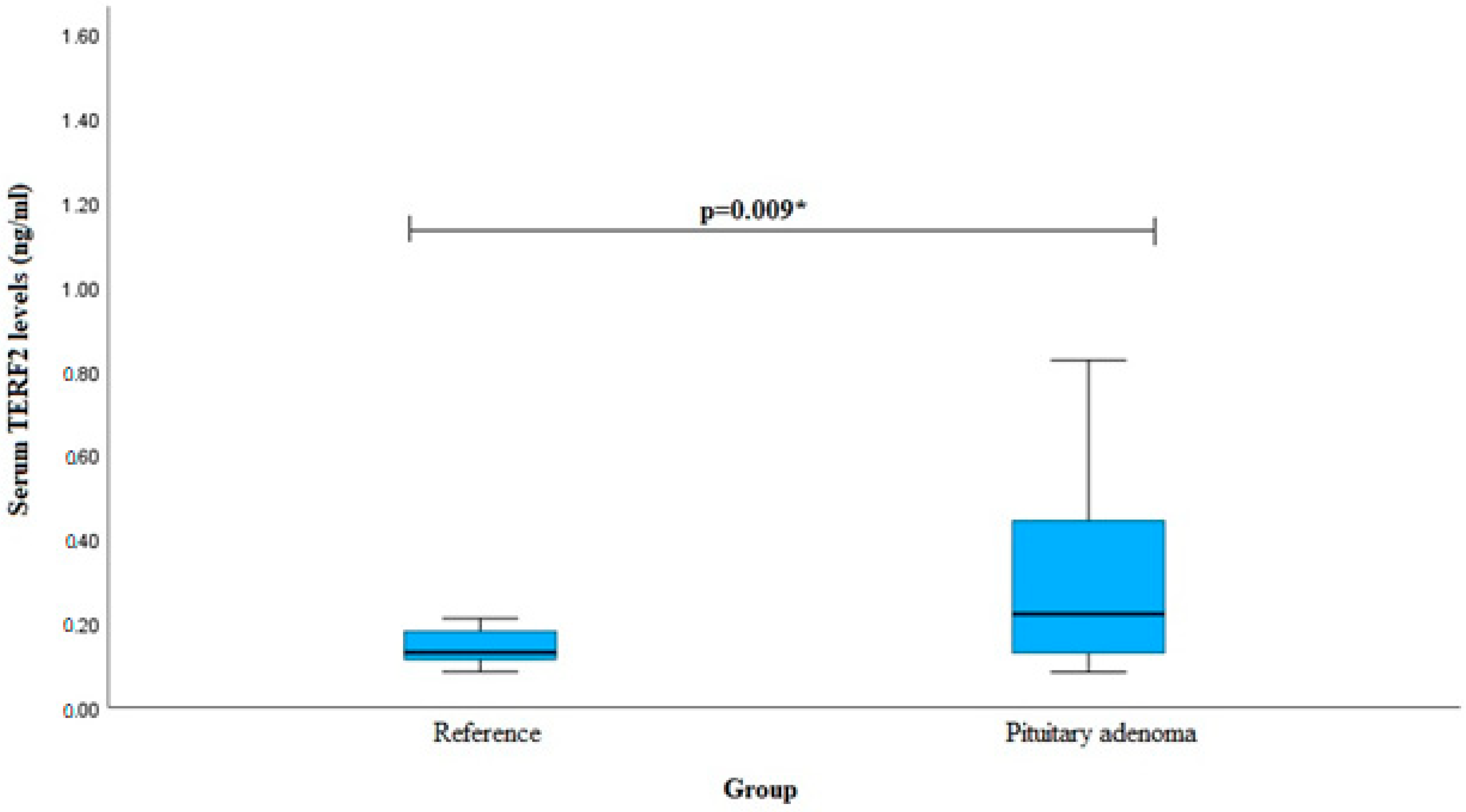
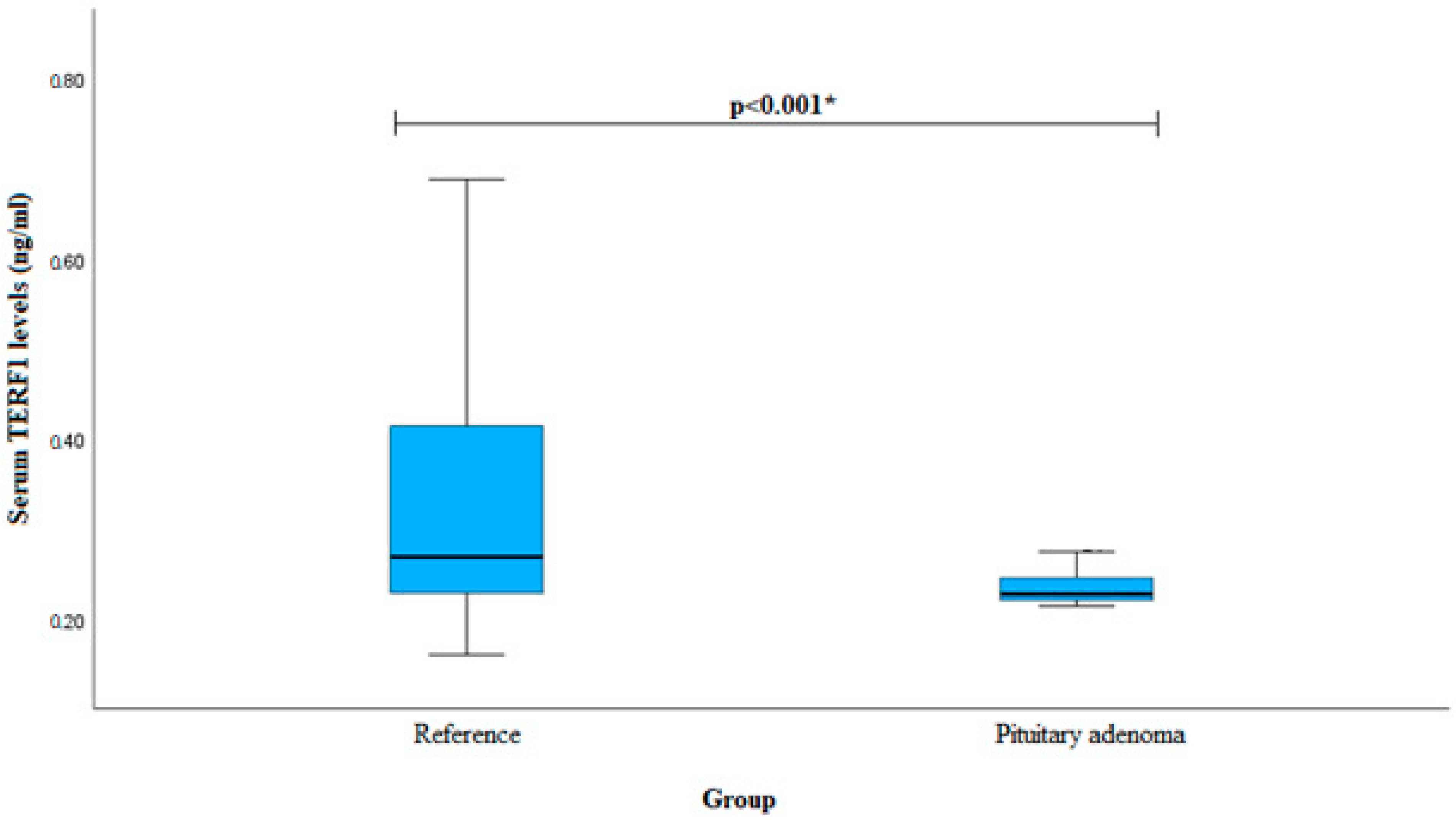

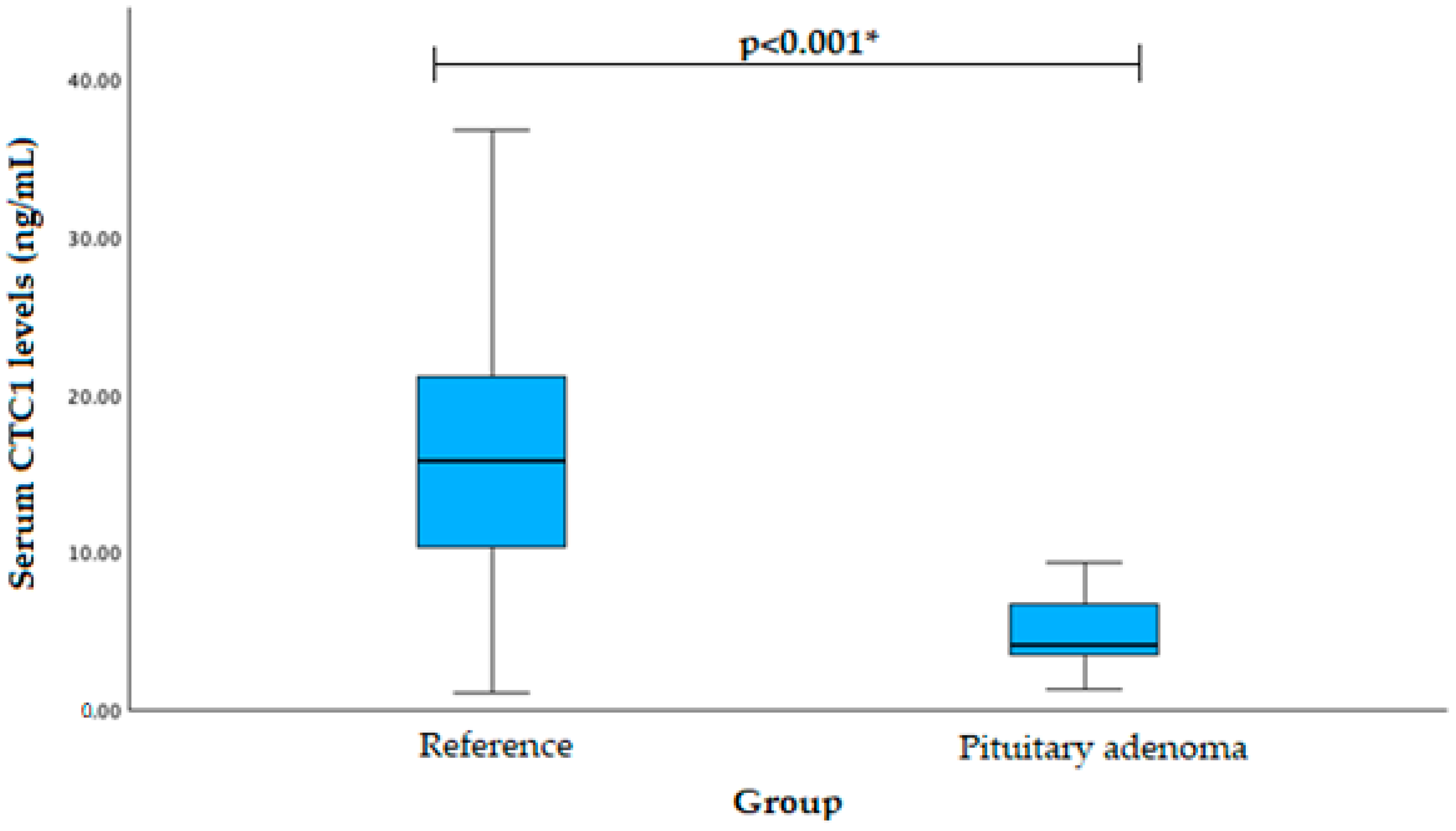

| Characteristics | Group | p-Value | ||
|---|---|---|---|---|
| PA Group | Reference Group | |||
| Gender | Males, n (%) | 50 (38.5) | 99 (30.9) | 0.124 |
| Females, n (%) | 80 (61.5) | 221 (69.1) | ||
| Age Mean (SD) | 52.73 (14.118) | 51.88 (21.325) | 0.620 * | |
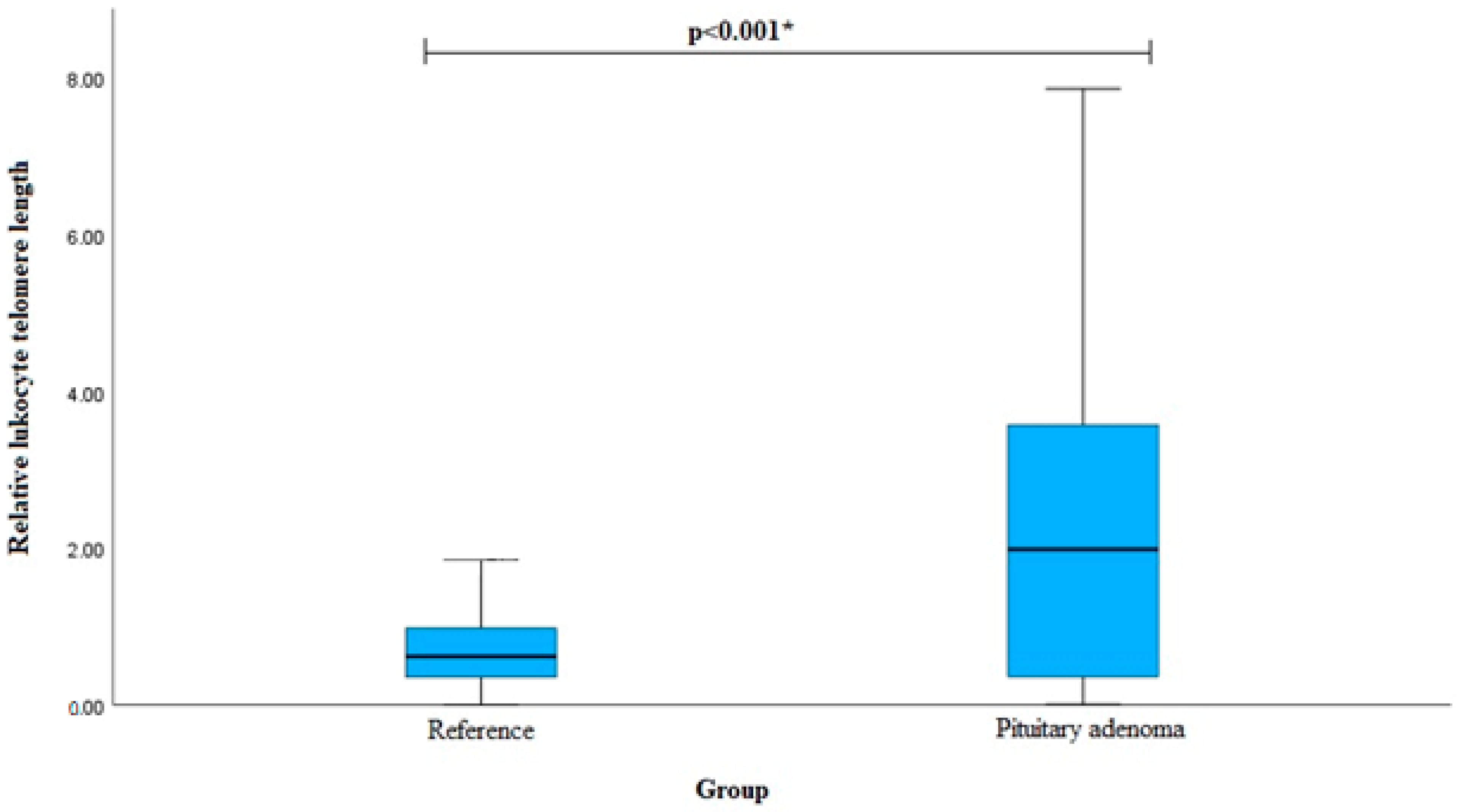
| Relative leukocyte telomere length Median (IQR) | 1.987 (3.225) | 0.619 (0.632) | <0.001 ** | |
| Gene, SNP | Genotype, Allele | PA Group, n (%) | Reference Group, n (%) | p-Value |
|---|---|---|---|---|
| TERF1 rs1545827 | CC | 81 (62.3) | 119 (37.2) | <0.001 |
| CT | 41 (31.5) | 162 (50.6) | ||
| TT | 8 (6.2) | 39 (12.2) | ||
| Total | 130 (100) | 320 (100) | ||
| Allele | ||||
| C | 203 (78.1) | 400 (62.5) | <0.001 | |
| T | 57 (21.9) | 240 (37.5) | ||
| TERF1 rs10107605 | AA | 118 (90.8) | 259 (80.9) | 0.007 |
| AC | 12 (9.2) | 42 (13.1) | ||
| CC | 0 (0.0) | 19 (5.9) | ||
| Total | 130 (100) | 320 (100) | ||
| Allele | ||||
| A | 248 (95.4) | 560 (87.5) | <0.001 | |
| C | 12 (4.6) | 80 (12.5) | ||
| TNKS2 rs10509637 | AA | 67 (51.6) | 219 (68.5) | <0.001 |
| AG | 41 (31.5) | 90 (28.1) | ||
| GG | 22 (16.8) | 11 (3.4) | ||
| Total | 130 (100) | 320 (100) | ||
| Allele | ||||
| A | 175 (67.3) | 528 (82.5) | <0.001 | |
| G | 85 (32.7) | 112 (17.5) | ||
| TNKS2 rs10509639 | AA | 107 (82.3) | 252 (78.8) | 0.513 |
| AG | 22 (16.9) | 67 (20.9) | ||
| GG | 1 (0.8) | 1 (0.3) | ||
| Total | 130 (100) | 320 (100) | ||
| Allele | ||||
| A | 236 (90.8) | 571 (89.2) | 0.489 | |
| G | 24 (9.2) | 69 (10.8) | ||
| TERF2 rs251796 | AA | 61 (46.9) | 154 (48.1) | 0.215 |
| AG | 59 (45.4) | 125 (39.1) | ||
| GG | 10 (7.7) | 41 (12.8) | ||
| Total | 130 (100) | 320 (100) | ||
| Allele | ||||
| A | 181 (69.6) | 433 (67.7) | 0.567 | |
| G | 79 (30.4) | 207 (32.3) | ||
| ZNF676 rs412658 | CC | 64 (49.2) | 135 (42.2) | 0.287 |
| CT | 50 (38.5) | 149 (46.6) | ||
| TT | 16 (12.3) | 36 (11.2) | ||
| Total | 130 (100) | 320 (100) | ||
| Allele | ||||
| C | 178 (68.5) | 419 (65.5) | 0.389 | |
| T | 82 (31.5) | 221 (34.5) | ||
| CTC1 rs3027234 | CC | 78 (60.0) | 199 (62.2) | 0.702 |
| CT | 46 (35.4) | 102 (31.9) | ||
| TT | 6 (4.6) | 19 (5.9) | ||
| Total | 130 (100) | 320 (100) | ||
| Allele | ||||
| C | 202 (76.9) | 500 (78.1) | 0.887 | |
| T | 58 (23.1) | 140 (21.9) |
| Gene and SNP | Allele Frequencies | Genotype Distribution | p-Value | |
|---|---|---|---|---|
| TERF1 rs1545827 | 0.62 C | 0.38 T | 39/162/119 | 0.152 |
| TERF1 rs10107605 | 0.87 A | 0.13 C | 19/42/259 | <0.0001 |
| TNKS2 rs10509637 | 0.82 A | 0.18 G | 11/90/219 | 0.642 |
| TNKS2 rs10509639 | 0.89 A | 0.11 G | 1/67/252 | 0.114 |
| TERF2 rs251796 | 0.68 A | 0.32 G | 41/125/154 | 0.055 |
| ZNF676 rs412658 | 0.65 C | 0.35 T | 36/149/135 | 0.594 |
| CTC1 rs3027234 | 0.78 C | 0.22 T | 19/102/199 | 0.228 |
| Model | Genotype/Allele | OR (95% CI) | p-Value | AIC |
|---|---|---|---|---|
| TERF1 rs1545827 | ||||
| Codominant | CT vs. CC TT vs. CC | 0.372 (0.239–0.580) 0.301 (0.134–0.678) | <0.001 0.004 | 521.1 |
| Dominant | CT + TT vs. CC | 0.358 (0.235–0.546) | <0.001 | 519.4 |
| Recessive | TT vs. CC + CT | 0.472 (0.214–1.041) | 0.063 | 519.1 |
| Overdominant | CT vs. CC + TT | 0.449 (0.292–0.691) | <0.001 | 529.2 |
| Additive | T | 0.453 (0.320–0.642) | <0.001 | 521.1 |
| TNKS2 rs10509637 | ||||
| Codominant | AG vs. AA GG vs. AA | 1.489 (0.940–2.358) 6.537 (3.015–14.172) | 0.090 <0.001 | 520.2 |
| Dominant | AG + GG vs. AA | 2.039 (1.344–3.094) | <0.001 | 531.9 |
| Recessive | AA vs. GG + AG | 5.722 (2.686–12.189) | <0.001 | 521.1 |
| Overdominant | AG vs. AA + GG | 1.177 (0.756–1.833) | 0.470 | 542.5 |
| Additive | G | 2.081 (1.516–2.857) | <0.001 | 522.3 |
| TNKS2 rs10509639 | ||||
| Codominant | AG vs. AA GG vs. AA | 0.773 (0.454–1.317) 2.355 (0.146–38.001) | 0.344 0.546 | 541.7 |
| Dominant | AG + GG vs. AA | 0.797 (0.472–1.345) | 0.395 | 542.3 |
| Recessive | AA vs. GG + AG | 2.473 (0.154–39.834) | 0.523 | 542.6 |
| Overdominant | AG vs. AA + GG | 0.769 (0.452–1.309) | 0.333 | 540.1 |
| Additive | G | 0.831 (0.502–1.378) | 0.474 | 542.5 |
| TERF2 rs251796 | ||||
| Codominant | AG vs. AA GG vs. AA | 1.192 (0.776–1.829) 0.616 (0.290–1.306) | 0.423 0.206 | 541.8 |
| Dominant | AG + GG vs. AA | 1.049 (0.698–1.579) | 0.817 | 543.0 |
| Recessive | AA vs. GG + AG | 0.567 (0.275–1.169) | 0.124 | 540.5 |
| Overdominant | AG vs. AA + GG | 1.296 (0.859–1.957) | 0.217 | 541.5 |
| Additive | G | 0.917 (0.677–1.243) | 0.578 | 542.7 |
| ZNF676 rs412658 | ||||
| Codominant | CT vs. CC TT vs. CC | 0.708 (0.457–1.096) 0.938 (0.485–1.813) | 0.121 0.848 | 542.5 |
| Dominant | CT + TT vs. CC | 0.753 (0.500–1.133) | 0.173 | 541.2 |
| Recessive | TT vs. CC + CT | 1.107 (0.591–2.074) | 0.750 | 542.9 |
| Overdominant | CT vs. CC + TT | 0.717 (0.473–1.087) | 0.117 | 540.6 |
| Additive | T | 0.874 (0.643–1.189) | 0.392 | 542.3 |
| CTC1 rs3027234 | ||||
| Codominant | CT vs. CC TT vs. CC | 1.151 (0.744–1.779) 0.806 (0.310–2.093) | 0.528 0.657 | 544.3 |
| Dominant | CT + TT vs. CC | 1.096 (0.722–1.664) | 0.666 | 542.9 |
| Recessive | TT vs. CC + CT | 0.767 (0.299–1.965) | 0.580 | 542.7 |
| Overdominant | CT vs. CC + TT | 1.170 (0.762–1.798) | 0.473 | 542.5 |
| Additive | T | 1.024 (0.729–1.439) | 0.889 | 543.0 |
| Gene, SNP | Genotype, Allele | PA Group Females, n (%) | Reference Group Females, n (%) | p-Value |
|---|---|---|---|---|
| TERF1 rs1545827 | CC | 52 (65.0) | 82 (37.1) | <0.001 |
| CT | 21 (26.3) | 108 (48.9) | ||
| TT | 7 (8.7) | 31 (14.0) | ||
| Total | 80 (100) | 221 (100) | ||
| Allele | ||||
| C | 125 (78.1) | 272 (61.5) | <0.001 | |
| T | 35 (21.9) | 170 (38.5) | ||
| TNKS2 rs10509637 | AA | 42 (52.5) | 152 (68.8) | <0.001 |
| AG | 25 (31.3) | 60 (27.1) | ||
| GG | 13 (16.2) | 9 (4.1) | ||
| Total | 80 (100) | 221 (100) | ||
| Allele | ||||
| A | 109 (68.1) | 364 (82.4) | <0.001 | |
| G | 51 (31.9) | 78 (17.6) |
| Model | Genotype/Allele | OR (95% CI) | p-Value | AIC |
|---|---|---|---|---|
| TERF1 rs1545827 | ||||
| Codominant | CT vs. CC TT vs. CC | 0.307 (0.171–0.549) 0.356 (0.146–0.868) | <0.001 0.023 | 333.9 |
| Dominant | CT + TT vs. CC | 0.318 (0.186–0.542) | <0.001 | 332.0 |
| Recessive | TT vs. CC + CT | 0.588 (0.248–1.394) | 0.228 | 349.0 |
| Overdominant | CT vs. CC + TT | 0.372 (0.212–0.654) | <0.001 | 337.8 |
| Additive | T | 0.455 (0.298–0.697) | <0.001 | 335.9 |
| TNKS2 rs10509637 | ||||
| Codominant | AG vs. AA GG vs. AA | 1.508 (0.846–2.689) 5.228 (2.092–3.065) | 0.164 <0.001 | 359.5 |
| Dominant | AG + GG vs. AA | 1.993 (1.181–3.362) | 0.010 | 343.9 |
| Recessive | AA vs. GG + AG | 4.579 (1.871–11.165) | <0.001 | 339.4 |
| Overdominant | AG vs. AA + GG | 1.220 (0.698–2.131) | 0.485 | 350.1 |
| Additive | G | 1.973 (1.334–2.920) | <0.001 | 339.0 |
| TERF2 rs251796 | ||||
| Codominant | AG vs. AA GG vs. AA | 0.917 (0.537–1.567) 0.322 (0.107–0.971) | 0.752 0.044 | 347.5 |
| Dominant | AG + GG vs. AA | 0.765 (0.458–1.277) | 0.306 | 349.5 |
| Recessive | AA vs. GG + AG | 0.335 (0.114–0.983) | 0.047 | 345.6 |
| Overdominant | AG vs. AA + GG | 1.081 (0.643–1.820) | 0.768 | 350.5 |
| Additive | G | 0.707 (0.476–1.051) | 0.086 | 347.5 |
| Gene, SNP | Genotype, Allele | PA Group Males, n (%) | Reference Group Males, n (%) | p-Value |
|---|---|---|---|---|
| TERF1 rs1545827 | CC | 29 (58.0) | 37 (37.4) | 0.036 |
| CT | 20 (40.0) | 54 (54.5) | ||
| TT | 1 (2.0) | 8 (8.1) | ||
| Total | 50 (100) | 99 (100) | ||
| Allele | ||||
| C | 78 (78.0) | 128 (64.6) | 0.018 | |
| T | 22 (22.0) | 70 (35.4) | ||
| TNKS2 rs10509637 | AA | 25 (50.0) | 67 (67.7) | 0.001 |
| AG | 16 (32.0) | 30 (30.3) | ||
| GG | 9 (18.0) | 2 (2.0) | ||
| Total | 50 (100) | 99 (100) | ||
| Allele | ||||
| A | 66 (66.0) | 164 (82.8) | 0.001 | |
| G | 34 (34.0) | 34 (17.2) |
| Model | Genotype/Allele | OR (95% CI) | p-Value | AIC |
|---|---|---|---|---|
| TERF1 rs1545827 | ||||
| Codominant | CT vs. CC TT vs. CC | 0.473 (0.233–0.958) 0.159 (0.019–1.349) | 0.038 0.092 | 187.2 |
| Dominant | CT + TT vs. CC | 0.432 (0.216–0.865) | 0.018 | 186.4 |
| Recessive | TT vs. CC + CT | 0.232 (0.028–1.910) | 0.174 | 189.6 |
| Overdominant | CT vs. CC + TT | 0.556 (0.279–1.108) | 0.095 | 189.3 |
| Additive | T | 0.450 (0.242–0.834) | 0.011 | 185.2 |
| TNKS2 rs10509637 | ||||
| Codominant | AG vs. AA GG vs. AA | 1.489 (0.668–3.059) 2.060 (2.436–59.707) | 0.358 0.002 | 181.5 |
| Dominant | AG + GG vs. AA | 2.094 (1.044–4.200) | 0.037 | 187.8 |
| Recessive | AA vs. GG + AG | 10.646 (2.204–51.433) | 0.003 | 180.3 |
| Overdominant | AG vs. AA + GG | 1.082 (0.520–2.252) | 0.832 | 192.1 |
| Additive | G | 2.298 (1.330–3.972) | 0.003 | 182.9 |
| Gene, SNP | Genotype, Allele | Reference Group, n (%) | PA Group with Relapse, n (%) | p-Value | PA Group without Relapse, n (%) | p-Value |
|---|---|---|---|---|---|---|
| TERF1 rs1545827 | CC | 119 (37.2) | 19 (50.0) | 0.290 | 62 (67.4) | <0.001 |
| CT | 162 (50.6) | 16 (42.1) | 25 (27.2) | |||
| TT | 39 (12.2) | 3 (7.9) | 5 (5.4) | |||
| Total | 320 (100) | 38 (100) | 92 (100) | |||
| Allele | ||||||
| C | 400 (62.5) | 54 (71.1) | 0.143 | 149 (81.0) | <0.001 | |
| T | 240 (37.5) | 22 (28.9) | 35 (19.0) | |||
| TNKS2 rs10509637 | AA | 219 (68.5) | 20 (52.6) | 0.012 | 47 (51.1) | <0.001 |
| AG | 90 (28.1) | 13 (34.2) | 28 (30.4) | |||
| GG | 11 (3.4) | 5 (13.2) | 17 (18.5) | |||
| Total | 320 (100) | 38 (100) | 92 (100) | |||
| Allele | ||||||
| A | 528 (82.5) | 53 (69.7) | 0.007 | 122 (66.3) | <0.001 | |
| G | 112 (17.5) | 23 (30.3) | 62 (33.7) |
| Model | Genotype/Allele | OR (95% CI) | p-Value | AIC |
|---|---|---|---|---|
| PA with Relapse | ||||
| TNKS2 rs10509637 | ||||
| Codominant | AG vs. AA GG vs. AA | 1.582 (0.755–3.316) 4.977 (1.573–15.751) | 0.225 0.006 | 239.5 |
| Dominant | AG + GG vs. AA | 1.951 (0.990–3.848) | 0.054 | 240.6 |
| Recessive | AA vs. GG + AG | 4.256 (1.394–12.998) | 0.011 | 238.9 |
| Overdominant | AG vs. AA + GG | 1.329 (0.651–2.711) | 0.435 | 243.7 |
| Additive | G | 1.968 (1.170–3.311) | 0.011 | 238.2 |
| PA without relapse | ||||
| TERF1 rs1545827 | ||||
| Codominant | CT vs. CC TT vs. CC | 0.296 (0.176–0.499) 0.246 (0.092–0.656) | <0.001 0.005 | 414.9 |
| Dominant | CT + TT vs. CC | 0.286 (0.175–0.468) | <0.001 | 413.1 |
| Recessive | TT vs. CC + CT | 0.414 (0.158–1.083) | 0.072 | 436.7 |
| Overdominant | CT vs. CC + TT | 0.364 (0.219–0.605) | <0.001 | 423.1 |
| Additive | T | 0.375 (0.247–0.570) | <0.001 | 415.2 |
| TNKS2 rs10509637 | ||||
| Codominant | AG vs. AA GG vs. AA | 1.450 (0.855–2.459) 7.201 (3.168–16.371) | 0.168 <0.001 | 418.9 |
| Dominant | AG + GG vs. AA | 2.076 (1.295–3.328) | 0.002 | 430.5 |
| Recessive | AA vs. GG + AG | 6.367 (2.863–14.160) | <0.001 | 418.8 |
| Overdominant | AG vs. AA + GG | 1.118 (0.674–1.855) | 0.666 | 439.4 |
| Additive | G | 2.173 (1.526–3.096) | <0.001 | 421.3 |
| Gene, SNP | Genotype, Allele | Long Telomeres | Short Telomeres | p-Value |
|---|---|---|---|---|
| TERF1 rs1545827 | CC | 108 (46.8) | 69 (36.9) | 0.043 |
| CT | 104 (45.0) | 91 (48.7) | ||
| TT | 19 (8.2) | 27 (14.4) | ||
| Total | 231 (100) | 187 (100) | ||
| Allele | ||||
| C | 320 (69.3) | 229 (61.2) | 0.015 | |
| T | 142 (30.1) | 145 (38.8) | ||
| CTC1 rs3027234 | CC | 132 (57.1) | 127 (67.9) | 0.006 |
| CT | 79 (34.2) | 56 (29.9) | ||
| TT | 20 (8.7) | 4 (2.1) | ||
| Total | 231 (100) | 187 (100) | ||
| Allele | ||||
| C | 343 (74.2) | 310 (82.9) | 0.003 | |
| T | 119 (25.8) | 64 (17.1) |
| Model | Genotype/Allele | OR (95% CI) | p-Value | AIC |
|---|---|---|---|---|
| TERF1 rs1545827 | ||||
| Codominant | CT vs. CC TT vs. CC | 0.730 (0.483–1.103) 0.450 (0.232–0.870) | 0.135 0.018 | 572.5 |
| Dominant | CT + TT vs. CC | 0.666 (0.449–0.987) | 0.043 | 572.7 |
| Recessive | TT vs. CC + CT | 0.531(0.285–0.989) | 0.046 | 572.8 |
| Overdominant | CT vs. CC + TT | 0.864 (0.587–1.272) | 0.458 | 576.3 |
| Additive | T | 0.690 (0.513–0.927) | 0.014 | 570.7 |
| CTC1 rs3027234 | ||||
| Codominant | CT vs. CC TT vs. CC | 1.357 (0.892–2.066) 4.811 (1.600–14.464) | 0.154 0.005 | 565.8 |
| Dominant | CT + TT vs. CC | 1.587 (1.061–2.375) | 0.024 | 571.7 |
| Recessive | TT vs. CC + CT | 4.336 (1.456–12.919) | 0.008 | 567.8 |
| Overdominant | CT vs. CC + TT | 1.216 (0.803–1.840) | 0.355 | 576.0 |
| Additive | T | 1.644 (1.174–2.302) | 0.004 | 568.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gedvilaite, G.; Kriauciuniene, L.; Tamasauskas, A.; Liutkeviciene, R. The Influence of Telomere-Related Gene Variants, Serum Levels, and Relative Leukocyte Telomere Length in Pituitary Adenoma Occurrence and Recurrence. Cancers 2024, 16, 643. https://doi.org/10.3390/cancers16030643
Gedvilaite G, Kriauciuniene L, Tamasauskas A, Liutkeviciene R. The Influence of Telomere-Related Gene Variants, Serum Levels, and Relative Leukocyte Telomere Length in Pituitary Adenoma Occurrence and Recurrence. Cancers. 2024; 16(3):643. https://doi.org/10.3390/cancers16030643
Chicago/Turabian StyleGedvilaite, Greta, Loresa Kriauciuniene, Arimantas Tamasauskas, and Rasa Liutkeviciene. 2024. "The Influence of Telomere-Related Gene Variants, Serum Levels, and Relative Leukocyte Telomere Length in Pituitary Adenoma Occurrence and Recurrence" Cancers 16, no. 3: 643. https://doi.org/10.3390/cancers16030643
APA StyleGedvilaite, G., Kriauciuniene, L., Tamasauskas, A., & Liutkeviciene, R. (2024). The Influence of Telomere-Related Gene Variants, Serum Levels, and Relative Leukocyte Telomere Length in Pituitary Adenoma Occurrence and Recurrence. Cancers, 16(3), 643. https://doi.org/10.3390/cancers16030643





