Navigating Precision Oncology: Insights from an Integrated Clinical Data and Biobank Repository Initiative across a Network Cancer Program
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
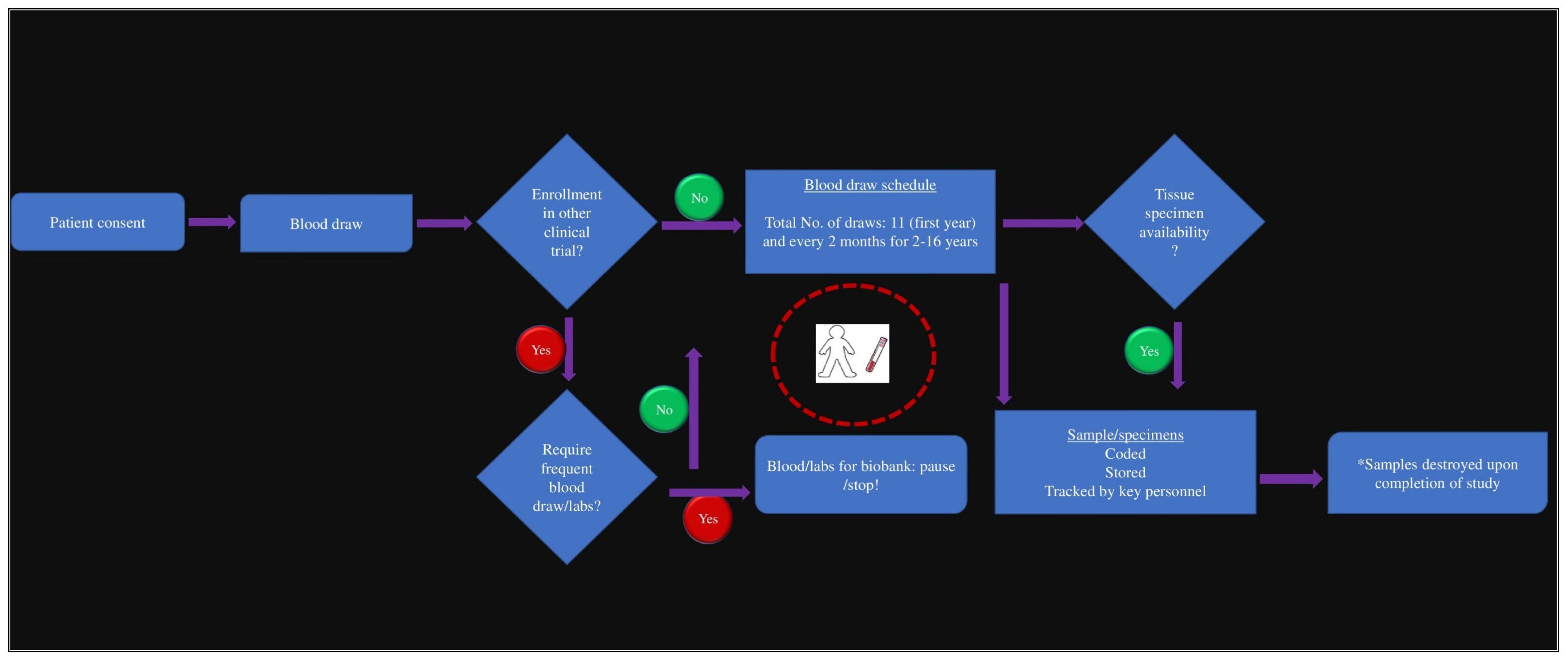
2.1. Building the Oncology Biobank and Data Repository
2.2. Developing the Clinical Data
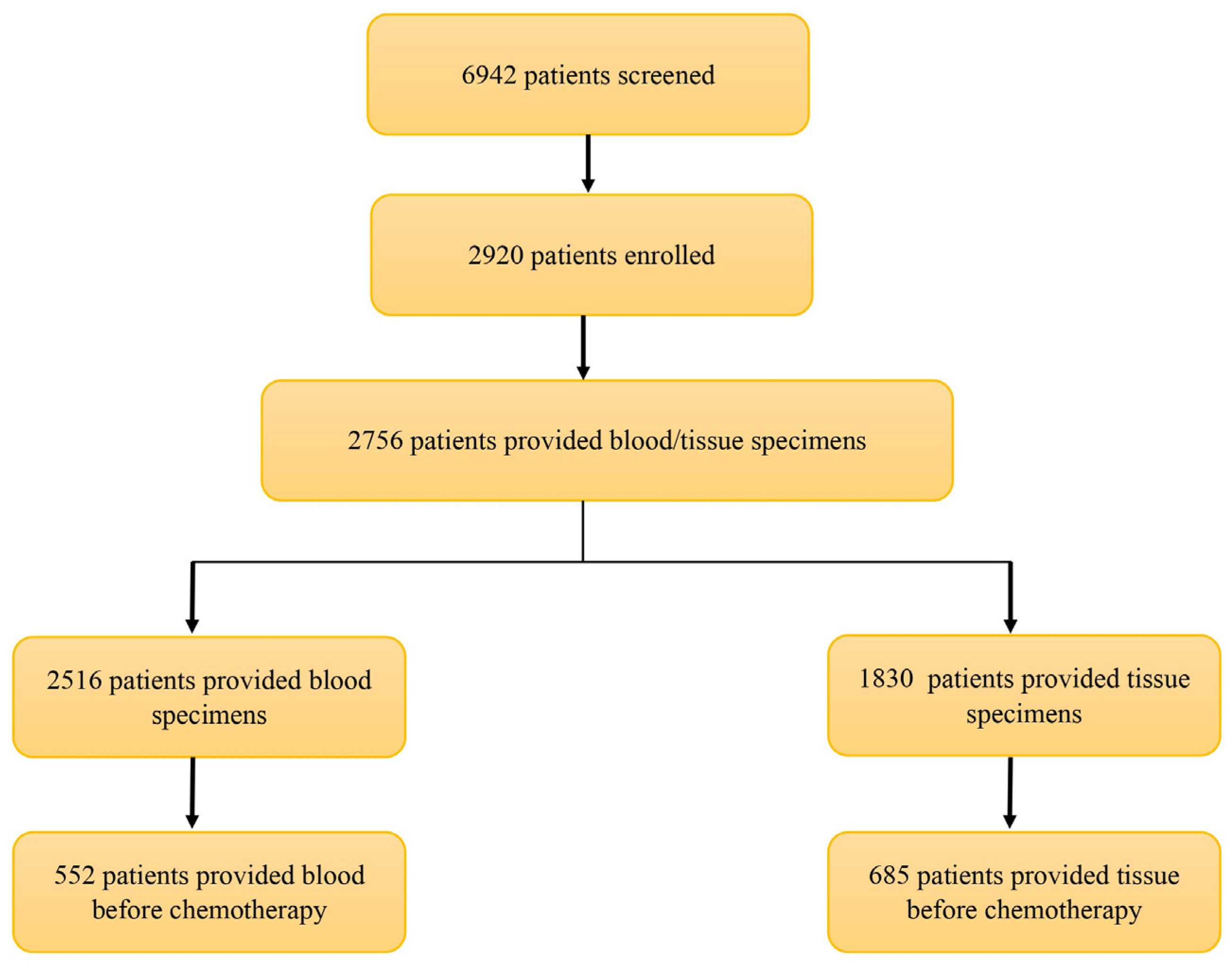
3. Results
4. Discussion
4.1. Data Standardization
4.2. Artificial Intelligence and Machine Learning
4.3. Next Steps
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- US Cancer Statistics Working Group. Cancer Statistics Data Visualizations Tool, Based on 2021 Submission Data (1999–2019): U.S. Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute. Available online: https://www.cdc.gov/cancer/dataviz (accessed on 20 September 2023).
- CDC. Cancer Deaths Among Men and Women, by Race and Ethnicity, United States, 2015–2019. National Center for Health Statistics. Available online: https://www.cdc.gov/nchs/hus/topics/cancer-deaths.htm (accessed on 5 January 2024).
- Ugai, T.; Sasamoto, N.; Lee, H.Y.; Ando, M.; Song, M.; Tamimi, R.M.; Kawachi, I.; Campbell, P.T.; Giovannucci, E.L.; Weiderpass, E.; et al. Is early-onset cancer an emerging global epidemic? Current evidence and future implications. Nat. Rev. Clin. Oncol. 2022, 19, 656–673. [Google Scholar] [CrossRef] [PubMed]
- Pankiw, M.; Brezden-Masley, C.; Charames, G.S. Comprehensive genomic profiling for oncological advancements by precision medicine. Med. Oncol. 2023, 41, 1. [Google Scholar] [CrossRef] [PubMed]
- Price, K.S.; Svenson, A.; King, E.; Ready, K.; Lazarin, G.A. Inherited Cancer in the Age of Next-Generation Sequencing. Biol. Res. Nurs. 2018, 20, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Tischler, J.; Crew, K.D.; Chung, W.K. Cases in Precision Medicine: The Role of Tumor and Germline Genetic Testing in Breast Cancer Management. Ann. Intern. Med. 2019, 171, 925–930. [Google Scholar] [CrossRef]
- Fiala, E.M.; Jayakumaran, G.; Mauguen, A.; Kennedy, J.A.; Bouvier, N.; Kemel, Y.; Fleischut, M.H.; Maio, A.; Salo-Mullen, E.E.; Sheehan, M.; et al. Prospective pan-cancer germline testing using MSK-IMPACT informs clinical translation in 751 patients with pediatric solid tumors. Nat. Cancer 2021, 2, 357–365. [Google Scholar] [CrossRef]
- Kraft, I.L.; Godley, L.A. Identifying potential germline variants from sequencing hematopoietic malignancies. Hematol. Am. Soc. Hematol. Educ. Program 2020, 2020, 219–227. [Google Scholar] [CrossRef]
- Mandelker, D.; Zhang, L.; Kemel, Y.; Stadler, Z.K.; Joseph, V.; Zehir, A.; Pradhan, N.; Arnold, A.; Walsh, M.F.; Li, Y.; et al. Mutation Detection in Patients With Advanced Cancer by Universal Sequencing of Cancer-Related Genes in Tumor and Normal DNA vs Guideline-Based Germline Testing. JAMA 2017, 318, 825–835. [Google Scholar] [CrossRef]
- Stadler, Z.K.; Maio, A.; Chakravarty, D.; Kemel, Y.; Sheehan, M.; Salo-Mullen, E.; Tkachuk, K.; Fong, C.J.; Nguyen, B.; Erakky, A.; et al. Therapeutic Implications of Germline Testing in Patients With Advanced Cancers. J. Clin. Oncol. 2021, 39, 2698–2709. [Google Scholar] [CrossRef]
- Subbiah, V.; Kurzrock, R. Universal Germline and Tumor Genomic Testing Needed to Win the War Against Cancer: Genomics Is the Diagnosis. J. Clin. Oncol. 2023, 41, 3100–3103. [Google Scholar] [CrossRef]
- Zhang, J.; Walsh, M.F.; Wu, G.; Edmonson, M.N.; Gruber, T.A.; Easton, J.; Hedges, D.; Ma, X.; Zhou, X.; Yergeau, D.A.; et al. Germline Mutations in Predisposition Genes in Pediatric Cancer. N. Engl. J. Med. 2015, 373, 2336–2346. [Google Scholar] [CrossRef]
- Post, A.R.; Burningham, Z.; Halwani, A.S. Electronic Health Record Data in Cancer Learning Health Systems: Challenges and Opportunities. JCO Clin. Cancer Inform. 2022, 6, e2100158. [Google Scholar] [CrossRef]
- American Society of Clinical Oncology. mCODE: Minimal Common Oncology Data Elements. Available online: https://mcodeinitiative.org (accessed on 20 September 2023).
- Osterman, T.J.; Terry, M.; Miller, R.S. Improving Cancer Data Interoperability: The Promise of the Minimal Common Oncology Data Elements (mCODE) Initiative. JCO Clin. Cancer Inform. 2020, 4, 993–1001. [Google Scholar] [CrossRef]
- Malm, J.; Sugihara, Y.; Szasz, M.; Kwon, H.J.; Lindberg, H.; Appelqvist, R.; Marko-Varga, G. Biobank integration of large-scale clinical and histopathology melanoma studies within the European Cancer Moonshot Lund Center. Clin. Transl. Med. 2018, 7, 28. [Google Scholar] [CrossRef] [PubMed]
- Mak, J.K.L.; McMurran, C.E.; Kuja-Halkola, R.; Hall, P.; Czene, K.; Jylhava, J.; Hagg, S. Clinical biomarker-based biological aging and risk of cancer in the UK Biobank. Br. J. Cancer 2023, 129, 94–103. [Google Scholar] [CrossRef] [PubMed]
- LaFramboise, W.; Zaidi, A.H.; Allen, C.J.; Bizhanova, Z.; Dalton, E.; Bapat, B.; Petrosko, P.; Gallo, P.; Gil, L.; Lam, J.T.; et al. Concordance of circulating and solid tumor DNA through comprehensive genomic profiling in a large integrated cancer network. J. Clin. Oncol. 2023, 41 (Suppl. 16), 3059. [Google Scholar] [CrossRef]
- Lyu, H.G.; Haider, A.H.; Landman, A.B.; Raut, C.P. The opportunities and shortcomings of using big data and national databases for sarcoma research. Cancer 2019, 125, 2926–2934. [Google Scholar] [CrossRef] [PubMed]
- Armitage, E.G.; Southam, A.D. Monitoring cancer prognosis, diagnosis and treatment efficacy using metabolomics and lipidomics. Metabolomics 2016, 12, 146. [Google Scholar] [CrossRef] [PubMed]
- Simpson, E.; Brown, R.; Sillence, E.; Coventry, L.; Lloyd, K.; Gibbs, J.; Tariq, S.; Durrant, A.C. Understanding the Barriers and Facilitators to Sharing Patient-Generated Health Data Using Digital Technology for People Living With Long-Term Health Conditions: A Narrative Review. Front Public Health 2021, 9, 641424. [Google Scholar] [CrossRef]
- Prictor, M.; Teare, H.J.A.; Kaye, J. Equitable Participation in Biobanks: The Risks and Benefits of a “Dynamic Consent” Approach. Front Public Health 2018, 6, 253. [Google Scholar] [CrossRef]
- Subbiah, V.; Kurzrock, R. Universal Genomic Testing Needed to Win the War Against Cancer: Genomics IS the Diagnosis. JAMA Oncol. 2016, 2, 719–720. [Google Scholar] [CrossRef] [PubMed]
- Berger, M.F.; Mardis, E.R. The emerging clinical relevance of genomics in cancer medicine. Nat. Rev. Clin. Oncol. 2018, 15, 353–365. [Google Scholar] [CrossRef] [PubMed]
- American Association for Cancer Research Project GENIE Consortium. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017, 7, 818–831. [Google Scholar] [CrossRef] [PubMed]
- Vasan, N.; Razavi, P.; Johnson, J.L.; Shao, H.; Shah, H.; Antoine, A.; Ladewig, E.; Gorelick, A.; Lin, T.Y.; Toska, E.; et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kalpha inhibitors. Science 2019, 366, 714–723. [Google Scholar] [CrossRef] [PubMed]
- Razavi, P.; Chang, M.T.; Xu, G.; Bandlamudi, C.; Ross, D.S.; Vasan, N.; Cai, Y.; Bielski, C.M.; Donoghue, M.T.A.; Jonsson, P.; et al. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell 2018, 34, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, P.; Lin, A.L.; Young, R.J.; DiStefano, N.M.; Hyman, D.M.; Li, B.T.; Berger, M.F.; Zehir, A.; Ladanyi, M.; Solit, D.B.; et al. Genomic Correlates of Disease Progression and Treatment Response in Prospectively Characterized Gliomas. Clin. Cancer Res. 2019, 25, 5537–5547. [Google Scholar] [CrossRef] [PubMed]
- Boehm, K.M.; Khosravi, P.; Vanguri, R.; Gao, J.; Shah, S.P. Harnessing multimodal data integration to advance precision oncology. Nat. Rev. Cancer 2022, 22, 114–126. [Google Scholar] [CrossRef]
- Im, Y.R.; Tsui, D.W.Y.; Diaz, L.A., Jr.; Wan, J.C.M. Next-Generation Liquid Biopsies: Embracing Data Science in Oncology. Trends Cancer 2021, 7, 283–292. [Google Scholar] [CrossRef]
- Senthil Kumar, K.; Miskovic, V.; Blasiak, A.; Sundar, R.; Pedrocchi, A.L.G.; Pearson, A.T.; Prelaj, A.; Ho, D. Artificial Intelligence in Clinical Oncology: From Data to Digital Pathology and Treatment. Am. Soc. Clin. Oncol. Educ. Book 2023, 43, e390084. [Google Scholar] [CrossRef]




| Site Code | Site Name |
|---|---|
| AHNCI—AGH | AHN Cancer Institute—AGH |
| AHNCI—B | AHN Cancer Institute—Beaver |
| AHNCI—BU | AHN Cancer Institute—Butler |
| AHNCI—C | AHN Cancer Institute—Canonsburg |
| AHNCI—F | AHN Cancer Institute—Forbes |
| AHNCI—GC | AHN Cancer Institute—Grove City |
| AHNCI—H | AHN Cancer Institute—Hempfield |
| AHNCI—J | AHN Cancer Institute—Jefferson |
| AHNCI—NC | AHN Cancer Institute—New Castle |
| AHNCI—SV | AHN Cancer Institute—Saint Vincent |
| AVH | Allegheny Valley Hospital |
| BPH&WP | Bethel Park Health & Wellness Pavilion |
| CH | Canonsburg Hospital |
| FN | Federal North |
| Forbes | Forbes |
| GCH | Grove City Hospital |
| H&WP Erie | Allegheny Health & Wellness Pavilion Erie |
| JH | Jefferson Hospital |
| PTH&WP | Peters Township Health & Wellness Pavilion |
| SG | Suburban General |
| SVH | Saint Vincent Hospital |
| WAGH | WPAON—Allegheny General |
| WAV | WPAON—Allegheny Valley |
| WBO | WPAON—Butler Office |
| WF | WPAON—Forbes |
| WH&WP | Wexford Health & Wellness Pavilion |
| WHO | WPAON—Hansen Office |
| WJO | WPAON—Jefferson Office |
| WNC | WPAON—New Castle |
| WPH | West Penn Hospital |
| WPO | WPAON—Peters Office |
| WPU | WPAON—Punxsutawney |
| WR | WPAON—Robinson |
| WWP | WPAON—West Penn |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aryal, B.; Bizhanova, Z.; Joseph, E.A.; Yin, Y.; Wagner, P.L.; Dalton, E.; LaFramboise, W.A.; Bartlett, D.L.; Allen, C.J. Navigating Precision Oncology: Insights from an Integrated Clinical Data and Biobank Repository Initiative across a Network Cancer Program. Cancers 2024, 16, 760. https://doi.org/10.3390/cancers16040760
Aryal B, Bizhanova Z, Joseph EA, Yin Y, Wagner PL, Dalton E, LaFramboise WA, Bartlett DL, Allen CJ. Navigating Precision Oncology: Insights from an Integrated Clinical Data and Biobank Repository Initiative across a Network Cancer Program. Cancers. 2024; 16(4):760. https://doi.org/10.3390/cancers16040760
Chicago/Turabian StyleAryal, Bibek, Zhadyra Bizhanova, Edward A. Joseph, Yue Yin, Patrick L. Wagner, Emily Dalton, William A. LaFramboise, David L. Bartlett, and Casey J. Allen. 2024. "Navigating Precision Oncology: Insights from an Integrated Clinical Data and Biobank Repository Initiative across a Network Cancer Program" Cancers 16, no. 4: 760. https://doi.org/10.3390/cancers16040760
APA StyleAryal, B., Bizhanova, Z., Joseph, E. A., Yin, Y., Wagner, P. L., Dalton, E., LaFramboise, W. A., Bartlett, D. L., & Allen, C. J. (2024). Navigating Precision Oncology: Insights from an Integrated Clinical Data and Biobank Repository Initiative across a Network Cancer Program. Cancers, 16(4), 760. https://doi.org/10.3390/cancers16040760






