Epigenetic Mechanisms in Latent Epstein-Barr Virus Infection and Associated Cancers
Abstract
Simple Summary
Abstract
1. Introduction
2. Epigenetic Regulation in Latent EBV Infection
2.1. DNA Methylation Is a Major Epigenetic Regulator of the EBV Genome
2.2. EBV Modulates Genome Accessibility by Associating with Histone Modifications
2.3. Chromatin Remodeling of Latent EBV Affects Viral Promoter Activity and Limits the Spread of Histone Modifications across the Genome
2.4. EBV microRNAs Modulate Host Gene Expression to Promote Viral Persistence
3. Host Virus Interactions and Epigenetic Hallmarks in EBV-Associated Tumorigenesis
3.1. Nasopharyngeal Carcinoma (NPC)
3.2. EBV-Associated Gastric Cancers (EBVaGC)
3.3. Burkitt Lymphoma (BL)
4. Future Research Directions
4.1. Genetic Evolution of EBV Latent Proteins and Polymerases
4.2. EBV in Epithelial Cells
4.3. The Road to EBV Therapeutics
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rous, P.; Murphy, J.B. On The Causation by Filterable Agents of Three Distinct Chicken Tumors. J. Exp. Med. 1914, 19, 52–68. [Google Scholar] [CrossRef][Green Version]
- Rous, P.; Murphy, J.B.; Tytler, W.H. A filterable agent the cause of a second chicken-tumor, an osteochondrosarcoma. J. Am. Med. Assoc. 1912, 59, 1793–1794. [Google Scholar] [CrossRef]
- Burkitt, D. A sarcoma involving the jaws in African children. Br. J. Surg. 1958, 46, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Epstein, M.A.; Achong, B.G.; Barr, Y.M. Virus particles in cultured lymphoblasts from burkitt’s lymphoma. Lancet 1964, 1, 702–703. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Kaul, R.; Murakami, M.; Robertson, E.S. Tumor viruses and cancer biology. Cancer Biol. Ther. 2010, 10, 961–978. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.C.; Robertson, E.S. Molecular biology and pathogenesis of Kaposi sarcoma-associated herpesvirus. FEMS Microbiol. Lett. 2003, 222, 155–163. [Google Scholar] [CrossRef]
- Davison, A.J.; Eberle, R.; Ehlers, B.; Hayward, G.S.; McGeoch, D.J.; Minson, A.C.; Pellett, P.E.; Roizman, B.; Studdert, M.J.; Thiry, E. The order Herpesvirales. Arch. Virol. 2009, 154, 171–177. [Google Scholar] [CrossRef]
- Santpere, G.; Darre, F.; Blanco, S.; Alcami, A.; Villoslada, P.; Mar Albà, M.; Navarro, A. Genome-Wide Analysis of Wild-Type Epstein–Barr Virus Genomes Derived from Healthy Individuals of the 1000 Genomes Project. Genome Biol. Evol. 2014, 6, 846–860. [Google Scholar] [CrossRef]
- Kwok, H.; Tong, A.H.Y.; Lin, C.H.; Lok, S.; Farrell, P.J.; Kwong, D.L.W.; Chiang, A.K.S. Genomic Sequencing and Comparative Analysis of Epstein-Barr Virus Genome Isolated from Primary Nasopharyngeal Carcinoma Biopsy. PLoS ONE 2012, 7, e36939. [Google Scholar] [CrossRef]
- Dolan, A.; Addison, C.; Gatherer, D.; Davison, A.J.; McGeoch, D.J. The genome of Epstein–Barr virus type 2 strain AG876. Virology 2006, 350, 164–170. [Google Scholar] [CrossRef]
- Smatti, M.K.; Al-Sadeq, D.W.; Ali, N.H.; Pintus, G.; Abou-Saleh, H.; Nasrallah, G.K. Epstein–Barr Virus Epidemiology, Serology, and Genetic Variability of LMP-1 Oncogene Among Healthy Population: An Update. Front. Oncol. 2018, 8, 211. [Google Scholar] [CrossRef]
- Zimmermann, J.; Hammerschmidt, W. Structure and role of tahe terminal repeats of Epstein-Barr virus in processing and packaging of virion DNA. J. Virol. 1995, 69, 3147–3155. [Google Scholar] [CrossRef]
- Rivailler, P.; Cho, Y.; Wang, F. Complete Genomic Sequence of an Epstein-Barr Virus-Related Herpesvirus Naturally Infecting a New World Primate: A Defining Point in the Evolution of Oncogenic Lymphocryptoviruses. J. Virol. 2002, 76, 12055–12068. [Google Scholar] [CrossRef] [PubMed]
- Kuri, A.; Jacobs, B.M.; Vickaryous, N.; Pakpoor, J.; Middeldorp, J.; Giovannoni, G.; Dobson, R. Epidemiology of Epstein-Barr virus infection and infectious mononucleosis in the United Kingdom. BMC Public Health 2020, 20, 912. [Google Scholar] [CrossRef]
- Fugl, A.; Andersen, C.L. Epstein-Barr virus and its association with disease—A review of relevance to general practice. BMC Fam. Pract. 2019, 20, 62. [Google Scholar] [CrossRef]
- Soldan, S.S.; Lieberman, P.M. Epstein–Barr virus and multiple sclerosis. Nat. Rev. Microbiol. 2023, 21, 51–64. [Google Scholar] [CrossRef]
- Bjornevik, K.; Münz, C.; Cohen, J.I.; Ascherio, A. Epstein–Barr virus as a leading cause of multiple sclerosis: Mechanisms and implications. Nat. Rev. Neurol. 2023, 19, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.D.; Alghareeb, R.; Hussain, A.; Maheshwari, M.V.; Khalid, N. The Association of Epstein-Barr Virus with Cancer. Cureus 2022, 14, e26314. [Google Scholar] [CrossRef]
- Rowe, M.; Lear, A.L.; Croom-Carter, D.; Davies, A.H.; Rickinson, A.B. Three pathways of Epstein-Barr virus gene activation from EBNA1-positive latency in B lymphocytes. J. Virol. 1992, 66, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Tempera, I.; Wiedmer, A.; Dheekollu, J.; Lieberman, P.M. CTCF Prevents the Epigenetic Drift of EBV Latency Promoter Qp. PLoS Pathog. 2010, 6, e1001048. [Google Scholar] [CrossRef]
- Price, A.M.; Luftig, M.A. Dynamic Epstein-Barr Virus Gene Expression on the Path to B-Cell Transformation. Adv. Virus Res. 2014, 88, 279–313. [Google Scholar]
- Sears, J.; Ujihara, M.; Wong, S.; Ott, C.; Middeldorp, J.; Aiyar, A. The Amino Terminus of Epstein-Barr Virus (EBV) Nuclear Antigen 1 Contains AT Hooks That Facilitate the Replication and Partitioning of Latent EBV Genomes by Tethering Them to Cellular Chromosomes. J. Virol. 2004, 78, 11487–11505. [Google Scholar] [CrossRef]
- Price, A.M.; Luftig, M.A. To Be or Not IIb: A Multi-Step Process for Epstein-Barr Virus Latency Establishment and Consequences for B Cell Tumorigenesis. PLoS Pathog. 2015, 11, e1004656. [Google Scholar] [CrossRef]
- Babcock, G.J.; Hochberg, D.; Thorley-Lawson, D.A. The Expression Pattern of Epstein-Barr Virus Latent Genes In Vivo is Dependent upon the Differentiation Stage of the Infected B Cell. Immunity 2000, 13, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Skalsky, R.L.; Cullen, B.R. EBV Noncoding RNAs. Curr. Top. Microbiol. Immunol. 2015, 391, 181–217. [Google Scholar] [PubMed]
- Henle, W.; Henle, G.; Lennette, E.T. The Epstein-Barr Virus. Sci. Am. 1979, 241, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Di Pietro, A. Epstein–Barr Virus Promotes B Cell Lymphomas by Manipulating the Host Epigenetic Machinery. Cancers 2020, 12, 3037. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.S.; Kieff, E. Epstein–Barr virus latent genes. Exp. Mol. Med. 2015, 47, e131. [Google Scholar] [CrossRef] [PubMed]
- Reisman, D.; Yates, J.; Sugden, B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol. Cell Biol. 1985, 5, 1822–1832. [Google Scholar] [PubMed]
- Yates, J.L.; Warren, N.; Sugden, B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 1985, 313, 812–815. [Google Scholar] [CrossRef]
- Harada, S.; Kieff, E. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J. Virol. 1997, 71, 6611–6618. [Google Scholar] [CrossRef]
- Henkel, T.; Ling, P.D.; Hayward, S.D.; Peterson, M.G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science 1994, 265, 92–95. [Google Scholar] [CrossRef]
- Zhao, B.; Zou, J.; Wang, H.; Johannsen, E.; Peng, C.; Quackenbush, J.; Mar, J.C.; Morton, C.C.; Freedman, M.L.; Blacklow, S.C.; et al. Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc. Natl. Acad. Sci. USA 2011, 108, 14902–14907. [Google Scholar] [CrossRef]
- Harth-Hertle, M.L.; Scholz, B.A.; Erhard, F.; Glaser, L.V.; Dölken, L.; Zimmer, R.; Kempkes, B. Inactivation of Intergenic Enhancers by EBNA3A Initiates and Maintains Polycomb Signatures across a Chromatin Domain Encoding CXCL10 and CXCL9. PLoS Pathog. 2013, 9, e1003638. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Sample, C.E. Epstein-barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an spi-1/Spi-B binding site. J. Virol. 2000, 74, 5151–5160. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Lu, J.; Cai, Q.; Saha, A.; Jha, H.C.; Dzeng, R.K.; Robertson, E.S. The EBV Latent Antigen 3C Inhibits Apoptosis through Targeted Regulation of Interferon Regulatory Factors 4 and 8. PLoS Pathog. 2013, 9, e1003314. [Google Scholar] [CrossRef]
- Mancao, C.; Altmann, M.; Jungnickel, B.; Hammerschmidt, W. Rescue of “crippled” germinal center B cells from apoptosis by Epstein-Barr virus. Blood 2005, 106, 4339–4344. [Google Scholar] [CrossRef] [PubMed]
- Incrocci, R.; McCormack, M.; Swanson-Mungerson, M. Epstein-Barr virus LMP2A increases IL-10 production in mitogen-stimulated primary B-cells and B-cell lymphomas. J. Gen. Virol. 2013, 94 Pt 5, 1127–1133. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.L.; Burkhardt, A.L.; Lee, J.H.; Stealey, B.; Longnecker, R.; Bolen, J.B.; Kieff, E. Integral membrane protein 2 of Epstein-Barr virus regulates reactivation from latency through dominant negative effects on protein-tyrosine kinases. Immunity 1995, 2, 155–166. [Google Scholar] [CrossRef]
- Wong, Y.; Meehan, M.T.; Burrows, S.R.; Doolan, D.L.; Miles, J.J. Estimating the global burden of Epstein–Barr virus-related cancers. J. Cancer Res. Clin. Oncol. 2022, 148, 31–46. [Google Scholar] [CrossRef]
- Cohen, J.I. Primary Immunodeficiencies Associated with EBV Disease. Curr. Top. Microbiol. Immunol. 2015, 390 Pt 1, 241–265. [Google Scholar] [PubMed]
- Tao, Q.; Robertson, K.D.; Manns, A.; Hildesheim, A.; Ambinder, R.F. Epstein-Barr virus (EBV) in endemic Burkitt’s lymphoma: Molecular analysis of primary tumor tissue. Blood 1998, 91, 1373–1381. [Google Scholar] [CrossRef] [PubMed]
- Coghill, A.E.; McGuire, A.; Sinha, S.; Homad, L.; Sinha, I.; Sholukh, A.; Koh, W.P.; Yuan, J.M. Epstein-Barr Virus Glycoprotein Antibody Titers and Risk of Nasopharyngeal Carcinoma. Open Forum Infect. Dis. 2022, 9, ofac635. [Google Scholar] [CrossRef] [PubMed]
- Hashmi, A.A.; Hussain, Z.F.; Hashmi, K.A.; Zafar, M.I.; Edhi, M.M.; Faridi, N.; Khan, M. Latent membrane protein 1 (LMP1) expression in Hodgkin lymphoma and its correlation with clinical and histologic parameters. World J. Surg. Oncol. 2017, 15, 89. [Google Scholar] [CrossRef] [PubMed]
- Levin, L.I.; Chang, E.T.; Ambinder, R.F.; Lennette, E.T.; Rubertone, M.V.; Mann, R.B.; Borowitz, M.; Weir, E.G.; Abbondanzo, S.L.; Mueller, N.E. Atypical prediagnosis Epstein-Barr virus serology restricted to EBV-positive Hodgkin lymphoma. Blood 2012, 120, 3750. [Google Scholar] [CrossRef] [PubMed]
- Tierney, R.J.; Kirby, H.E.; Nagra, J.K.; Desmond, J.; Bell, A.I.; Rickinson, A.B. Methylation of Transcription Factor Binding Sites in the Epstein-Barr Virus Latent Cycle Promoter Wp Coincides with Promoter Down-Regulation during Virus-Induced B-Cell Transformation. J. Virol. 2000, 74, 10468–10479. [Google Scholar] [CrossRef][Green Version]
- Zhang, L.; Wang, R.; Xie, Z. The roles of DNA methylation on the promotor of the Epstein–Barr virus (EBV) gene and the genome in patients with EBV-associated diseases. Appl. Microbiol. Biotechnol. 2022, 106, 4413. [Google Scholar] [CrossRef]
- Kempkes, B.; Ling, P.D. EBNA2 and Its Coactivator EBNA-LP. Curr. Top. Microbiol. Immunol. 2015, 391, 35–59. [Google Scholar]
- Alfieri, C.; Birkenbach, M.; Kieff, E. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 1991, 181, 595–608. [Google Scholar] [CrossRef]
- Kirby, H.; Rickinson, A.; Bell, A. The activity of the Epstein-Barr virus BamHI W promoter in B cells is dependent on the binding of CREB/ATF factors. J. Gen. Virol. 2000, 81 Pt 4, 1057–1066. [Google Scholar] [CrossRef]
- Tierney, R.; Kirby, H.; Nagra, J.; Rickinson, A.; Bell, A. The Epstein-Barr Virus Promoter Initiating B-Cell Transformation is Activated by RFX Proteins and the B-Cell-Specific Activator Protein BSAP/Pax5. J. Virol. 2000, 74, 10458. [Google Scholar] [CrossRef][Green Version]
- Schaefer, B.C.; Strominger, J.L.; Speck, S.H. Host-cell-determined methylation of specific Epstein-Barr virus promoters regulates the choice between distinct viral latency programs. Mol. Cell Biol. 1997, 17, 364–377. [Google Scholar] [CrossRef][Green Version]
- Salamon, D.; Takacs, M.; Ujvari, D.; Uhlig, J.; Wolf, H.; Minarovits, J.; Niller, H.H. Protein-DNA Binding and CpG Methylation at Nucleotide Resolution of Latency-Associated Promoters Qp, Cp, and LMP1p of Epstein-Barr Virus. J. Virol. 2001, 75, 2584–2596. [Google Scholar] [CrossRef]
- Robertson, K.D.; Hayward, S.D.; Ling, P.D.; Samid, D.; Ambinder, R.F. Transcriptional Activation of the Epstein-Barr Virus Latency C Promoter after 5-Azacytidine Treatment: Evidence that Demethylation at a Single CpG Site Is Crucial. Mol. Cell Biol. 1995, 15, 6150–6159. [Google Scholar] [CrossRef]
- Lu, F.; Wiedmer, A.; Martin, K.A.; Wickramasinghe, P.J.M.S.; Kossenkov, A.V.; Lieberman, P.M. Coordinate Regulation of TET2 and EBNA2 Controls the DNA Methylation State of Latent Epstein-Barr Virus. J. Virol. 2017, 91, e00804-17. [Google Scholar] [CrossRef]
- Hughes, D.J.; Marendy, E.M.; Dickerson, C.A.; Yetming, K.D.; Sample, C.E.; Sample, J.T. Contributions of CTCF and DNA Methyltransferases DNMT1 and DNMT3B to Epstein-Barr Virus Restricted Latency. J. Virol. 2012, 86, 1034–1045. [Google Scholar] [CrossRef] [PubMed]
- Tao, Q.; Robertson, K.D.; Manns, A.; Hildesheim, A.; Ambinder, R.F. The Epstein-Barr Virus Major Latent Promoter Qp is Constitutively Active, Hypomethylated, and Methylation Sensitive. J. Virol. 1998, 72, 7075–7083. [Google Scholar] [CrossRef] [PubMed]
- Paulson, E.J.; Speck, S.H. Differential Methylation of Epstein-Barr Virus Latency Promoters Facilitates Viral Persistence in Healthy Seropositive Individuals. J. Virol. 1999, 73, 9959–9968. [Google Scholar] [CrossRef]
- Takacs, M.; Banati, F.; Koroknai, A.; Segesdi, J.; Salamon, D.; Wolf, H.; Niller, H.H.; Minarovits, J. Epigenetic regulation of latent Epstein–Barr virus promoters. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2010, 1799, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Sample, J.; Henson, E.B.; Sample, C. The Epstein-Barr virus nuclear protein 1 promoter active in type I latency is autoregulated. J. Virol. 1992, 66, 4654–4661. [Google Scholar] [CrossRef] [PubMed]
- Bristol, J.A.; Djavadian, R.; Albright, E.R.; Coleman, C.B.; Ohashi, M.; Hayes, M.; Romero-Masters, J.C.; Barlow, E.A.; Farrell, P.J.; Rochford, R.; et al. A cancer-associated Epstein-Barr virus BZLF1 promoter variant enhances lytic infection. PLoS Pathog. 2018, 14, e1007179. [Google Scholar] [CrossRef]
- Brink, A.A.T.P.; Meijer, C.J.L.M.; Nicholls, J.M.; Middeldorp, J.M.; van den Brule, A.J.C. Activity of the EBNA1 promoter associated with lytic replication (Fp) in Epstein-Barr virus associated disorders. Mol. Pathol. 2001, 54, 98–102. [Google Scholar] [CrossRef]
- Zetterberg, H.; Stenglein, M.; Jansson, A.; Ricksten, A.; Rymo, L. Relative levels of EBNA1 gene transcripts from the C/W, F and Q promoters in Epstein-Barr virus-transformed lymphoid cells in latent and lytic stages of infection. J. Gen. Virol. 1999, 80, 457–466. [Google Scholar] [CrossRef][Green Version]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 2016, 32, 42–56. [Google Scholar] [CrossRef]
- Miller, J.L.; Grant, P.A. The Role of DNA Methylation and Histone Modifications in Transcriptional Regulation in Humans. Subcell. Biochem. 2013, 61, 289. [Google Scholar]
- Millán-Zambrano, G.; Burton, A.; Bannister, A.J.; Schneider, R. Histone post-translational modifications—Cause and consequence of genome function. Nat. Rev. Genet. 2022, 23, 563–580. [Google Scholar] [CrossRef]
- Cheung, A.K.L.; Ip, J.C.Y.; Chu, A.C.H.; Cheng, Y.; Leong, M.M.L.; Ko, J.M.Y.; Shuen, W.H.; Lung, H.L.; Lung, M.L. PTPRG suppresses tumor growth and invasion via inhibition of Akt signaling in nasopharyngeal carcinoma. Oncotarget 2015, 6, 13434–13447. [Google Scholar] [CrossRef] [PubMed]
- Leong, M.M.L.; Lung, M.L. The Impact of Epstein-Barr Virus Infection on Epigenetic Regulation of Host Cell Gene Expression in Epithelial and Lymphocytic Malignancies. Front. Oncol. 2021, 11, 629780. [Google Scholar] [CrossRef] [PubMed]
- Funata, S.; Matsusaka, K.; Yamanaka, R.; Yamamoto, S.; Okabe, A.; Fukuyo, M.; Aburatani, H.; Fukayama, M.; Kaneda, A. Histone modification alteration coordinated with acquisition of promoter DNA methylation during Epstein-Barr virus infection. Oncotarget 2017, 8, 55265–55279. [Google Scholar] [CrossRef]
- Buschle, A.; Hammerschmidt, W. Epigenetic lifestyle of Epstein-Barr virus. Semin. Immunopathol. 2020, 42, 131–142. [Google Scholar] [CrossRef]
- Caruso, L.B.; Maestri, D.; Tempera, I. Three-Dimensional Chromatin Structure of the EBV Genome: A Crucial Factor in Viral Infection. Viruses 2023, 15, 1088. [Google Scholar] [CrossRef]
- Arvey, A.; Tempera, I.; Tsai, K.; Chen, H.S.; Tikhmyanova, N.; Klichinsky, M.; Leslie, C.; Lieberman, P.M. An Atlas of the Epstein-Barr Virus Transcriptome and Epigenome Reveals Host-Virus Regulatory Interactions. Cell Host Microbe 2012, 12, 233–245. [Google Scholar] [CrossRef]
- Tempera, I.; Lieberman, P.M. Epigenetic Regulation of EBV Persistence and Oncogenesis. Semin. Cancer Biol. 2014, 26, 22–29. [Google Scholar] [CrossRef]
- Arvey, A.; Tempera, I.; Lieberman, P.M. Interpreting the Epstein-Barr Virus (EBV) Epigenome Using High-Throughput Data. Viruses 2013, 5, 1042–1054. [Google Scholar] [CrossRef]
- Gerle, B.; Koroknai, A.; Fejer, G.; Bakos, A.; Banati, F.; Szenthe, K.; Wolf, H.; Niller, H.H.; Minarovits, J.; Salamon, D. Acetylated Histone H3 and H4 Mark the Upregulated LMP2A Promoter of Epstein-Barr Virus in Lymphoid Cells. J. Virol. 2007, 81, 13242–13247. [Google Scholar] [CrossRef]
- Day, L.; Chau, C.M.; Nebozhyn, M.; Rennekamp, A.J.; Showe, M.; Lieberman, P.M. Chromatin Profiling of Epstein-Barr Virus Latency Control Region. J. Virol. 2007, 81, 6389. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Kim, K.D.; Cho, M.; Huh, S.; An, S.H.; Seo, D.; Kang, K.; Lee, M.; Tanizawa, H.; Jung, I.; et al. Characterization of a new CCCTC-binding factor binding site as a dual regulator of Epstein-Barr virus latent infection. PLoS Pathog. 2023, 19, e1011078. [Google Scholar] [CrossRef] [PubMed]
- Tempera, I.; Klichinsky, M.; Lieberman, P.M. EBV Latency Types Adopt Alternative Chromatin Conformations. PLoS Pathog. 2011, 7, 1002180. [Google Scholar] [CrossRef]
- Chong, M.M.W.; Zhang, G.; Cheloufi, S.; Neubert, T.A.; Hannon, G.J.; Littman, D.R. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010, 24, 1951–1960. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Y.; Pfuhl, T.; Motsch, N.; Barth, S.; Nicholls, J.; Grässer, F.; Meister, G. Identification of Novel Epstein-Barr Virus MicroRNA Genes from Nasopharyngeal Carcinomas. J. Virol. 2009, 83, 3333–3341. [Google Scholar] [CrossRef]
- Grundhoff, A.; Sullivan, C.S.; Ganem, D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. Rna 2006, 12, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Lo, A.K.F.; To, K.F.; Lo, K.W.; Lung, R.W.M.; Hui, J.W.Y.; Liao, G.; Hayward, S.D. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc. Natl. Acad. Sci. USA 2007, 104, 16164–16169. [Google Scholar] [CrossRef] [PubMed]
- Amoroso, R.; Fitzsimmons, L.; Thomas, W.A.; Kelly, G.L.; Rowe, M.; Bell, A.I. Quantitative Studies of Epstein-Barr Virus-Encoded MicroRNAs Provide Novel Insights into Their Regulation. J. Virol. 2011, 85, 996–1010. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.J.; Chen, G.H.; Chen, Y.H.; Liu, C.Y.; Chang, K.P.; Chang, Y.S.; Chen, H.C. Characterization of Epstein-Barr Virus miRNAome in Nasopharyngeal Carcinoma by Deep Sequencing. PLoS ONE 2010, 5, e12745. [Google Scholar] [CrossRef] [PubMed]
- Barth, S.; Meister, G.; Grässer, F.A. EBV-encoded miRNAs. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2011, 1809, 631–640. [Google Scholar] [CrossRef]
- Fan, C.; Tang, Y.; Wang, J.; Xiong, F.; Guo, C.; Wang, Y.; Xiang, B.; Zhou, M.; Li, X.; Wu, X.; et al. The emerging role of Epstein-Barr virus encoded microRNAs in nasopharyngeal carcinoma. J. Cancer 2018, 9, 2852–2864. [Google Scholar] [CrossRef]
- De Re, V.; Caggiari, L.; De Zorzi, M.; Fanotto, V.; Miolo, G.; Puglisi, F.; Cannizzaro, R.; Canzonieri, V.; Steffan, A.; Farruggia, P.; et al. Epstein-Barr virus BART microRNAs in EBV- associated Hodgkin lymphoma and gastric cancer. Infect. Agents Cancer 2020, 15, 42. [Google Scholar] [CrossRef]
- Gorbea, C.; Elhakiem, A.; Cazalla, D. Allosteric regulation of noncoding RNA function by microRNAs. Nucleic Acids Res. 2022, 50, 6511–6520. [Google Scholar] [CrossRef]
- Pernitzsch, S.R.; Tirier, S.M.; Beier, D.; Sharma, C.M. A variable homopolymeric G-repeat defines small RNA-mediated posttranscriptional regulation of a chemotaxis receptor in Helicobacter pylori. Proc. Natl. Acad. Sci. USA 2014, 111, E501–E510. [Google Scholar] [CrossRef]
- Forte, E.; Luftig, M.A. The role of microRNAs in Epstein-Barr virus latency and lytic reactivation. Microbes Infect. Inst. Pasteur. 2011, 13, 1156–1167. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; O’Hara, A.; Araujo, I.; Barreto, J.; Carvalho, E.; Sapucaia, J.B.; Ramos, J.C.; Luz, E.; Pedroso, C.; Manrique, M.; et al. EBV MicroRNAs in Primary Lymphomas and Targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008, 68, 1436. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Schäfer, A.; Lu, S.; Bilello, J.P.; Desrosiers, R.C.; Edwards, R.; Raab-Traub, N.; Cullen, B.R. Epstein–Barr Virus MicroRNAs Are Evolutionarily Conserved and Differentially Expressed. PLoS Pathog. 2006, 2, e23. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Cosmopoulos, K.; Pegtel, M.; Hopmans, E.; Murray, P.; Middeldorp, J.; Shapiro, M.; Thorley-Lawson, D.A. A Novel Persistence Associated EBV miRNA Expression Profile is Disrupted in Neoplasia. PLoS Pathog. 2011, 7, e1002193. [Google Scholar] [CrossRef] [PubMed]
- Cosmopoulos, K.; Pegtel, M.; Hawkins, J.; Moffett, H.; Novina, C.; Middeldorp, J.; Thorley-Lawson, D.A. Comprehensive Profiling of Epstein-Barr Virus MicroRNAs in Nasopharyngeal Carcinoma. J. Virol. 2009, 83, 2357–2367. [Google Scholar] [CrossRef]
- Klinke, O.; Feederle, R.; Delecluse, H.J. Genetics of Epstein–Barr virus microRNAs. Semin. Cancer Biol. 2014, 26, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Motsch, N.; Alles, J.; Imig, J.; Zhu, J.; Barth, S.; Reineke, T.; Tinguely, M.; Cogliatti, S.; Dueck, A.; Meister, G.; et al. MicroRNA Profiling of Epstein-Barr Virus-Associated NK/T-Cell Lymphomas by Deep Sequencing. PLoS ONE 2012, 7, e42193. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Okuno, Y.; Sato, Y.; Watanabe, T.; Murata, T. Deletion of Viral microRNAs in the Oncogenesis of Epstein–Barr Virus-Associated Lymphoma. Front. Microbiol. 2021, 12, 667968. [Google Scholar] [CrossRef]
- Edwards, R.H.; Marquitz, A.R.; Raab-Traub, N. Epstein-Barr Virus BART MicroRNAs Are Produced from a Large Intron prior to Splicing. J. Virol. 2008, 82, 9094–9106. [Google Scholar] [CrossRef]
- Pfeffer, S.; Zavolan, M.; Grässer, F.A.; Chien, M.; Russo, J.J.; Ju, J.; John, B.; Enright, A.J.; Marks, D.; Sander, C.; et al. Identification of Virus-Encoded MicroRNAs. Science 2004, 304, 734–736. [Google Scholar] [CrossRef]
- Lung, R.W.M.; Tong, J.H.M.; Sung, Y.M.; Leung, P.S.; Ng, D.C.H.; Chau, S.L.; Chan, A.W.H.; Ng, E.K.O.; Lo, K.W.; To, K.F. Modulation of LMP2A Expression by a Newly Identified Epstein-Barr Virus-Encoded MicroRNA miR-BART22. Neoplasia 2009, 11, 1174–1184. [Google Scholar] [CrossRef]
- Nachmani, D.; Stern-Ginossar, N.; Sarid, R.; Mandelboim, O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 2009, 5, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Skalsky, R.L.; Corcoran, D.L.; Gottwein, E.; Frank, C.L.; Kang, D.; Hafner, M.; Nusbaum, J.D.; Feederle, R.; Delecluse, H.J.; Luftig, M.A.; et al. The Viral and Cellular MicroRNA Targetome in Lymphoblastoid Cell Lines. PLoS Pathog. 2012, 8, e1002484. [Google Scholar] [CrossRef] [PubMed]
- Skalsky, R.L.; Kang, D.; Linnstaedt, S.D.; Cullen, B.R. Evolutionary Conservation of Primate Lymphocryptovirus MicroRNA Targets. J. Virol. 2014, 88, 1617–1635. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Zeng, Z.; Gong, Z.; Zhang, W.; Li, X.; He, B.; Song, Y.; Li, Q.; Zeng, Y.; Liao, Q.; et al. EBV-miR-BART10-3p facilitates epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma by targeting BTRC. Oncotarget 2015, 6, 41766–41782. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Baccianti, F.; Delecluse, S.; Tsai, M.H.; Shumilov, A.; Cheng, X.; Ma, S.; Hoffmann, I.; Poirey, R.; Delecluse, H.J. The Epstein–Barr virus noncoding RNA EBER2 transactivates the UCHL1 deubiquitinase to accelerate cell growth. Proc. Natl. Acad. Sci. USA 2021, 118, e2115508118. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, D.; Kieffer-Kwon, P.; Ziegelbauer, J.M. Emerging Themes from EBV and KSHV microRNA Targets. Viruses 2012, 4, 1687–1710. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fachko, D.; Ivanov, N.S.; Skinner, C.M.; Skalsky, R.L. Epstein-Barr virus microRNAs regulate B cell receptor signal transduction and lytic reactivation. PLoS Pathog. 2019, 15, e1007535. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Qin, Z.; Wang, J.; Zheng, X.; Lu, J.; Zhang, X.; Wei, L.; Peng, Q.; Zheng, Y.; Ou, C.; et al. Epstein-Barr Virus miR-BART6-3p Inhibits the RIG-I Pathway. J. Innate Immun. 2017, 9, 574–586. [Google Scholar] [CrossRef]
- Huang, W.T.; Lin, C.W. EBV-Encoded miR-BART20-5p and miR-BART8 Inhibit the IFN-γ–STAT1 Pathway Associated with Disease Progression in Nasal NK-Cell Lymphoma. Am. J. Pathol. 2014, 184, 1185–1197. [Google Scholar] [CrossRef]
- Hooykaas, M.J.G.; van Gent, M.; Soppe, J.A.; Kruse, E.; Boer, I.G.J.; van Leenen, D.; Groot Koerkamp, M.J.A.; Holstege, F.C.P.; Ressing, M.E.; Wiertz, E.J.H.J.; et al. EBV MicroRNA BART16 Suppresses Type I IFN Signaling. J. Immunol. 2017, 198, 4062–4073. [Google Scholar] [CrossRef] [PubMed]
- Haneklaus, M.; Gerlic, M.; Kurowska-Stolarska, M.; Rainey, A.A.; Pich, D.; McInnes, I.B.; Hammerschmidt, W.; O’Neill, L.A.J.; Masters, S.L. Cutting Edge: miR-223 and EBV miR-BART15 Regulate the NLRP3 Inflammasome and IL-1β Production. J. Immunol. 2012, 189, 3795–3799. [Google Scholar] [CrossRef] [PubMed]
- Marquitz, A.R.; Mathur, A.; Nam, C.S.; Raab-Traub, N. The Epstein-Barr Virus BART microRNAs target the pro-apoptotic protein Bim. Virology 2011, 412, 392–400. [Google Scholar] [CrossRef]
- Czabotar, P.E.; Lessene, G.; Strasser, A.; Adams, J.M. Control of apoptosis by the BCL-2 protein family: Implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 2014, 15, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Zhang, M.; Li, Q.; Xiong, H.; Liu, S.; Fang, W.; Zhang, Q.; Liu, Z.; Xu, X.; Jiang, Q. The Epstein-Barr Virus-encoded miR-BART22 targets MAP3K5 to promote host cell proliferative and invasive abilities in nasopharyngeal carcinoma. J. Cancer 2017, 8, 305–313. [Google Scholar] [CrossRef]
- Harold, C.; Cox, D.; Riley, K.J. Epstein-Barr viral microRNAs target caspase 3. Virol. J. 2016, 13, 145. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Zheng, H.; Cheung, A.K.L.; Lung, M.L. Genetic and epigenetic landscape of nasopharyngeal carcinoma. Chin. Clin. Oncol. 2016, 5, 16. [Google Scholar] [CrossRef]
- Nishikawa, J.; Iizasa, H.; Yoshiyama, H.; Nakamura, M.; Saito, M.; Sasaki, S.; Shimokuri, K.; Yanagihara, M.; Sakai, K.; Suehiro, Y.; et al. The Role of Epigenetic Regulation in Epstein-Barr Virus-Associated Gastric Cancer. Int. J. Mol. Sci. 2017, 18, 1606. [Google Scholar] [CrossRef]
- Marques-Piubelli, M.L.; Salas, Y.I.; Pachas, C.; Becker-Hecker, R.; Vega, F.; Miranda, R.N. Epstein-Barr virus-associated B-cell lymphoproliferative disorders and lymphomas: A review. Pathology 2020, 52, 40–52. [Google Scholar] [CrossRef]
- Syrykh, C.; Péricart, S.; Lamaison, C.; Escudié, F.; Brousset, P.; Laurent, C. Epstein-Barr Virus-Associated T- and NK-Cell Lymphoproliferative Diseases: A Review of Clinical and Pathological Features. Cancers 2021, 13, 3315. [Google Scholar] [CrossRef]
- Tse, E.; Kwong, Y.L. Epstein Barr virus-associated lymphoproliferative diseases: The virus as a therapeutic target. Exp. Mol. Med. 2015, 47, e136. [Google Scholar] [CrossRef]
- Wong, K.C.W.; Hui, E.P.; Lo, K.W.; Lam, W.K.J.; Johnson, D.; Li, L.; Tao, Q.; Chan, K.C.A.; To, K.F.; King, A.D.; et al. Nasopharyngeal carcinoma: An evolving paradigm. Nat. Rev. Clin. Oncol. 2021, 18, 679–695. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Bei, J.X.; Jia, W.H.; Zeng, Y.X. Familial and large-scale case–control studies identify genes associated with nasopharyngeal carcinoma. Semin. Cancer Biol. 2012, 22, 96–106. [Google Scholar] [CrossRef]
- Xu, M.; Yao, Y.; Chen, H.; Zhang, S.; Cao, S.M.; Zhang, Z.; Luo, B.; Liu, Z.; Li, Z.; Xiang, T.; et al. Genome sequencing analysis identifies Epstein–Barr virus subtypes associated with high risk of nasopharyngeal carcinoma. Nat. Genet. 2019, 51, 1131–1136. [Google Scholar] [CrossRef]
- Huang, S.Y.; Wu, C.C.; Cheng, Y.J.; Chou, S.P.; Jiang, Y.J.; Chu, K.C.; Tsai, C.H.; Lin, S.F.; Chen, J.Y. Epstein–Barr virus BRLF1 induces genomic instability and progressive malignancy in nasopharyngeal carcinoma cells. Oncotarget 2017, 8, 78948–78964. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.Y.; Siak, P.Y.; Leong, C.O.; Cheah, S.C. The role of Epstein–Barr virus in nasopharyngeal carcinoma. Front. Microbiol. 2023, 14, 1116143. [Google Scholar] [CrossRef] [PubMed]
- Choy, E.Y.W.; Siu, K.L.; Kok, K.H.; Lung, R.W.M.; Tsang, C.M.; To, K.F.; Kwong, D.L.W.; Tsao, S.W.; Jin, D.Y. An Epstein-Barr virus–encoded microRNA targets PUMA to promote host cell survival. J. Exp. Med. 2008, 205, 2551–2560. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.C.; Hsiao, J.R.; Chen, C.W.; Wu, S.Y.; Lee, C.H.; Su, I.J.; Takada, K.; Chang, Y. Endogenous latent membrane protein 1 in Epstein-Barr virus-infected nasopharyngeal carcinoma cells attracts T lymphocytes through upregulation of multiple chemokines. Virology 2010, 405, 464–473. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Duan, Y.; Cheng, S.; Chen, Y.; Hu, Y.; Zhang, L.; He, J.; Liao, Q.; Yang, L.; Sun, L.Q. EBV-encoded RNA via TLR3 induces inflammation in nasopharyngeal carcinoma. Oncotarget 2015, 6, 24291–24303. [Google Scholar] [CrossRef] [PubMed]
- Huo, S.; Luo, Y.; Deng, R.; Liu, X.; Wang, J.; Wang, L.; Zhang, B.; Wang, F.; Lu, J.; Li, X. EBV-EBNA1 constructs an immunosuppressive microenvironment for nasopharyngeal carcinoma by promoting the chemoattraction of Treg cells. J. Immunother. Cancer 2020, 8, e001588. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Luo, Y.; Bi, P.; Lu, J.; Wang, F.; Liu, X.; Zhang, B.; Li, X. Mechanisms of Epstein-Barr virus nuclear antigen 1 favor Tregs accumulation in nasopharyngeal carcinoma. Cancer Med. 2020, 9, 5598–5608. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Feng, L.; Zhang, S.; Zhang, H.; Zhang, X.; Qi, X.; Zhang, Y.; Feng, Q.; Xiang, T.; Zeng, Y. Induction of chemokine (C-C motif) ligand 5 by Epstein–Barr virus infection enhances tumor angiogenesis in nasopharyngeal carcinoma. Cancer Sci. 2018, 109, 1710–1722. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, J.D.; Owen, T.J.; Wood, V.H.J.; Date, K.L.; Valentine, R.; Chukwuma, M.B.; Arrand, J.R.; Dawson, C.W.; Young, L.S. Epstein-Barr virus-encoded EBNA1 modulates the AP-1 transcription factor pathway in nasopharyngeal carcinoma cells and enhances angiogenesis in vitro. J. Gen. Virol. 2008, 89 Pt 11, 2833–2842. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Li, Z.; He, J.; Fu, S.; Duan, Y.; Zhou, Q.; Yan, Y.; Liu, X.; Liu, L.; Feng, C.; et al. Epstein-Barr virus noncoding RNAs from the extracellular vesicles of nasopharyngeal carcinoma (NPC) cells promote angiogenesis via TLR3/RIG-I-mediated VCAM-1 expression. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 1201–1213. [Google Scholar] [CrossRef]
- Xiang, T.; Lin, Y.X.; Ma, W.; Zhang, H.J.; Chen, K.M.; He, G.P.; Zhang, X.; Xu, M.; Feng, Q.S.; Chen, M.Y.; et al. Vasculogenic mimicry formation in EBV-associated epithelial malignancies. Nat. Commun. 2018, 9, 5009. [Google Scholar] [CrossRef]
- Xu, S.; Bai, J.; Zhuan, Z.; Li, B.; Zhang, Z.; Wu, X.; Luo, X.; Yang, L. EBV-LMP1 is involved in vasculogenic mimicry formation via VEGFA/VEGFR1 signaling in nasopharyngeal carcinoma. Oncol. Rep. 2018, 40, 377–384. [Google Scholar] [CrossRef]
- Wang, J.; Ge, J.; Wang, Y.; Xiong, F.; Guo, J.; Jiang, X.; Zhang, L.; Deng, X.; Gong, Z.; Zhang, S.; et al. EBV miRNAs BART11 and BART17-3p promote immune escape through the enhancer-mediated transcription of PD-L1. Nat. Commun. 2022, 13, 866. [Google Scholar] [CrossRef]
- Fang, W.; Zhang, J.; Hong, S.; Zhan, J.; Chen, N.; Qin, T.; Tang, Y.; Zhang, Y.; Kang, S.; Zhou, T.; et al. EBV-driven LMP1 and IFN-γ up-regulate PD-L1 in nasopharyngeal carcinoma: Implications for oncotargeted therapy. Oncotarget 2014, 5, 12189–12202. [Google Scholar] [CrossRef]
- Hau, P.M.; Lung, H.L.; Wu, M.; Tsang, C.M.; Wong, K.L.; Mak, N.K.; Lo, K.W. Targeting Epstein-Barr Virus in Nasopharyngeal Carcinoma. Front. Oncol. 2020, 10, 600. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, C.; Li, H.; Li, H.; Li, Y. Identifying Key Genes for Nasopharyngeal Carcinoma by Prioritized Consensus Differentially Expressed Genes Caused by Aberrant Methylation. J. Cancer 2021, 12, 874–884. [Google Scholar] [CrossRef]
- Challouf, S.; Ziadi, S.; Zaghdoudi, R.; Ksiaa, F.; Ben Gacem, R.; Trimeche, M. Patterns of aberrant DNA hypermethylation in nasopharyngeal carcinoma in Tunisian patients. Clin. Chim. Acta 2012, 413, 795–802. [Google Scholar] [CrossRef]
- Rotondo, J.C.; Borghi, A.; Selvatici, R.; Mazzoni, E.; Bononi, I.; Corazza, M.; Kussini, J.; Montinari, E.; Gafà, R.; Tognon, M.; et al. Association of Retinoic Acid Receptor β Gene With Onset and Progression of Lichen Sclerosus–Associated Vulvar Squamous Cell Carcinoma. JAMA Dermatol. 2018, 154, 819–823. [Google Scholar] [CrossRef]
- Ka-Yue Chow, L.; Lai-Shun Chung, D.; Tao, L.; Chan, K.F.; Tung, S.Y.; Cheong Ngan, R.K.; Ng, W.T.; Wing-Mui Lee, A.; Yau, C.C.; Lai-Wan Kwong, D.; et al. Epigenomic landscape study reveals molecular subtypes and EBV-associated regulatory epigenome reprogramming in nasopharyngeal carcinoma. EBioMedicine 2022, 86, 104357. [Google Scholar] [CrossRef]
- Cai, M.Y.; Tong, Z.T.; Zhu, W.; Wen, Z.Z.; Rao, H.L.; Kong, L.L.; Guan, X.Y.; Kung, H.F.; Zeng, Y.X.; Xie, D. H3K27me3 Protein Is a Promising Predictive Biomarker of Patients’ Survival and Chemoradioresistance in Human Nasopharyngeal Carcinoma. Mol. Med. 2011, 17, 1137–1145. [Google Scholar] [CrossRef]
- Naseem, M.; Barzi, A.; Brezden-Masley, C.; Puccini, A.; Berger, M.D.; Tokunaga, R.; Battaglin, F.; Soni, S.; McSkane, M.; Zhang, W.; et al. Outlooks on Epstein-Barr virus associated gastric cancer. Cancer Treat. Rev. 2018, 66, 15–22. [Google Scholar] [CrossRef]
- Murphy, G.; Pfeiffer, R.; Camargo, M.C.; Rabkin, C.S. Meta-analysis Shows That Prevalence of Epstein–Barr Virus-Positive Gastric Cancer Differs Based on Sex and Anatomic Location. Gastroenterology 2009, 137, 824–833. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Gao, Y.; Luo, B.; Zhao, Y. Construction and Antiapoptosis Activities of Recombinant Adenoviral Expression Vector Carrying EBV Latent Membrane Protein 2A. Gastroenterol. Res. Pract. 2011, 2011, e182832. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, M.; Imai, S.; Tokunaga, M.; Koizumi, S.; Uchizawa, M.; Okamoto, K.; Osato, T. Transcriptional analysis of Epstein-Barr virus gene expression in EBV-positive gastric carcinoma: Unique viral latency in the tumour cells. Br. J. Cancer 1996, 74, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Hino, R.; Uozaki, H.; Inoue, Y.; Shintani, Y.; Ushiku, T.; Sakatani, T.; Takada, K.; Fukayama, M. Survival Advantage of EBV-Associated Gastric Carcinoma: Survivin Up-regulation by Viral Latent Membrane Protein 2A. Cancer Res. 2008, 68, 1427–1435. [Google Scholar] [CrossRef]
- Mittal, V. Epithelial Mesenchymal Transition in Tumor Metastasis. Annu. Rev. Pathol. Mech. Dis. 2018, 13, 395–412. [Google Scholar] [CrossRef]
- Tsai, C.Y.; Liu, Y.Y.; Liu, K.H.; Hsu, J.T.; Chen, T.C.; Chiu, C.T.; Yeh, T.S. Comprehensive profiling of virus microRNAs of Epstein–Barr virus-associated gastric carcinoma: Highlighting the interactions of ebv-Bart9 and host tumor cells. J. Gastroenterol. Hepatol. 2017, 32, 82–91. [Google Scholar] [CrossRef]
- Park, M.C.; Kim, H.; Choi, H.; Chang, M.S.; Lee, S.K. Epstein-Barr Virus miR-BART1-3p Regulates the miR-17-92 Cluster by Targeting E2F3. Int. J. Mol. Sci. 2021, 22, 10936. [Google Scholar] [CrossRef]
- Liu, T.; Zhou, X.; Zhang, Z.; Qin, Y.; Wang, R.; Qin, Y.; Huang, Y.; Mo, Y.; Huang, T. The role of EBV-encoded miRNA in EBV-associated gastric cancer. Front. Oncol. 2023, 13, 1204030. [Google Scholar] [CrossRef]
- Wang, J.; Zheng, X.; Qin, Z.; Wei, L.; Lu, Y.; Peng, Q.; Gao, Y.; Zhang, X.; Zhang, X.; Li, Z.; et al. Epstein–Barr virus miR-BART3-3p promotes tumorigenesis by regulating the senescence pathway in gastric cancer. J. Biol. Chem. 2019, 294, 4854–4866. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.S.; Uozaki, H.; Chong, J.M.; Ushiku, T.; Sakuma, K.; Ishikawa, S.; Hino, R.; Barua, R.R.; Iwasaki, Y.; Arai, K.; et al. CpG island methylation status in gastric carcinoma with and without infection of Epstein-Barr virus. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2006, 12, 2995–3002. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.H.; Lee, S.; Kim, W.H.; Lee, H.W.; Kim, J.C.; Rhyu, M.G.; Ro, J.Y. Epstein-Barr Virus-Positive Gastric Carcinoma Demonstrates Frequent Aberrant Methylation of Multiple Genes and Constitutes CpG Island Methylator Phenotype-Positive Gastric Carcinoma. Am. J. Pathol. 2002, 160, 787–794. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Kook, M.C.; Kim, Y.W.; Cho, N.Y.; Jung, N.; Kwon, H.J.; Kim, T.Y.; Kang, G.H. CpG island hypermethylator phenotype in gastric carcinoma and its clinicopathological features. Virchows Arch. Int. J. Pathol. 2010, 457, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Toyota, M.; Ahuja, N.; Suzuki, H.; Itoh, F.; Ohe-Toyota, M.; Imai, K.; Baylin, S.B.; Issa, J.P. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res. 1999, 59, 5438–5442. [Google Scholar]
- Matsusaka, K.; Kaneda, A.; Nagae, G.; Ushiku, T.; Kikuchi, Y.; Hino, R.; Uozaki, H.; Seto, Y.; Takada, K.; Aburatani, H.; et al. Classification of Epstein–Barr Virus–Positive Gastric Cancers by Definition of DNA Methylation Epigenotypes. Cancer Res. 2011, 71, 7187–7197. [Google Scholar] [CrossRef]
- Zhang, M.; Sun, W.; You, X.; Xu, D.; Wang, L.; Yang, J.; Li, E.; He, S. LINE-1 repression in Epstein–Barr virus-associated gastric cancer through viral–host genome interaction. Nucleic Acids Res. 2023, 51, 4867–4880. [Google Scholar] [CrossRef]
- Grywalska, E.; Rolinski, J. Epstein-Barr Virus–Associated Lymphomas. Semin. Oncol. 2015, 42, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Knoepfler, P.S. Myc goes global: New tricks for an old oncogene. Cancer Res. 2007, 67, 5061–5063. [Google Scholar] [CrossRef]
- Dang, C.V.; O’Donnell, K.A.; Zeller, K.I.; Nguyen, T.; Osthus, R.C.; Li, F. The c-Myc target gene network. Semin. Cancer Biol. 2006, 16, 253–264. [Google Scholar] [CrossRef]
- Scott, R.S. Epstein-Barr Virus: A Master Epigenetic Manipulator. Curr. Opin. Virol. 2017, 26, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Erikson, J.; Ar-Rushdi, A.; Drwinga, H.L.; Nowell, P.C.; Croce, C.M. Transcriptional activation of the translocated c-myc oncogene in Burkitt lymphoma. Proc. Natl. Acad. Sci. USA 1983, 80, 820–824. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.; Papenhausen, P.; Shao, H. The Role of c-MYC in B-Cell Lymphomas: Diagnostic and Molecular Aspects. Genes 2017, 8, 116. [Google Scholar] [CrossRef]
- Fujita, S.; Buziba, N.; Kumatori, A.; Senba, M.; Yamaguchi, A.; Toriyama, K. Early stage of Epstein-Barr virus lytic infection leading to the “starry sky” pattern formation in endemic Burkitt lymphoma. Arch. Pathol. Lab. Med. 2004, 128, 549–552. [Google Scholar] [CrossRef]
- Whittle, H.C.; Brown, J.; Marsh, K.; Greenwood, B.M.; Seidelin, P.; Tighe, H.; Wedderburn, L. T-cell control of Epstein-Barr virus-infected B cells is lost during P. falciparum malaria. Nature 1984, 312, 449–450. [Google Scholar] [CrossRef]
- Wilmore, J.R.; Asito, A.S.; Wei, C.; Piriou, E.; Sumba, P.O.; Sanz, I.; Rochford, R. AID expression in peripheral blood of children living in a malaria holoendemic region is associated with changes in B cell subsets and Epstein-Barr virus. Int. J. Cancer 2015, 136, 1371–1380. [Google Scholar] [CrossRef]
- Grande, B.M.; Gerhard, D.S.; Jiang, A.; Griner, N.B.; Abramson, J.S.; Alexander, T.B.; Allen, H.; Ayers, L.W.; Bethony, J.M.; Bhatia, K.; et al. Genome-wide discovery of somatic coding and noncoding mutations in pediatric endemic and sporadic Burkitt lymphoma. Blood 2019, 133, 1313–1324. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.D.; Tanizawa, H.; De Leo, A.; Vladimirova, O.; Kossenkov, A.; Lu, F.; Showe, L.C.; Noma, K.; Lieberman, P.M. Epigenetic specifications of host chromosome docking sites for latent Epstein-Barr virus. Nat. Commun. 2020, 11, 877. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Vargas, H.; Gruffat, H.; Cros, M.P.; Diederichs, A.; Sirand, C.; Vargas-Ayala, R.C.; Jay, A.; Durand, G.; Le Calvez-Kelm, F.; Herceg, Z.; et al. Viral driven epigenetic events alter the expression of cancer-related genes in Epstein-Barr-virus naturally infected Burkitt lymphoma cell lines. Sci. Rep. 2017, 7, 5852. [Google Scholar] [CrossRef] [PubMed]
- Gewurz, B.E.; Mar, J.C.; Padi, M.; Zhao, B.; Shinners, N.P.; Takasaki, K.; Bedoya, E.; Zou, J.Y.; Cahir-Mcfarland, E.; Quackenbush, J.; et al. Canonical NF-κB Activation Is Essential for Epstein-Barr Virus Latent Membrane Protein 1 TES2/CTAR2 Gene Regulation. J. Virol. 2011, 85, 6764–6773. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.D.; Sabunciyan, S.; Langmead, B.; Nagy, N.; Curley, R.; Klein, G.; Klein, E.; Salamon, D.; Feinberg, A.P. Large-scale hypomethylated blocks associated with Epstein-Barr virus–induced B-cell immortalization. Genome Res. 2014, 24, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Jha, H.C.; Upadhyay, S.K.; Robertson, E.S. Epigenetic silencing of tumor suppressor genes during in vitro Epstein-Barr virus infection. Proc. Natl. Acad. Sci. USA 2015, 112, E5199–E5207. [Google Scholar] [CrossRef]
- Kretzmer, H.; Bernhart, S.H.; Wang, W.; Haake, A.; Weniger, M.A.; Bergmann, A.K.; Betts, M.J.; Carrillo-de-Santa-Pau, E.; Doose, G.; Gutwein, J.; et al. DNA-methylome analysis in Burkitt and follicular lymphomas identifies differentially methylated regions linked to somatic mutation and transcriptional control. Nat. Genet. 2015, 47, 1316–1325. [Google Scholar] [CrossRef]
- Khasnis, S.; Veenstra, H.; McClellan, M.J.; Ojeniyi, O.; Wood, C.D.; West, M.J. Regulation of B cell receptor signalling by Epstein–Barr virus nuclear antigens. Biochem. J. 2022, 479, 2395–2417. [Google Scholar] [CrossRef]
- Asakawa, Y.; Okabe, A.; Fukuyo, M.; Li, W.; Ikeda, E.; Mano, Y.; Funata, S.; Namba, H.; Fujii, T.; Kita, K.; et al. Epstein-Barr virus-positive gastric cancer involves enhancer activation through activating transcription factor 3. Cancer Sci. 2020, 111, 1818–1828. [Google Scholar] [CrossRef]
- Li, W.; Okabe, A.; Usui, G.; Fukuyo, M.; Matsusaka, K.; Rahmutulla, B.; Mano, Y.; Hoshii, T.; Funata, S.; Hiura, N.; et al. Activation of EHF via STAT3 phosphorylation by LMP2A in Epstein-Barr virus–positive gastric cancer. Cancer Sci. 2021, 112, 3349–3362. [Google Scholar] [CrossRef]
- Mancao, C.; Hammerschmidt, W. Epstein-Barr virus latent membrane protein 2A is a B-cell receptor mimic and essential for B-cell survival. Blood 2007, 110, 3715–3721. [Google Scholar] [CrossRef]
- Ersing, I.; Bernhardt, K.; Gewurz, B.E. NF-κB and IRF7 Pathway Activation by Epstein-Barr Virus Latent Membrane Protein 1. Viruses 2013, 5, 1587–1606. [Google Scholar] [CrossRef]
- Correia, S.; Bridges, R.; Wegner, F.; Venturini, C.; Palser, A.; Middeldorp, J.M.; Cohen, J.I.; Lorenzetti, M.A.; Bassano, I.; White, R.E.; et al. Sequence Variation of Epstein-Barr Virus: Viral Types, Geography, Codon Usage, and Diseases. J. Virol. 2018, 92, e01132-18. [Google Scholar] [CrossRef] [PubMed]
- Banko, A.V.; Lazarevic, I.B.; Folic, M.M.; Djukic, V.B.; Cirkovic, A.M.; Karalic, D.Z.; Cupic, M.D.; Jovanovic, T.P. Characterization of the Variability of Epstein-Barr Virus Genes in Nasopharyngeal Biopsies: Potential Predictors for Carcinoma Progression. PLoS ONE 2016, 11, e0153498. [Google Scholar] [CrossRef]
- Mainou, B.A.; Raab-Traub, N. LMP1 Strain Variants: Biological and Molecular Properties. J. Virol. 2006, 80, 6458–6468. [Google Scholar] [CrossRef] [PubMed]
- Shannon-Lowe, C.D.; Neuhierl, B.; Baldwin, G.; Rickinson, A.B.; Delecluse, H.J. Resting B cells as a transfer vehicle for Epstein–Barr virus infection of epithelial cells. Proc. Natl. Acad. Sci. USA 2006, 103, 7065–7070. [Google Scholar] [CrossRef] [PubMed]
- Tsao, S.W.; Tsang, C.M.; Lo, K.W. Epstein–Barr virus infection and nasopharyngeal carcinoma. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160270. [Google Scholar] [CrossRef] [PubMed]
- Stanland, L.J.; Luftig, M.A. The Role of EBV-Induced Hypermethylation in Gastric Cancer Tumorigenesis. Viruses 2020, 12, 1222. [Google Scholar] [CrossRef] [PubMed]
- Young, L.S.; Dawson, C.W. Epstein-Barr virus and nasopharyngeal carcinoma. Chin. J. Cancer 2014, 33, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Amini, M.A.; Karimi, J.; Talebi, S.S.; Piri, H. The Association of COVID-19 and Reactive Oxygen Species Modulator 1 (ROMO1) with Oxidative Stress. Chonnam Med. J. 2022, 58, 1–5. [Google Scholar] [CrossRef]
- Luo, B.; Wang, Y.; Wang, X.F.; Liang, H.; Yan, L.P.; Huang, B.H.; Zhao, P. Expression of Epstein-Barr virus genes in EBV-associated gastric carcinomas. World J. Gastroenterol. WJG 2005, 11, 629–633. [Google Scholar] [CrossRef]
- Trivedi, P.; Spinsanti, P.; Cuomo, L.; Volpe, M.; Takada, K.; Frati, L.; Faggioni, A. Differential Regulation of Epstein-Barr Virus (EBV) Latent Gene Expression in Burkitt Lymphoma Cells Infected with a Recombinant EBV Strain. J. Virol. 2001, 75, 4929–4935. [Google Scholar] [CrossRef][Green Version]
- Kimura, H. Pathogenesis of chronic active Epstein-Barr virus infection: Is this an infectious disease, lymphoproliferative disorder, or immunodeficiency? Rev. Med. Virol. 2006, 16, 251–261. [Google Scholar] [CrossRef]
- Cohen, J.I. Optimal Treatment for Chronic Active Epstein-Barr Virus Disease. Pediatr. Transplant. 2009, 13, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Krummenacher, C.; Zhang, W.; Hong, J.; Feng, Q.; Chen, Y.; Zhao, Q.; Zeng, M.S.; Zeng, Y.X.; Xu, M.; et al. Urgency and necessity of Epstein-Barr virus prophylactic vaccines. Npj Vaccines 2022, 7, 159. [Google Scholar] [CrossRef]
- Cox, C.; Naylor, B.A.; Mackett, M.; Arrand, J.R.; Griffin, B.E.; Wedderburn, N. Immunization of common marmosets with Epstein-Barr virus (EBV) envelope glycoprotein gp340: Effect on viral shedding following EBV challenge. J. Med. Virol. 1998, 55, 255–261. [Google Scholar] [CrossRef]
- Kanekiyo, M.; Bu, W.; Joyce, M.G.; Meng, G.; Whittle, J.R.R.; Baxa, U.; Yamamoto, T.; Narpala, S.; Todd, J.P.; Rao, S.S.; et al. Rational Design of an Epstein-Barr Virus Vaccine Targeting the Receptor-Binding Site. Cell 2015, 162, 1090–1100. [Google Scholar] [CrossRef] [PubMed]
- Mackett, M.; Arrand, J. Recombinant vaccinia virus induces neutralising antibodies in rabbits against Epstein-Barr virus membrane antigen gp340. EMBO J. 1985, 4, 3229–3234. [Google Scholar] [CrossRef]
- Morgan, A.J.; Mackett, M.; Finerty, S.; Arrand, J.R.; Scullion, F.T.; Epstein, M.A. Recombinant vaccinia virus expressing epstein-barr virus glycoprotein gp340 protects cottontop tamarins against EB virus-induced malignant lymphomas. J. Med. Virol. 1988, 25, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Münz, C. Humanized mouse models for Epstein Barr virus infection. Curr. Opin. Virol. 2017, 25, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, A.; Rosenzweig, M.; Lee-Parritz, D.; Annis, B.; Johnson, R.P.; Wang, F. An Animal Model for Acute and Persistent Epstein-Barr Virus Infection. Science 1997, 276, 2030–2033. [Google Scholar] [CrossRef] [PubMed]


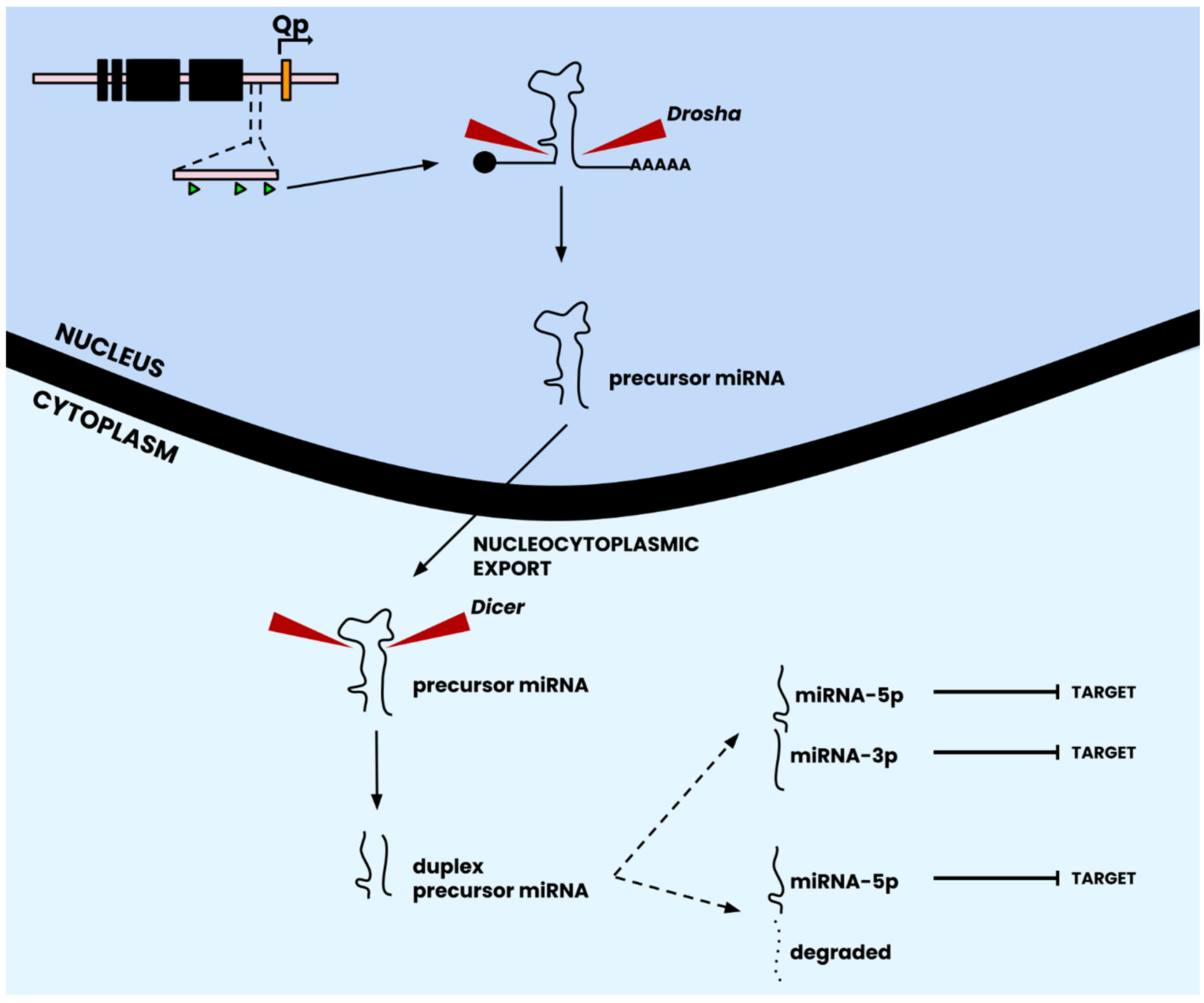
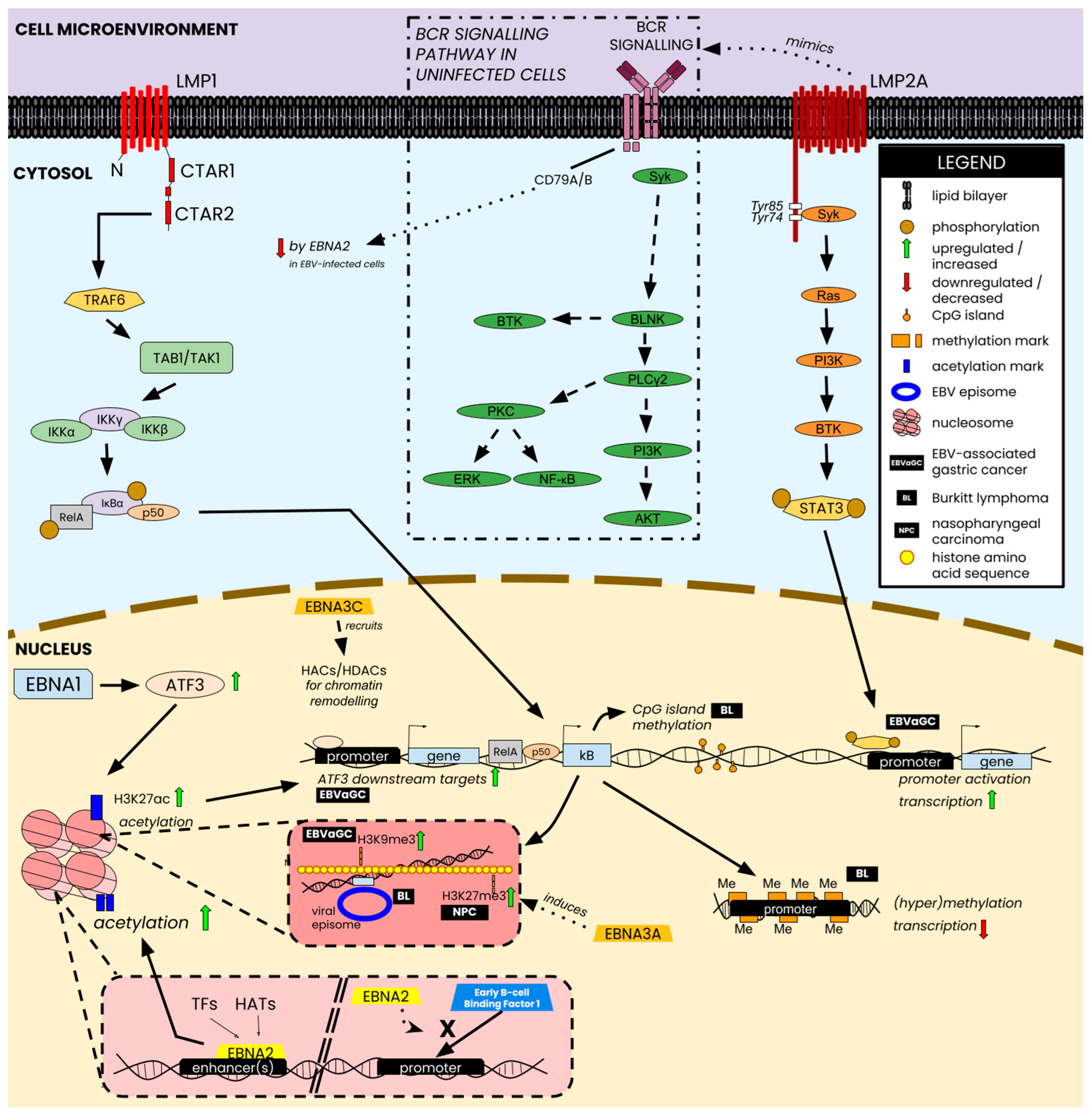
| Protein | Functions | Refs. |
|---|---|---|
| EBNA1 | Required for efficient viral genome replication and persistence in proliferating infected cells | [29,30] |
| EBNA-LP | Reduces EBNA2 binding site occupancy by eliminating repressors; coactivator of EBNA2 transcription | [28,31] |
| EBNA2 | Transcriptional activator; deposits H3K4me1 epigenetic marks on histones and depletes nucleosomes | [32,33] |
| EBNA3A | Engages in polycomb group-mediated epigenetic silencing of CXCL9/10 on host genome | [34] |
| EBNA3B | Inhibitory role in growth through upregulation of CXCL10 chemokines; putative tumor suppressor | [28] |
| EBNA3C | Coactivates LMP1 promoter with EBNA2; regulates chromatin remodeling via histone deacetyltransferase recruitment; inhibits apoptosis by modulating IRFs | [28,35,36] |
| LMP1 | Mimics CD40 signaling; activates NF-kB and p38 pathways; essential for EBV-mediated cell transformation | [28,37] |
| LMP2A | Mimics BCR signaling; promotes growth and cell cycle induction; upregulates IL10 and other anti-apoptotic chemokines and factors in B-cells | [28,38,39] |
| LMP2B | Negatively regulates the function of LMP2A; lowers BCR crosslinking threshold needed for lytic reactivation | [28] |
| miRbase Accession | miRNA | Mature Sequence 1 | Coordinates (bp) 2 | Refs. |
|---|---|---|---|---|
| MI0001064 | BHRF1-1 | 4—UAACCUGAUCAGCCCCGGAGUU—25 | 41,471–41,536 | [84] |
| MI0001065 | BHRF1-2-5p BHRF1-2-3p | 6—AAAUUCUGUUGCAGCAGAUAGC—27 41—UAUCUUUUGCGGCAGAAAUUGA—62 | 42,848–42,912 | |
| MI0001066 | BHRF1-3 | 3—UAACGGGAAGUGUGUAAGCACA—24 | 42,966–43,030 | |
| MI0001067 | BART1-5p BART1-3p |
6—UCUUAGUGGAAGUGACGUGCUGUG—29 42—UAGCACCGCUAUCCACUAUGUC—63 | 139,346–139,415 | [84,85] |
| MI0001068 | BART2-5p BART2-3p | 3—UAUUUUCUGCAUUCGCCCUUGC—24 39—AAGGAGCGAUUUGGAGAAAAUAAA—62 | 152,745–152,806 | [84] |
| MI0003725 | BART3-5p BART3-3p |
12—ACCUAGUGUUAGUGUUGUGCU—32 49—CGCACCACUAGUCACCAGGUGU—70 | 139,076–139,154 | [85,86] |
| MI0003726 | BART4-5p BART4-3p | 9—GACCUGAUGCUGCUGGUGUGCU—30 47—CACAUCACGUAGGCACCAGGUGU—69 | 139,220–139,295 | |
| MI0003727 | BART5-5p BART5-3p | 15—CAAGGUGAAUAUAGCUGCCCAUCG—38 57—GUGGGCCGCUGUUCACCU—74 | 139,661–139,749 | |
| MI0003728 | BART6-5p BART6-3p | 18—UAAGGUUGGUCCAAUCCAUAGG—39 57—CGGGGAUCGGACUAGCCUUAGA—78 | 140,016–140,107 | |
| MI0003729 | BART7-5p BART7-3p |
15—CCUGGACCUUGACUAUGAAACA—36 51—CAUCAUAGUCCAGUGUCCAGGG—72 | 146,425–146,510 | |
| MI0003730 | BART8-5p BART8-3p | 14—UACGGUUUCCUAGAUUGUACAG—35 49—GUCACAAUCUAUGGGGUCGUAGA—71 | 146,759–146,840 | |
| MI0003731 | BART9-5p BART9-3p | 14—UACUGGACCCUGAAUUGGAAAC—35 52—UAACACUUCAUGGGUCCCGUAGU—74 | 146,946–147,032 | |
| MI0003732 | BART10-5p BART10-3p | 18—GCCACCUCUUUGGUUCUGUACA—39 53—UACAUAACCAUGGAGUUGGCUGU—75 | 147,304–147,393 | |
| MI0003733 | BART11-5p BART11-3p |
14—UCAGACAGUUUGGUGCGCUAGUUG—37 52—ACGCACACCAGGCUGACUGCC—72 | 147,524–147,609 | |
| MI0003734 | BART12 | 49—UCCUGUGGUGUUUGGUGUGGUU—70 | 147,888–147,970 | |
| MI0003735 | BART13-5p BART13-3p | 15—AACCGGCUCGUGGCUCGUACAG—36 52—UGUAACUUGCCAGGGACGGCUGA—74 | 148,512–148,597 | |
| MI0003736 | BART14-5p BART14-3p | 14—UACCCUACGCUGCCGAUUUACA—35 48—UAAAUGCUGCAGUAGUAGGGAU—69 | 148,731–148,815 | |
| MI0004988 | BART15 | 47—GUCAGUGGUUUUGUUUCCUUGA—68 | 139,507–139,584 | [86] |
| MI0004989 | BART16 | 20—UUAGAUAGAGUGGGUGUGUGCUCU—43 | 139,776–139,874 | |
| MI0004990 | BART17-5p BART17-3p |
22—UAAGAGGACGCAGGCAUACAAG—43 60—UGUAUGCCUGGUGUCCCCUUAGU—82 | 139,894–139,995 | |
| MI0004991 | BART18-5p BART18-3p | 31—UCAAGUUCGCACUUCCUAUACA—52 67—UAUCGGAAGUUUGGGCUUCGUC—88 | 145,932–146,050 | |
| MI0004992 | BART19-5p BART19-3p | 18—ACAUUCCCCGCAAACAUGACAUG—40 57—UUUUGUUUGCUUGGGAAUGCU—77 | 148,198–148,290 | |
| MI0004993 | BART20-5p BART20-3p | 21—UAGCAGGCAUGUCUUCAUUCC—41 56—CAUGAAGGCACAGCCUGUUACC—77 | 148,319–148,417 | |
| MI0010627 | BART21-5p BART21-3p | 12—UCACUAGUGAAGGCAACUAAC—32 46—CUAGUUGUGCCCACUGGUGUUU—67 | 145,503–145,578 | [83] |
| MI0010628 | BART22 | 43—UUACAAAGUCAUGGUCUAGUAGU—65 | 147,161–147,231 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torne, A.S.; Robertson, E.S. Epigenetic Mechanisms in Latent Epstein-Barr Virus Infection and Associated Cancers. Cancers 2024, 16, 991. https://doi.org/10.3390/cancers16050991
Torne AS, Robertson ES. Epigenetic Mechanisms in Latent Epstein-Barr Virus Infection and Associated Cancers. Cancers. 2024; 16(5):991. https://doi.org/10.3390/cancers16050991
Chicago/Turabian StyleTorne, Atharva S., and Erle S. Robertson. 2024. "Epigenetic Mechanisms in Latent Epstein-Barr Virus Infection and Associated Cancers" Cancers 16, no. 5: 991. https://doi.org/10.3390/cancers16050991
APA StyleTorne, A. S., & Robertson, E. S. (2024). Epigenetic Mechanisms in Latent Epstein-Barr Virus Infection and Associated Cancers. Cancers, 16(5), 991. https://doi.org/10.3390/cancers16050991







