Evaluating Outcome Prediction via Baseline, End-of-Treatment, and Delta Radiomics on PET-CT Images of Primary Mediastinal Large B-Cell Lymphoma
Abstract
:Simple Summary
Abstract
1. Introduction
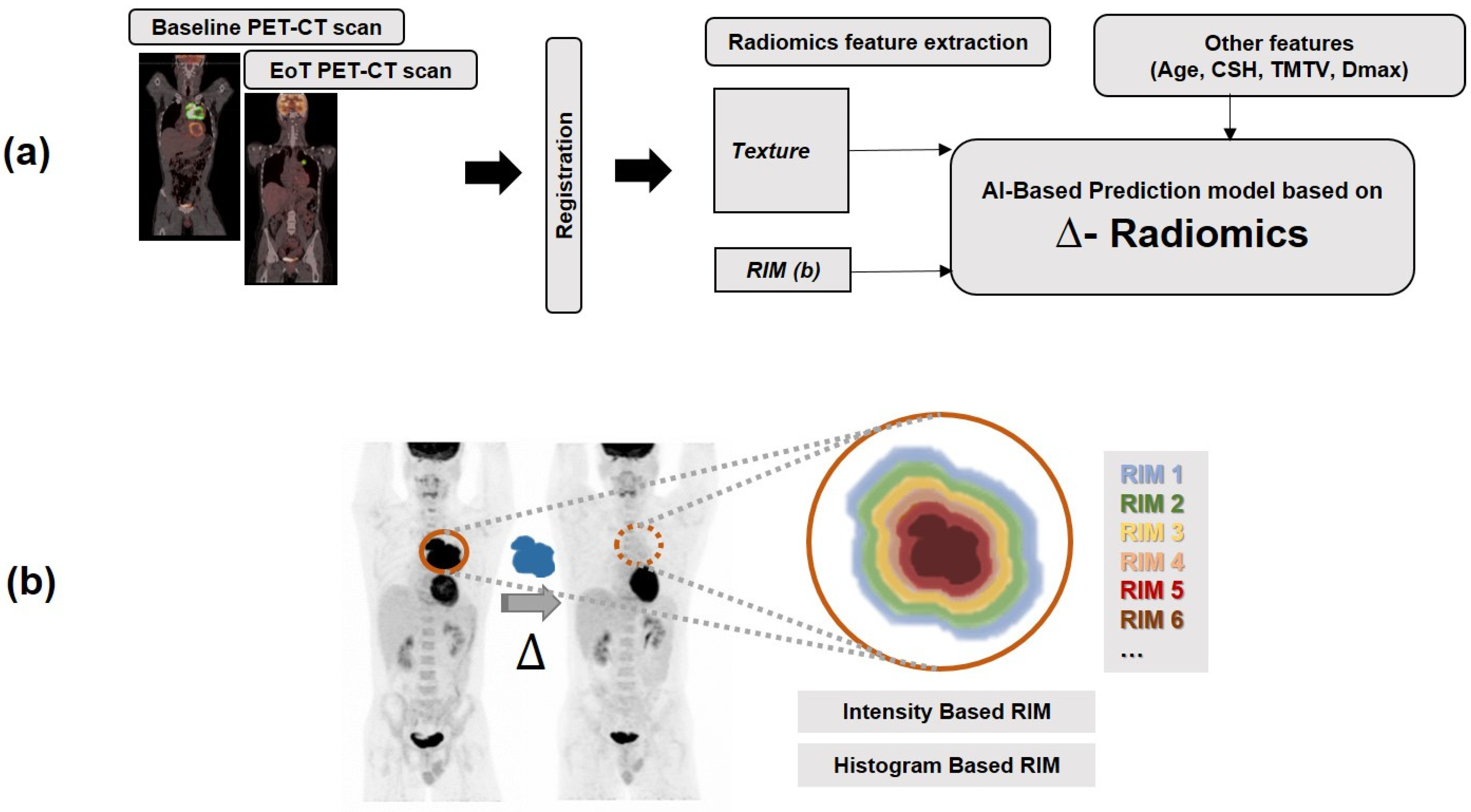
2. Materials and Methods
2.1. Patient Data
2.2. Manual Segmentation
2.3. Extraction of Radiomics Features and Computation of Delta Radiomics
2.3.1. Missing Data Imputation
2.3.2. Harmonization
2.4. Prediction Tasks
2.4.1. Progression Prediction
2.4.2. Time to Progression Survival Analysis
2.4.3. Prediction of Recurrence (Subsequent) TMTV Values on EoT
3. Results
3.1. Progression Prediction
3.2. TTP Analysis
3.3. Prediction of Recurrence Volume on EoT Scans
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Savage, K.J. Primary Mediastinal Large B-Cell Lymphoma. Oncologist 2006, 11, 488–495. [Google Scholar] [CrossRef]
- Savage, K.J. Primary mediastinal large B-cell lymphoma. Blood 2022, 140, 955–970. [Google Scholar] [CrossRef] [PubMed]
- Hayden, A.R.; Tonseth, P.; Lee, D.G.; Villa, D.; Gerrie, A.S.; Scott, D.W.; Freeman, C.L.; Slack, G.W.; Farinha, P.; Skinnider, B.; et al. Outcome of primary mediastinal large B-cell lymphoma using R-CHOP: Impact of a PET-adapted approach. Blood 2020, 136, 2803–2811. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef]
- Liu, P.P.; Wang, K.F.; Xia, Y.; Bi, X.W.; Sun, P.; Wang, Y.; Li, Z.M.; Jiang, W.Q. Racial patterns of patients with primary mediastinal large B-cell lymphoma: SEER analysis. Medicine 2016, 95, e4054. [Google Scholar] [CrossRef]
- Martelli, M.; Ferreri, A.; Di Rocco, A.; Ansuinelli, M.; Johnson, P.W.M. Primary mediastinal large B-cell lymphoma. Crit. Rev. Oncol. Hematol. 2017, 113, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Pfau, D.; Smith, D.A.; Beck, R.; Gilani, K.A.; Gupta, A.; Caimi, P.; Ramaiya, N.H. Primary Mediastinal Large B-Cell Lymphoma: A Review for Radiologists. Am. J. Roentgenol. 2019, 213, W194–W210. [Google Scholar] [CrossRef]
- Martelli, M.; Ceriani, L.; Zucca, E.; Zinzani, P.L.; Ferreri, A.J.; Vitolo, U.; Stelitano, C.; Brusamolino, E.; Cabras, M.G.; Rigacci, L.; et al. [18F]fluorodeoxyglucose positron emission tomography predicts survival after chemoimmunotherapy for primary mediastinal large B-cell lymphoma: Results of the International Extranodal Lymphoma Study Group IELSG-26 Study. J. Clin. Oncol. 2014, 32, 1769–1775. [Google Scholar] [CrossRef] [PubMed]
- Ceriani, L.; Martelli, M.; Zinzani, P.L.; Ferreri, A.J.M.; Botto, B.; Stelitano, C.; Gotti, M.; Cabras, M.G.; Rigacci, L.; Gargantini, L.; et al. Utility of baseline 18FDG-PET/CT functional parameters in defining prognosis of primary mediastinal (thymic) large B-cell lymphoma. Blood 2015, 126, 950–956. [Google Scholar] [CrossRef]
- Ceriani, L.; Milan, L.; Martelli, M.; Ferreri, A.J.M.; Cascione, L.; Zinzani, P.L.; Di Rocco, A.; Conconi, A.; Stathis, A.; Cavalli, F.; et al. Metabolic heterogeneity on baseline 18FDG-PET/CT scan is a predictor of outcome in primary mediastinal B-cell lymphoma. Blood 2018, 132, 179–186. [Google Scholar] [CrossRef]
- Cottereau, A.-S.; Versari, A.; Loft, A.; Casasnovas, O.; Bellei, M.; Ricci, R.; Bardet, S.; Castagnoli, A.; Brice, P.; Raemaekers, J.; et al. Prognostic value of baseline metabolic tumor volume in early-stage Hodgkin lymphoma in the standard arm of the H10 trial. Blood 2018, 131, 1456–1463. [Google Scholar] [CrossRef]
- Song, M.-K.; Chung, J.-S.; Shin, H.-J.; Lee, S.-M.; Lee, S.-E.; Lee, H.-S.; Lee, G.-W.; Kim, S.-J.; Lee, S.-M.; Chung, D.-S. Clinical significance of metabolic tumor volume by PET/CT in stages II and III of diffuse large B cell lymphoma without extranodal site involvement. Ann. Hematol. 2012, 91, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Vercellino, L.; Cottereau, A.-S.; Casasnovas, O.; Tilly, H.; Feugier, P.; Chartier, L.; Fruchart, C.; Roulin, L.; Oberic, L.; Pica, G.M.; et al. High total metabolic tumor volume at baseline predicts survival independent of response to therapy. Blood 2020, 135, 1396–1405. [Google Scholar] [CrossRef] [PubMed]
- Kostakoglu, L.; Martelli, M.; Sehn, L.H.; Belada, D.; Carella, A.-M.; Chua, N.; Gonzalez-Barca, E.; Hong, X.; Pinto, A.; Shi, Y.; et al. Baseline PET-Derived Metabolic Tumor Volume Metrics Predict Progression-Free and Overall Survival in DLBCL after First-Line Treatment: Results from the Phase 3 GOYA Study. Blood 2017, 130, 824. [Google Scholar] [CrossRef]
- Mikhaeel, N.G.; Smith, D.; Dunn, J.T.; Phillips, M.; Møller, H.; Fields, P.A.; Wrench, D.; Barrington, S.F. Combination of baseline metabolic tumour volume and early response on PET/CT improves progression-free survival prediction in DLBCL. Eur. J. Nucl. Med. Mol. Imaging 2016, 43, 1209–1219. [Google Scholar] [CrossRef] [PubMed]
- Cottereau, A.S.; Rebaud, L.; Trotman, J.; Feugier, P.; Nastoupil, L.J.; Bachy, E.; Flinn, I.W.; Haioun, C.; Ysebaert, L.; Bartlett, N.L.; et al. Metabolic tumor volume predicts outcome in patients with advanced stage follicular lymphoma from the RELEVANCE trial. Ann. Oncol. 2023, 35, 130–137. [Google Scholar] [CrossRef]
- Yousefirizi, F.; Klyuzhin, I.S.; Harsini, S.; Tie, X.; Shiri, I.; Shin, M.; Lee, C.; Cho, S.Y.; Bradshaw, T.J.; Zaidi, H.; et al. TMTV-Net: Fully automated total metabolic tumor volume segmentation in lymphoma PET/CT images—A multi-center generalizability analysis. Eur. J. Nucl. Med. Mol. Imaging 2024, 1–18. [Google Scholar] [CrossRef]
- Adams, H.J.; de Klerk, J.M.; Fijnheer, R.; Heggelman, B.G.; Dubois, S.V.; Nievelstein, R.A.; Kwee, T.C. Prognostic superiority of the National Comprehensive Cancer Network International Prognostic Index over pretreatment whole-body volumetric–metabolic FDG-PET/CT metrics in diffuse large B-cell lymphoma. Eur. J. Haematol. 2015, 94, 532–539. [Google Scholar] [CrossRef]
- Cottereau, A.-S.; Meignan, M.; Nioche, C.; Capobianco, N.; Clerc, J.; Chartier, L.; Vercellino, L.; Casasnovas, O.; Thieblemont, C.; Buvat, I. Risk stratification in diffuse large B-cell lymphoma using lesion dissemination and metabolic tumor burden calculated from baseline PET/CT†. Ann. Oncol. 2021, 32, 404–411. [Google Scholar] [CrossRef]
- Angelopoulou, M.K.; Mosa, E.; Pangalis, G.A.; Rondogianni, P.; Chatziioannou, S.; Prassopoulos, V.; Moschogianni, M.; Tsirkinidis, P.; Asimakopoulos, J.V.; Konstantinou, I.; et al. The Significance of PET/CT in the Initial Staging of Hodgkin Lymphoma: Experience Outside Clinical Trials. Anticancer. Res. 2017, 37, 5727–5736. [Google Scholar] [CrossRef]
- Akhtari, M.; Milgrom, S.A.; Pinnix, C.C.; Reddy, J.P.; Dong, W.; Smith, G.L.; Mawlawi, O.; Yehia, Z.A.; Gunther, J.; Osborne, E.M.; et al. Reclassifying patients with early-stage Hodgkin lymphoma based on functional radiographic markers at presentation. Blood 2018, 131, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Ceriani, L.; Milan, L.; Cascione, L.; Di Rocco, A.; Kryachok, I.; Davies, A.J.; Stathis, A.; Johnson, P.; Ciccone, G.; Martelli, M.; et al. Baseline pet radiomics outperforms clinical risk scores in predicting primary mediastinal b-cell lymphoma outcome: Insights from the ielsg37 study. Hematol. Oncol. 2023, 41, 90–91. [Google Scholar] [CrossRef]
- Gillies, R.; Anderson, A.; Gatenby, R.; Morse, D. The biology underlying molecular imaging in oncology: From genome to anatome and back again. Clin. Radiol. 2010, 65, 517–521. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Orlhac, F.; Nioche, C.; Klyuzhin, I.; Rahmim, A.; Buvat, I. Radiomics in PET Imaging: A Practical Guide for Newcomers. PET Clin. 2021, 16, 597–612. [Google Scholar] [CrossRef]
- Bradshaw, T.; Boellaard, R.; Dutta, J.; Jha, A.; Jacobs, P.; Li, Q.; Liu, C.; Sitek, A.; Saboury, B.; Scott, P.; et al. Pitfalls in the development of artificial intelligence algorithms in nuclear medicine and how to avoid them. J. Nucl. Med. 2022, 63, 2724. [Google Scholar] [CrossRef]
- Milgrom, S.A.; Elhalawani, H.; Lee, J.; Wang, Q.; Mohamed, A.S.R.; Dabaja, B.S.; Pinnix, C.C.; Gunther, J.R.; Court, L.; Rao, A.; et al. A PET Radiomics Model to Predict Refractory Mediastinal Hodgkin Lymphoma. Sci. Rep. 2019, 9, 1322. [Google Scholar] [CrossRef]
- Eertink, J.J.; Zwezerijnen, G.J.C.; Cysouw, M.C.F.; Wiegers, S.E.; Pfaehler, E.A.G.; Lugtenburg, P.J.; van der Holt, B.; Hoekstra, O.S.; de Vet, H.C.W.; Zijlstra, J.M.; et al. Comparing lesion and feature selections to predict progression in newly diagnosed DLBCL patients with FDG PET/CT radiomics features. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 4642–4651. [Google Scholar] [CrossRef]
- Kostakoglu, L.; Dalmasso, F.; Berchialla, P.; Pierce, L.A.; Vitolo, U.; Martelli, M.; Sehn, L.H.; Trněný, M.; Nielsen, T.G.; Bolen, C.R.; et al. A prognostic model integrating PET-derived metrics and image texture analyses with clinical risk factors from GOYA. eJHaem 2022, 3, 406–414. [Google Scholar] [CrossRef]
- Sun, Y.; Qiao, X.; Jiang, C.; Liu, S.; Zhou, Z. Texture Analysis Improves the Value of Pretreatment 18F-FDG PET/CT in Predicting Interim Response of Primary Gastrointestinal Diffuse Large B-Cell Lymphoma. Contrast Media Mol. Imaging 2020, 2020, 2981585. [Google Scholar] [CrossRef]
- Ahrari, S.; Zaragori, T.; Zinsz, A.; Oster, J.; Imbert, L.; Verger, A. Application of PET imaging delta radiomics for predicting progression-free survival in rare high-grade glioma. Sci. Rep. 2024, 14, 3256. [Google Scholar] [CrossRef]
- Barabino, E.; Rossi, G.; Pamparino, S.; Fiannacca, M.; Caprioli, S.; Fedeli, A.; Zullo, L.; Vagge, S.; Cittadini, G.; Genova, C. Exploring Response to Immunotherapy in Non-Small Cell Lung Cancer Using Delta-Radiomics. Cancers 2022, 14, 350. [Google Scholar] [CrossRef]
- Chelala, L.; Hossain, R.; Kazerooni, E.A.; Christensen, J.D.; Dyer, D.S.; White, C.S. Lung-RADS Version 1.1: Challenges and a Look Ahead, From the AJR Special Series on Radiology Reporting and Data Systems. AJR Am. J. Roentgenol. 2021, 216, 1411–1422. [Google Scholar] [CrossRef]
- Fave, X.; Zhang, L.; Yang, J.; Mackin, D.; Balter, P.; Gomez, D.; Followill, D.; Jones, A.K.; Stingo, F.; Liao, Z.; et al. Delta-radiomics features for the prediction of patient outcomes in non–small cell lung cancer. Sci. Rep. 2017, 7, 588. [Google Scholar] [CrossRef]
- Mazzei, M.A.; Di Giacomo, L.; Bagnacci, G.; Nardone, V.; Gentili, F.; Lucii, G.; Tini, P.; Marrelli, D.; Morgagni, P.; Mura, G.; et al. Delta-radiomics and response to neoadjuvant treatment in locally advanced gastric cancer—A multicenter study of GIRCG (Italian Research Group for Gastric Cancer). Quant. Imaging Med. Surg. 2021, 11, 2376–2387. [Google Scholar] [CrossRef]
- Wang, L.; Gao, Z.; Li, C.; Sun, L.; Li, J.; Yu, J.; Meng, X. Computed Tomography-Based Delta-Radiomics Analysis for Discriminating Radiation Pneumonitis in Patients With Esophageal Cancer After Radiation Therapy. Int. J. Radiat. Oncol. Biol. Phys. 2021, 111, 443–455. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, H.; Huang, S.; Chen, X.; Zhou, H.; Chang, H.; Xia, Y.; Wang, G.; Yang, X. Early prediction of acute xerostomia during radiation therapy for nasopharyngeal cancer based on delta radiomics from CT images. Quant. Imaging Med. Surg. 2019, 9, 1288–1302. [Google Scholar] [CrossRef]
- Avanzo, M.; Wei, L.; Stancanello, J.; Vallières, M.; Rao, A.; Morin, O.; Mattonen, S.A.; El Naqa, I. Machine and deep learning methods for radiomics. Med. Phys. 2020, 47, e185–e202. [Google Scholar] [CrossRef]
- Nardone, V.; Reginelli, A.; Grassi, R.; Boldrini, L.; Vacca, G.; D’ippolito, E.; Annunziata, S.; Farchione, A.; Belfiore, M.P.; Desideri, I.; et al. Delta radiomics: A systematic review. La Radiol. Medica 2021, 126, 1571–1583. [Google Scholar] [CrossRef] [PubMed]
- Cousin, F.; Louis, T.; Dheur, S.; Aboubakar, F.; Ghaye, B.; Occhipinti, M.; Vos, W.; Bottari, F.; Paulus, A.; Sibille, A.; et al. Radiomics and Delta-Radiomics Signatures to Predict Response and Survival in Patients with Non-Small-Cell Lung Cancer Treated with Immune Checkpoint Inhibitors. Cancers 2023, 15, 1968. [Google Scholar] [CrossRef]
- Cui, Y.; Jiang, Y.; Deng, X.; Long, W.; Liu, B.; Fan, W.; Li, Y.; Zhang, X. 18F-FDG PET-Based Combined Baseline and End-Of-Treatment Radiomics Model Improves the Prognosis Prediction in Diffuse Large B Cell Lymphoma after First-Line Therapy. Acad. Radiol. 2023, 30, 1408–1418. [Google Scholar] [CrossRef] [PubMed]
- Nardone, V.; Reginelli, A.; Guida, C.; Belfiore, M.P.; Biondi, M.; Mormile, M.; Buonamici, F.B.; Di Giorgio, E.; Spadafora, M.; Tini, P.; et al. Delta-radiomics increases multicentre reproducibility: A phantom study. Med. Oncol. 2020, 37, 38. [Google Scholar] [CrossRef] [PubMed]
- Plautz, T.E.; Zheng, C.; Noid, G.; Li, X.A. Time stability of delta-radiomics features and the impact on patient analysis in longitudinal CT images. Med. Phys. 2019, 46, 1663–1676. [Google Scholar] [CrossRef]
- Nakamoto, T.; Yamashita, H.; Jinnouchi, H.; Nawa, K.; Imae, T.; Takenaka, S.; Aoki, A.; Ohta, T.; Ozaki, S.; Nozawa, Y.; et al. Cone-beam computed-tomography-based delta-radiomic analysis for investigating prognostic power for esophageal squamous cell cancer patients undergoing concurrent chemoradiotherapy. Phys. Medica 2024, 117, 103182. [Google Scholar] [CrossRef]
- Lin, P.; Yang, P.-F.; Chen, S.; Shao, Y.-Y.; Xu, L.-M.; Wu, Y.; Teng, W.; Zhou, X.-Z.; Li, B.-H.; Luo, C.; et al. A Delta-radiomics model for preoperative evaluation of Neoadjuvant chemotherapy response in high-grade osteosarcoma. Cancer Imaging 2020, 20, 7. [Google Scholar] [CrossRef]
- Kim, S.J.; Choi, J.Y.; Ahn, Y.C.; Ahn, M.-J.; Moon, S.H. The prognostic value of radiomic features from pre- and post-treatment 18F-FDG PET imaging in patients with nasopharyngeal carcinoma. Sci. Rep. 2023, 13, 8462. [Google Scholar] [CrossRef]
- Rebaud, L.; Escobar, T.; Khalid, F.; Girum, K.; Buvat, I. Simplicity Is All You Need: Out-of-the-Box nnUNet Followed by Binary-Weighted Radiomic Model for Segmentation and Outcome Prediction in Head and Neck PET/CT. In Head and Neck Tumor Segmentation and Outcome Prediction; Springer Nature: Cham, Switzerland, 2023; pp. 121–134. [Google Scholar]
- Yousefirizi, F.; Bloise, I.; Martineau, P.; Wilson, D.; Benard, F.; Bradshaw, T.; Rahmim, A.; Uribe, C. Reproducibility of a semi-automatic gradient-based segmentation approach for lymphoma PET. Eur. J. Nucl. Med. Mol. Imaging (EJNMMI) 2021, 48, S507. [Google Scholar]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Rahmim, A.; Toosi, A.; Salmanpour, M.R.; Dubljevic, N.; Janzen, I.; Shiri, I.; Ramezani, M.A.; Yuan, R.; Ho, C.; Zaidi, H.; et al. Tensor radiomics: Paradigm for systematic incorporation of multi-flavoured radiomics features. Quant. Imaging Med. Surg. 2023, 13, 7680–7694. [Google Scholar] [CrossRef]
- Buizza, G.; Toma-Dasu, I.; Lazzeroni, M.; Paganelli, C.; Riboldi, M.; Chang, Y.; Smedby, Ö.; Wang, C. Early tumor response prediction for lung cancer patients using novel longitudinal pattern features from sequential PET/CT image scans. Phys. Medica 2018, 54, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Astaraki, M.; Wang, C.; Buizza, G.; Toma-Dasu, I.; Lazzeroni, M.; Smedby, Ö. Early survival prediction in non-small cell lung cancer from PET/CT images using an intra-tumor partitioning method. Phys. Medica 2019, 60, 58–65. [Google Scholar] [CrossRef]
- Chang, R.; Qi, S.; Wu, Y.; Yue, Y.; Zhang, X.; Guan, Y.; Qian, W. Deep radiomic model based on the sphere–shell partition for predicting treatment response to chemotherapy in lung cancer. Transl. Oncol. 2023, 35, 101719. [Google Scholar] [CrossRef] [PubMed]
- Taquia, J.P. Comparison of Statistical Methods for Missing Data Imputation in MIR-Radiomics. 2020. Available online: https://www.politesi.polimi.it/handle/10589/154564 (accessed on 28 October 2023).
- Beer, J.C.; Tustison, N.J.; Cook, P.A.; Davatzikos, C.; Sheline, Y.I.; Shinohara, R.T.; Linn, K.A. Longitudinal ComBat: A method for harmonizing longitudinal multi-scanner imaging data. Neuroimage 2020, 220, 117129. [Google Scholar] [CrossRef] [PubMed]
- Eertink, J.J.; Heymans, M.W.; Zwezerijnen, G.J.C.; Zijlstra, J.M.; de Vet, H.C.W.; Boellaard, R. External validation: A simulation study to compare cross-validation versus holdout or external testing to assess the performance of clinical prediction models using PET data from DLBCL patients. EJNMMI Res. 2022, 12, 58. [Google Scholar] [CrossRef] [PubMed]
- Fernández, A.; García, S.; Galar, M.; Prati, R.C.; Krawczyk, B.; Herrera, F. Learning from Imbalanced Data Sets; Springer International Publishing: Cham, Switzerland, 2018. [Google Scholar] [CrossRef]
- Yousefirizi, F.; Harsini, S.; Holloway, C.; Tonseth, P.; Alexander, A.; Saboury, B.; Uribe, C.; Rahmim, A. Pretreatment 18F-FDG PET/CT radiomics predict recurrence in patients treated with radiotherapy for cervical cancer. J. Nucl. Med. 2023, 64, 1248. [Google Scholar]
- Casasnovas, R.-O.; Ysebaert, L.; Thieblemont, C.; Bachy, E.; Feugier, P.; Delmer, A.; Tricot, S.; Gabarre, J.; Andre, M.; Fruchart, C.; et al. FDG-PET–driven consolidation strategy in diffuse large B-cell lymphoma: Final results of a randomized phase 2 study. Blood 2017, 130, 1315–1326. [Google Scholar] [CrossRef] [PubMed]
- Dührsen, U.; Müller, S.; Hertenstein, B.; Thomssen, H.; Kotzerke, J.; Mesters, R.; Berdel, W.E.; Franzius, C.; Kroschinsky, F.; Weckesser, M.; et al. Positron Emission Tomography-Guided Therapy of Aggressive Non-Hodgkin Lymphomas (PETAL): A Multicenter, Randomized Phase III Trial. J. Clin. Oncol. 2018, 36, 2024–2034. [Google Scholar] [CrossRef]
- Orton, M.R.; Hann, E.; Doran, S.J.; Shepherd, S.T.C.; Ap Dafydd, D.; Spencer, C.E.; López, J.I.; Albarrán-Artahona, V.; Comito, F.; Warren, H.; et al. Interpretability of radiomics models is improved when using feature group selection strategies for predicting molecular and clinical targets in clear-cell renal cell carcinoma: Insights from the TRACERx Renal study. Cancer Imaging 2023, 23, 76. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, T.J.; Huemann, Z.; Hu, J.; Rahmim, A. A Guide to Cross-Validation for Artificial Intelligence in Medical Imaging. Radiol. Artif. Intell. 2023, 5, e220232. [Google Scholar] [CrossRef]
- Camus, V.; Viennot, M.; Lévêque, E.; Viailly, P.-J.; Tonnelet, D.; Veresezan, E.-L.; Drieux, F.; Etancelin, P.; Dubois, S.; Stamatoullas, A.; et al. Circulating tumor DNA in primary mediastinal large B-cell lymphoma versus classical Hodgkin lymphoma: A retrospective study. Leuk. Lymphoma 2022, 63, 834–844. [Google Scholar] [CrossRef]
| Scan Time-Point | # of Cases | Progression (Percentage) | Average Follow-Up (y) (±std) |
|---|---|---|---|
| EoT | 50 | 24.0% | 5.56 ± 3.74 |
| Baseline and EoT | 31 | 15.6% | 3.73 ± 2.17 |
| Number of Cases | Scan Time Points | Features Set (PET and CT) | Task | Prediction Approach | ||
|---|---|---|---|---|---|---|
| 31 | Single time point scan | baseline | Progression prediction (Section 2.4.1) | Time to Progression (Section 2.4.2) | ICARE (Individual Coefficient Approximation for Risk Estimation) [47] | Machine Learning (RF, KNN, LDA) |
| EoT | ||||||
| 31 | Two time points scans | baseline + EoT | ||||
| 31 | Delta | Relative | ||||
| Absolute | ||||||
| 31 | Baseline + Delta | baseline + Relative | ||||
| baseline + Absolute | ||||||
| 50 | Single time point scan | EoT | ||||
| Features | ICARE | KNN | LDA | Random Forest | ||||
|---|---|---|---|---|---|---|---|---|
| Accuracy | F1 Score | Accuracy | F1 Score | Accuracy | F1 Score | Accuracy | F1 Score | |
| PET EoT | 0.61 ± 0.07 | 0.56 ± 0.12 | 0.80 ± 0.03 | 0.83 ± 0.02 | 0.85 ± 0.04 | 0.85 ± 0.04 | 0.87 ± 0.03 | 0.87 ± 0.03 |
| PET-CT EoT | 0.79 ± 0.09 | 0.81 ± 0.08 | 0.81 ± 0.02 | 0.84 ± 0.02 | 0.85 ± 0.02 | 0.86 ± 0.02 | 0.92 ± 0.02 | 0.91 ± 0.02 |
| Features Set (PET) | Accuracy | F1 Score | Recall | Precision | ROC AUC |
|---|---|---|---|---|---|
| Baseline | 0.56 ± 0.16 | 0.57 ± 0.19 | 0.58 ± 0.12 | 0.65 ± 0.15 | 0.77 ± 0.14 |
| EoT | 0.56 ± 0.18 | 0.56 ± 0.18 | 0.47 ± 0.14 | 0.62 ± 0.12 | 0.63 ± 0.10 |
| Baseline + EoT | 0.64 ± 0.12 | 0.67 ± 0.14 | 0.59 ± 0.13 | 0.63 ± 0.13 | 0.77 ± 0.13 |
| Relative Delta | 0.65 ± 0.18 | 0.67 ± 0.19 | 0.53 ± 0.11 | 0.61 ± 0.11 | 0.67 ± 0.15 |
| Absolute Delta | 0.69 ± 0.25 | 0.66 ± 0.24 | 0.55 ± 0.14 | 0.56 ± 0.06 | 0.63 ± 0.19 |
| Baseline + Relative Delta | 0.65 ± 0.16 | 0.67 ± 0.14 | 0.63 ± 0.15 | 0.63 ± 0.13 | 0.81 ± 0.08 |
| Baseline + Absolute Delta | 0.64 ± 0.19 | 0.63 ± 0.21 | 0.58 ± 0.12 | 0.59 ± 0.09 | 0.69 ± 0.14 |
| Features Set (PET-CT) | Accuracy | F1 Score | Recall | Precision | ROC AUC |
|---|---|---|---|---|---|
| Baseline | 0.60 ± 0.16 | 0.65 ± 0.15 | 0.58 ± 0.24 | 0.66 ± 0.16 | 0.79 ± 0.09 |
| EoT | 0.78 ± 0.14 | 0.76 ± 0.19 | 0.50 ± 0.14 | 0.75 ± 0.25 | 0.75 ± 0.12 |
| Baseline + EoT | 0.69 ± 0.17 | 0.65 ± 0.31 | 0.68 ± 0.24 | 0.75 ± 0.25 | 0.88 ± 0.13 |
| Relative Delta | 0.66 ± 0.16 | 0.60 ± 0.19 | 0.62 ± 0.14 | 0.71 ± 0.21 | 0.88 ± 0.14 |
| Absolute Delta | 0.81 ± 0.15 | 0.77 ± 0.18 | 0.63 ± 0.15 | 0.68 ± 0.18 | 0.87 ± 0.17 |
| Baseline + Relative Delta | 0.84 ± 0.11 | 0.82 ± 0.13 | 0.50 ± 0.14 | 0.75 ± 0.25 | 0.75 ± 0.15 |
| Baseline + Absolute Delta | 0.70 ± 0.13 | 0.60 ± 0.24 | 0.57 ± 0.13 | 0.75 ± 0.25 | 0.74 ± 0.09 |
| Features Set (PET-CT) | KNN | LDA | Random Forest | |||
|---|---|---|---|---|---|---|
| Accuracy | F1 Score | Accuracy | F1 Score | Accuracy | F1 Score | |
| Baseline | 0.67 ± 0.14 | 0.65 ± 0.13 | 0.68 ± 0.10 | 0.69 ± 0.09 | 0.69 ± 0.18 | 0.69 ± 0.04 |
| EoT | 0.78 ± 0.04 | 0.77 ± 0.04 | 0.77 ± 0.02 | 0.74 ± 0.03 | 0.83 ± 0.05 | 0.84 ± 0.05 |
| Relative Delta | 0.77 ± 0.05 | 0.82 ± 0.03 | 0.82 ± 0.03 | 0.80 ± 0.3 | 0.89 ± 0.04 | 0.87 ± 0.05 |
| Absolute Delta | 0.73 ± 0.02 | 0.77 ± 0.02 | 0.89 ± 0.03 | 0.89 ± 0.03 | 0.87 ± 0.04 | 0.86 ± 0.05 |
| Baseline + Relative Delta | 0.86 ± 0.03 | 0.87 ± 0.03 | 0.75 ± 0.04 | 0.75 ± 0.04 | 0.88 ± 0.03 | 0.87 ± 0.03 |
| Features Set (PET-CT) | #Cases | ICARE | CoxNet (SFS) | CoxNet (Chi-Square) | CoxNet (MI) | CoxNet (RF) | CoxNet (LASSO) |
|---|---|---|---|---|---|---|---|
| Baseline + Delta (absolute) | 31 | 0.61 ± 0.11 | 0.65 ± 0.17 | 0.60 ± 0.08 | 0.60 ± 0.04 | 0.57 ± 0.10 | 0.67 ± 0.06 |
| EoT | 31 | 0.60 ± 0.24 | 0.54 ± 0.04 | 0.45 ± 0.10 | 0.37 ± 0.10 | 0.58 ± 0.09 | 0.65 ± 0.07 |
| EoT | 50 | 0.65 ± 0.23 | 0.68 ± 0.09 | 0.56 ± 0.03 | 0.64 ± 0.05 | 0.58 ± 0.04 | 0.67 ± 0.09 |
| PET-CT Features Set (n = 31 Cases) | Gradient Boosting Regressor | R-squared (R2) | Mean Absolute Error | Mean Absolute Percentage Error |
|---|---|---|---|---|
| Baseline | Tuned with Grid Search | 0.86 ± 0.09 | 0.29 ± 0.12 | 0.39 ± 0.15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yousefirizi, F.; Gowdy, C.; Klyuzhin, I.S.; Sabouri, M.; Tonseth, P.; Hayden, A.R.; Wilson, D.; Sehn, L.H.; Scott, D.W.; Steidl, C.; et al. Evaluating Outcome Prediction via Baseline, End-of-Treatment, and Delta Radiomics on PET-CT Images of Primary Mediastinal Large B-Cell Lymphoma. Cancers 2024, 16, 1090. https://doi.org/10.3390/cancers16061090
Yousefirizi F, Gowdy C, Klyuzhin IS, Sabouri M, Tonseth P, Hayden AR, Wilson D, Sehn LH, Scott DW, Steidl C, et al. Evaluating Outcome Prediction via Baseline, End-of-Treatment, and Delta Radiomics on PET-CT Images of Primary Mediastinal Large B-Cell Lymphoma. Cancers. 2024; 16(6):1090. https://doi.org/10.3390/cancers16061090
Chicago/Turabian StyleYousefirizi, Fereshteh, Claire Gowdy, Ivan S. Klyuzhin, Maziar Sabouri, Petter Tonseth, Anna R. Hayden, Donald Wilson, Laurie H. Sehn, David W. Scott, Christian Steidl, and et al. 2024. "Evaluating Outcome Prediction via Baseline, End-of-Treatment, and Delta Radiomics on PET-CT Images of Primary Mediastinal Large B-Cell Lymphoma" Cancers 16, no. 6: 1090. https://doi.org/10.3390/cancers16061090






