NDRGs in Breast Cancer: A Review and In Silico Analysis
Abstract
Simple Summary
Abstract
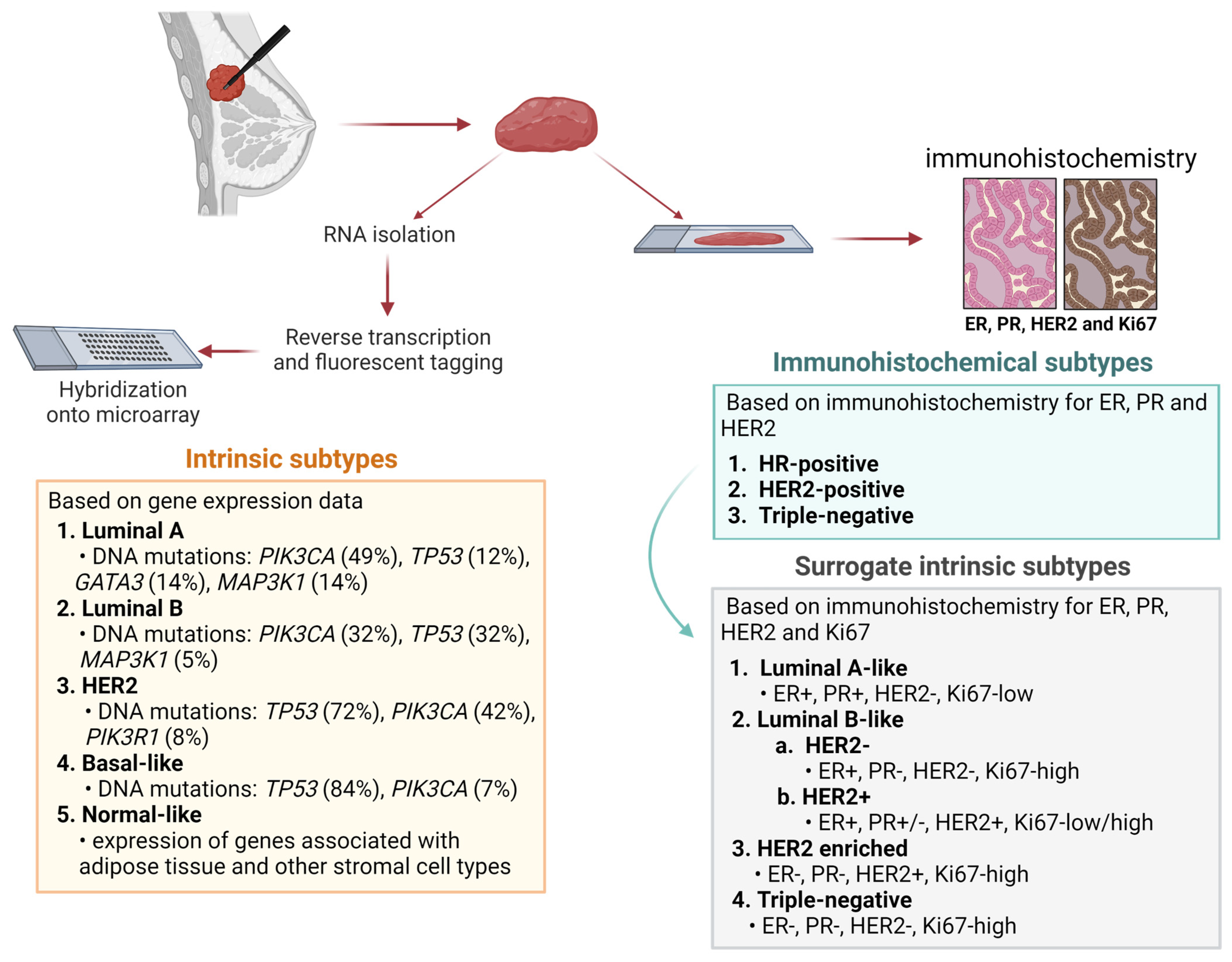
1. Introduction
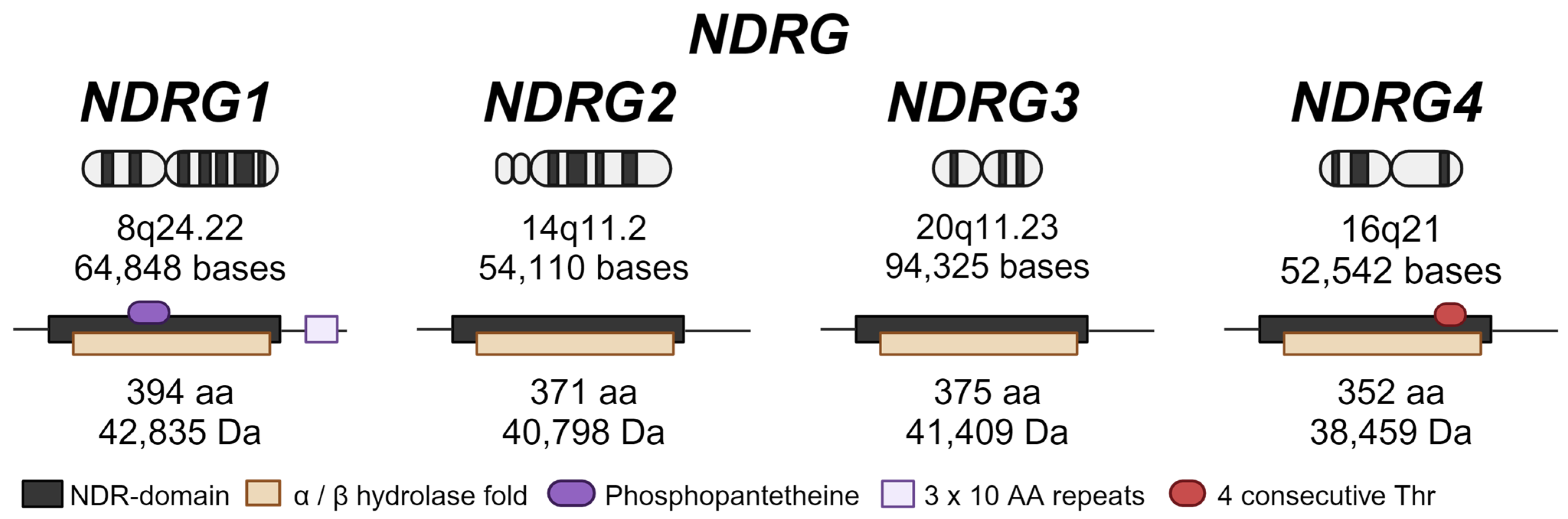
2. NDRG Family Proteins
3. NDRGs and Breast Cancer
3.1. NDRG1
3.2. NDRG2
3.3. NDRG3
3.4. NDRG4
| Function | References | Cell Lines Analyzed | No. of Patients Analyzed | Experimental Conditions | Major Conclusions | ||
|---|---|---|---|---|---|---|---|
| In Vitro | In Vivo | ||||||
| NDRG1 | Tumor suppressor | Bandyopadhyay, 2004 [38] | MDA-MB-468, MDA-MB-435, MDA-MB-231, MCF-7 | 85 | Yes | No | 1. Overexpression of NDRG1 reduced invasion of MDA-468 breast cancer cells in vitro. |
| 2. Treatment with 5-azacytidine suppressed breast cancer cell invasion in vitro. | |||||||
| 3. Patients with NDRG1-negative tumors had worse disease-free survival than those with NDRG1 -positive tumors. | |||||||
| Bandyopadhyay, 2004 [37] | MDA-468, BT-549 | 85 | Yes | No | 1. PTEN regulated NDRG1 expression. | ||
| 2. Patients with PTEN-negative and NDRG1-negative tumors had better outcomes than patients with PTEN-negative and NDRG1-positive tumors. | |||||||
| Fotovati, 2006 [39] | SK-BR-3, MDA-MB-231, T47D, MCF-7, ZR75-1, R-27 (MCF-7 | 96 | Yes | No | 1. Cells that were resistant to tamoxifen expressed higher levels of NDRG1 than parental cells. | ||
| tamoxifen-resistant cell line) | 2. NDRG1 expression was inversely correlated with estrogen receptor alpha (ER-α) expression. | ||||||
| 3. Treatment with 17β-estradiol E2 reduced NDRG1 levels in ERα-positive cell lines, but not in ERα-negative cell lines. | |||||||
| Redmond, 2010 [42] | MCF-7, BT474, MDA-MB-157, MDA-MB-453, | Not reported | Yes | No | 1. NDRG1 expression was repressed by TBX2 through EGR1. | ||
| MDA-MB-231, MDA-MB-468, T47D, ZR75-1 | 2. TBX2 had a tumor promoter function and inhibited NDRG1 to promote cell growth. | ||||||
| Fotovati, 2011 [50] | SK-BR-3, MDA-MB-231, T47D, MCF-7 | 45 | Yes | Yes | 1. NDRG1 expression was upregulated during the differentiation of breast cancer cells in vitro and could be used as a marker for differentiation of breast cancer. | ||
| 2. Induction of NDRG1 could be a strategy for cancer treatment. | |||||||
| Lai, 2011 [43] | MCF-7 | Not reported | Yes | No | 1. NDRG1 was highly expressed upon reoxygenation, and reoxygenated cells showed increased levels of migration. | ||
| Liu, 2011 [48] | MCF-7 | 33 | Yes | Yes [a] | [a] Used WB1-1 cells (cell line isolated tumor cells from the mammary tumor of MMTV-Wnt mouse model). | ||
| 1. NDRG1 repressed Wnt-β pathway by interacting with LRP6 (Wnt co-receptor) and re-activating GSK3β. | |||||||
| 2. NDRG1 correlated with epithelial traits in breast cancer cell lines (high E-cadherin, low vimentin). | |||||||
| 3. NDRG1 functioned as a metastasis suppressor by inhibiting Wnt signaling. | |||||||
| Han, 2013 [44] | MDA-MB-231, T47D | 389 | Yes | No | 1. NDRG1 methylation status could be involved in tumorigenesis in breast cancer. | ||
| Chiang, 2015 [46] | MCF-7, MDA-MB-231 | Not reported | Yes | Yes [b] | [b] In vivo studies are focused on WISP1 not NDRG1 | ||
| 1. Overexpression of WISP1 induced epithelial-mesenchymal–transition, migration, and invasion. | |||||||
| 2. NDRG1 expression was inhibited in WISP1-overexpressing cells. | |||||||
| 3. Overexpression of NDRG1 reduced the effect of WISP1 in proliferation and invasion of breast cancer cells. | |||||||
| Salis, 2016 [41] | MCF-7 | Not reported | Yes | No | 1. NDRG1 mRNA expression levels were reduced after treatment with TGF-β1. | ||
| 2. Treatment with fluvastatin inhibited migration of TGF-β1-treated cells and induced an increase in NDRG1 expression. | |||||||
| Tian, 2017 [40] | MDA-MB-231, MDA-MB-453, MCF-7 | Not reported | Yes | No | 1. siNDRG1 promoted migration/invasion of MDA-231 breast cancer cells, an effect that was inhibited by treatment with an SGK1 inhibitor. | ||
| Godbole, 2018 [45] | T47D, BT474, MDA-MB-231, ZR-75-1, MCF-7 | Not reported | Yes | No | 1. NDRG1 expression was inhibited after knocking down SGK1, increasing cell migration and invasion. | ||
| 2. Silencing of NDRG1 increased levels of phosphorylated EGFR (pEGFR), AKT (pAKT), and ERK1/2 (pERK1/2) in T47D and MDA-MB-231 cells. | |||||||
| Abascal, 2022 [49] | T47D, MDA-231 | Not reported | Yes | No | 1. NDRG1 expression is higher in Luminal B than in Luminal A tumors | ||
| 2. Metastatic potential of breast cancer was influenced by progesterone receptor isoforms (A or B) regulating NDRG1 | |||||||
| Tumor promoter | Nagai, 2011 [56] | Not reported | 596 | No | No | 1. NDRG1-positive tumors were associated with worse outcomes. | |
| 2. The 10-year overall survival rates were 67% for NDRG1-negative versus 34% for NDRG1-positive tumors. | |||||||
| Mao, 2011 [58] | Not reported | 215 + 20 [c] | No | No | [c] Involved 215 samples of different subtypes of breast cancer; 20 tumor tissues had a paired non-tumor portion. | ||
| 1. NDRG1 expression was associated with a progression of breast cancer, from atypia to carcinoma development. | |||||||
| 2. NDRG1 expression correlated with a high tumor category in invasive breast cancer. | |||||||
| Sommer, 2013 [53] | BT-474, CAMA-1, ZR-75-1, T47D, HCC-1187, SUM-52-PE, | Not reported | Yes | No | 1. NDRG1 and SGK1 expression were increased in AKT inhibitor-resistant cell lines. | ||
| HCC-1937, MDA-MB-436, BT-549, MDA-MB-157, | 2. High levels of SGK1 were one means of predicting resistance to AKT inhibitors. | ||||||
| MDA-MB-231, HCC-1806, JIMT-1 | 3. Levels of phosphorylated NDRG1 could serve as a marker for response to AKT inhibitors. | ||||||
| Parris, 2014 [55] | Not reported | 229 | No | No | 1. NDRG1 was hypomethylated and highly expressed in breast cancer samples. | ||
| Li, 2016 [59] | MCF-7 | Not reported | Yes | No | 1. NDRG1 knockdown inhibited proliferation and migration; it induced cell cycle arrest under hypoxia. | ||
| Sevinsky, 2018 [54] | SKBR3, MCF-7, HCC1569, BT474, MDA-MB-231, | 3554 [d] | Yes | No | [d] Meta-analysis of 23 distinct breast cancer cohorts. | ||
| MDA-MB-468 | 1. Patients with NDRG1-high tumors had worse recurrence- and metastasis-free survival. | ||||||
| 2. High expression of NDRG1 correlated with hypoxia and glycolytic pathways. | |||||||
| 3. NDRG1 knockdown reduced proliferation and led to dysfunction in lipid metabolism. | |||||||
| Mishra, 2020 [57] | MDA-231, SUM159 | Not reported | Yes | No | 1. NDRG1 expression reduced in cybrids with benign mitochondria. | ||
| 2. NDRG1 knockdown reduced proliferation of SUM159 cells. | |||||||
| Villodre, 2020 [51] | Not reported | 64 | No | No | 1. NDRG1 was an independent predictor of worse outcomes in inflammatory breast cancer. | ||
| 2. NDRG1, together with estrogen receptor status and disease stage, could be used to further stratify patient outcomes. | |||||||
| Berghoff, 2021 [61] | JIMT1, MDA-231 | 74 + 61 [e] | Yes | Yes | [e] Involved 75 primary breast cancer and 61 breast cancer brain metastasis specimens. | ||
| 1. Slow-cycling cells efficiently formed brain metastasis and extracranial metastasis and expressed high levels of NDRG1. | |||||||
| 2. Silencing NDRG1 reduced the ability of cells to develop brain metastasis. | |||||||
| 3. Patients with high NDRG1-expressing tumors had worse metastasis-free survival. | |||||||
| Villodre, 2022 [52] | SUM149, BCX010, MDA-IBC3 | 216 | Yes | Yes | 1. NDRG1 knockdown inhibited migration, invasion, and cancer-stem cell features in aggressive breast cancer cell lines. | ||
| 2. Silencing of NDRG1 inhibited primary tumor growth and brain metastasis. | |||||||
| 3. Patents with breast cancer and high NDRG1 expression had worse outcomes, and NDRG1 was an independent prognostic factor. | |||||||
| de Nonneville, 2022 [60] | Not reported | 7850 [f] | Not reported | Not reported | [f] Involved 5929 ER+/HER2- and 1936 TN cases. | ||
| 1. Patients with NDRG1 -high tumors had worse overall survival. | |||||||
| 2. The 10-year overall survival rates were 68% for NDRG1-high tumors versus 78% for NDRG1-low tumors. | |||||||
| 3. High expression of NDRG1 was associated with aggressive tumor features. | |||||||
| 4. NDRG1 was an independent predictor of overall survival in patients with ER+/HER2- disease. | |||||||
| López-Tejada, 2023 [63] | BT549, Hs578T, MDA-MB-231, MDA-MB-436, MDA-MB-468, SUM159 | 83 | Yes | No | 1. High NDRG1 expression was associated with poor cumulative survival. | ||
| 2. Negative nuclear phospho-NDRG1 expression was associated with poor cumulative survival. | |||||||
| 3. Cellular expression and subcellular localization of NDRG1 and phospho-NDRG1 in TNBC correlated with patient survival. | |||||||
| 4. TGFβ governed the activity of NDRG1 in tumor progression to modulate epithelial–mesenchymal transition, metastasis, and the tumor-initiating capacity of cancer cells. | |||||||
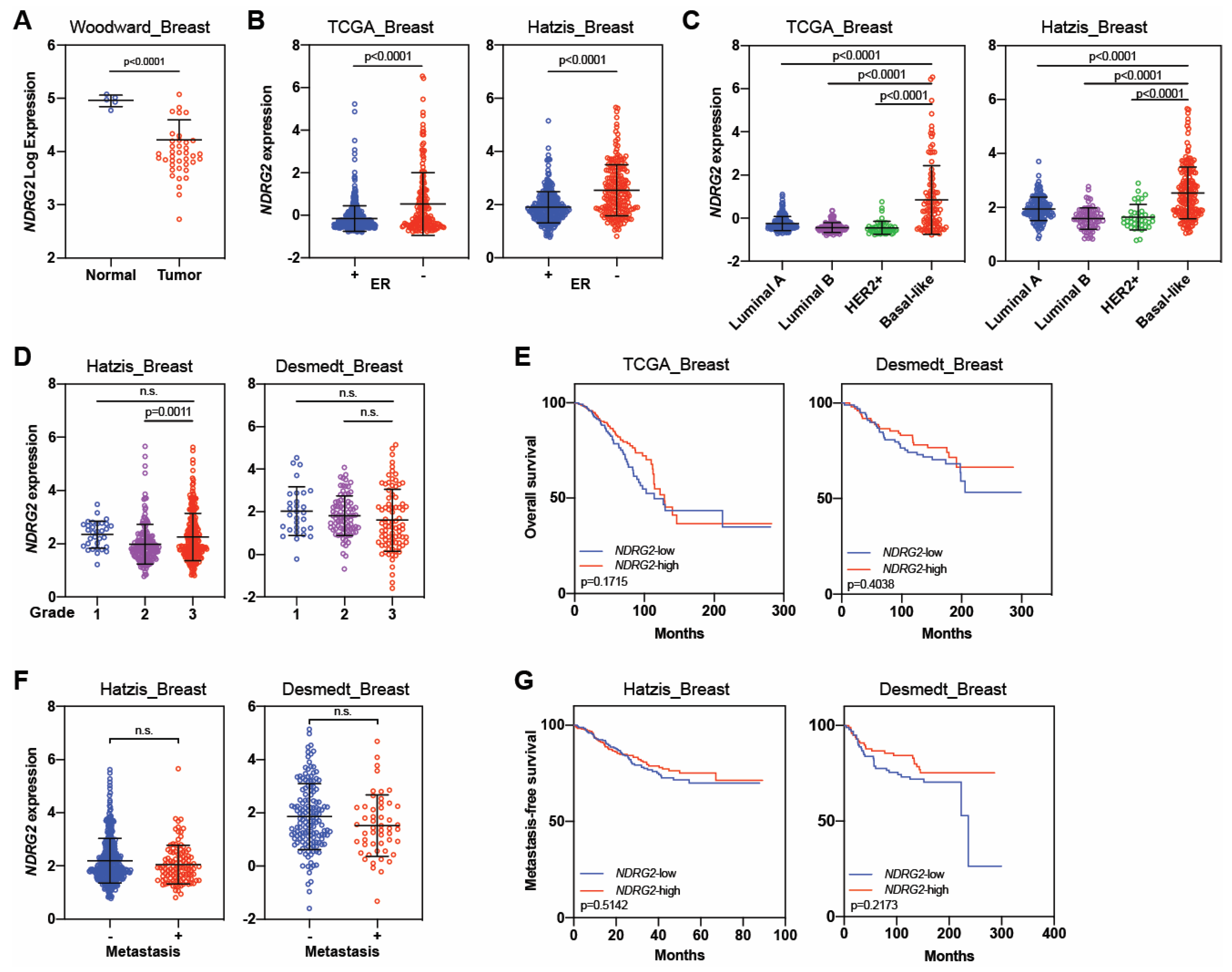
| NDRG2 | Tumor suppressor | Liu, 2007 [26] | MCF-7, MDA-MB-231, SK-BR-3 | 21 | Yes | No | 1. Low NDRG2 levels were observed in breast cancer cell lines and in 5 of 21 breast cancer tissues samples. |
| Park, 2007 [67] | T47D, MCF-7, MDA-MB-453, MDA-MB-231 | Not reported | Yes | No | 1. High expression of NDRG2 reduced phospho-AKT and induced phosphorylation of p38 MAP kinase. | ||
| 2. T47D and MCF7 cells (less malignant) had strong expressions of NDRG2, but NDRG2 was not detected in MDA-MB-453 and MDA-MB-231 cells (highly malignant). | |||||||
| Shon, 2009 [66] | MDA-MB-231, MCF-7 | Not reported | Yes | No | 1. NDRG2 induced BMP-4 and suppressed MMP-9 activity. | ||
| 2. NDRG2 expression inhibited the in vitro migration and invasion potential of breast cancer cells. | |||||||
| Zheng, 2010 [77] | MCF-7, Bcap-37 | Not reported | Yes | No | 1. NDRG2 suppressed adhesion and invasion of breast cancer cells. | ||
| Lorentzen, 2011 [69] | Not reported | 48 | Not reported | Not reported | 1. NDRG2 mRNA expression was reduced in breast cancer relative to normal tissue. | ||
| Oh, 2012 [68] | 4T1 | 189 | Yes | Yes | 1. High levels of NDRG2 correlated with better disease-free survival but not with overall survival. | ||
| 2. NDRG2 overexpression reduced migration and invasion in vitro. | |||||||
| 3. High levels of NDRG2 reduced tumor growth in vivo. | |||||||
| Kim, 2014 [73] | MDA-MB-231 | Not reported | Yes | No | 1. Overexpression of NDRG2 induced apoptosis. | ||
| Kim, 2014 [71] | MDA-MB-231 | Not reported | Yes | No | 1. Overexpression of NDRG2 inhibited the epithelial–mesenchymal transition through STAT3/Snail signaling. | ||
| Kim, 2014 [72] | MDA-MB-231, MCF-7 | Not reported | 1. Overexpression of NDRG2 downregulated COX-2 through NF-kB signaling. | ||||
| 2. Overexpression of NDRG2 reduced migration and invasion of MDA-MB-231 cells. | |||||||
| Kim, 2016 [74] | 4T1 | Not reported | Yes | No | 1. NDRG2 expression in breast cancer cells inhibited osteoclast differentiation. | ||
| Wei, 2017 [65] | MCF-7, MDA-MB-231, T47D | Not reported | Yes | No | 1. Doxorubicin-resistant breast cancer cells expressed reduced levels of NDRG2. | ||
| Lee, 2021 [70] | MDA-231, MCF-7, 4T1 | Yes | No | 1. NDRG2 negatively regulated PDL1 expression in malignant breast cancer cells by suppressing NF-kB signaling. | |||
| 2. NDRG2 expression was inversely correlated with PDL1 expression, mainly in TNBC. | |||||||
| Zhai, 2022 [64] | MDA-231, SK-BR-3, HCC2157, BT474, HCC1569, T47D | 120 | Yes | Yes [g] | [g] In vivo studies focused on miR-181a-5p, not NDRG2. | ||
| 1. NDRG2 expression was high in normal tissue compared with breast cancer. | |||||||
| 2. Patients with NDRG2 -low tumors had worse outcomes than those with NDRG2 -high tumors. | |||||||
| 3. MiR-181a-5p inhibited NDRG2 to promote proliferation and invasion via activation of the PTEN/AKT pathway. | |||||||
| Tumor promoter | Kloten, 2016 [75] | HCC1806, BT20, MCF-7 | 62 + 211 [h] | Yes | No | [h] Involved 62 tissue samples, 45 from breast tumors and 17 from adjacent normal tissues, and tissue microarray with 211 patient samples. | |
| 1. Basal-like tumors had abundant NDRG2 expression compared with luminal tumors. | |||||||
| 2. Basal-like tumors had positive correlation with NDRG2 expression. | |||||||
| 3. Tumor suppressor function could be limited to luminal and basal-B subtypes, but NDRG2 acted as a tumor promoter in basal-A subtype. | |||||||
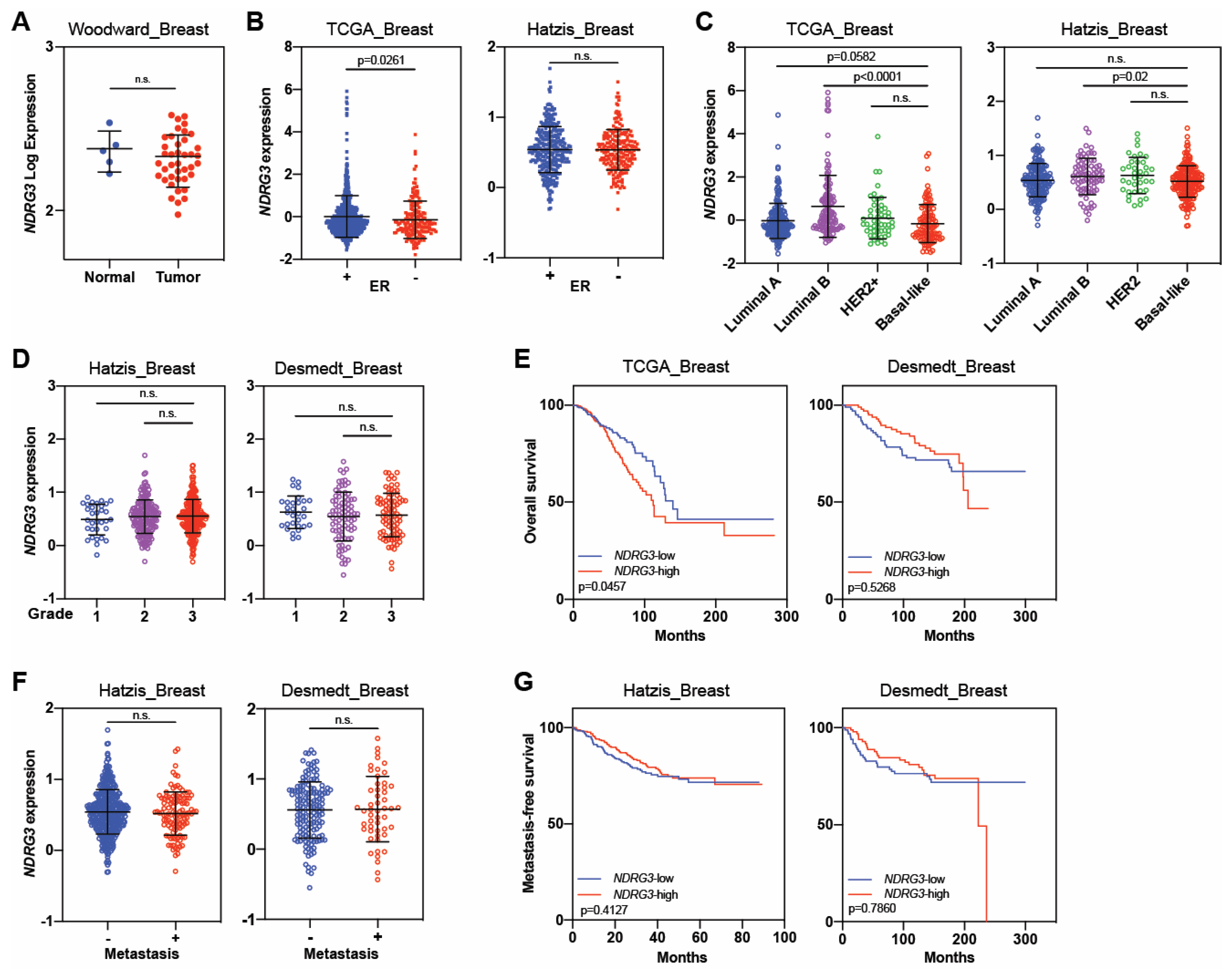
| NDRG3 | Tumor suppressor | Estiar, 2017 [79] | Not reported | 88 | Not reported | Not reported | 1. NDRG3 was downregulated in patients with breast cancer, particularly those with advanced disease. |
| Tumor promoter | Kim, 2019 [78] | Not reported | 1339 | Not reported | Not reported | 1. Patients with NDRG3-positive invasive breast cancer had worse overall survival than those with NDRG3-negative tumors. | |
| 2. NDRG3 independently predicted worse overall survival and disease-free survival. | |||||||
| NDRG4 | Tumor suppressor | Jandrey, 2019 [80] | MCF-7, T47D, MDA-MB-231, MDA-MB-435 | 61 | Yes | No | 1. NDRG4 was highly methylated in breast cancer samples relative to normal breast. |
| 2. Patients with NDRG4-methylated tumors had worse overall survival and distant metastasis-free survival. | |||||||
| 3. NDRG4 methylation status was an independent predictor of distant metastasis-free survival. | |||||||
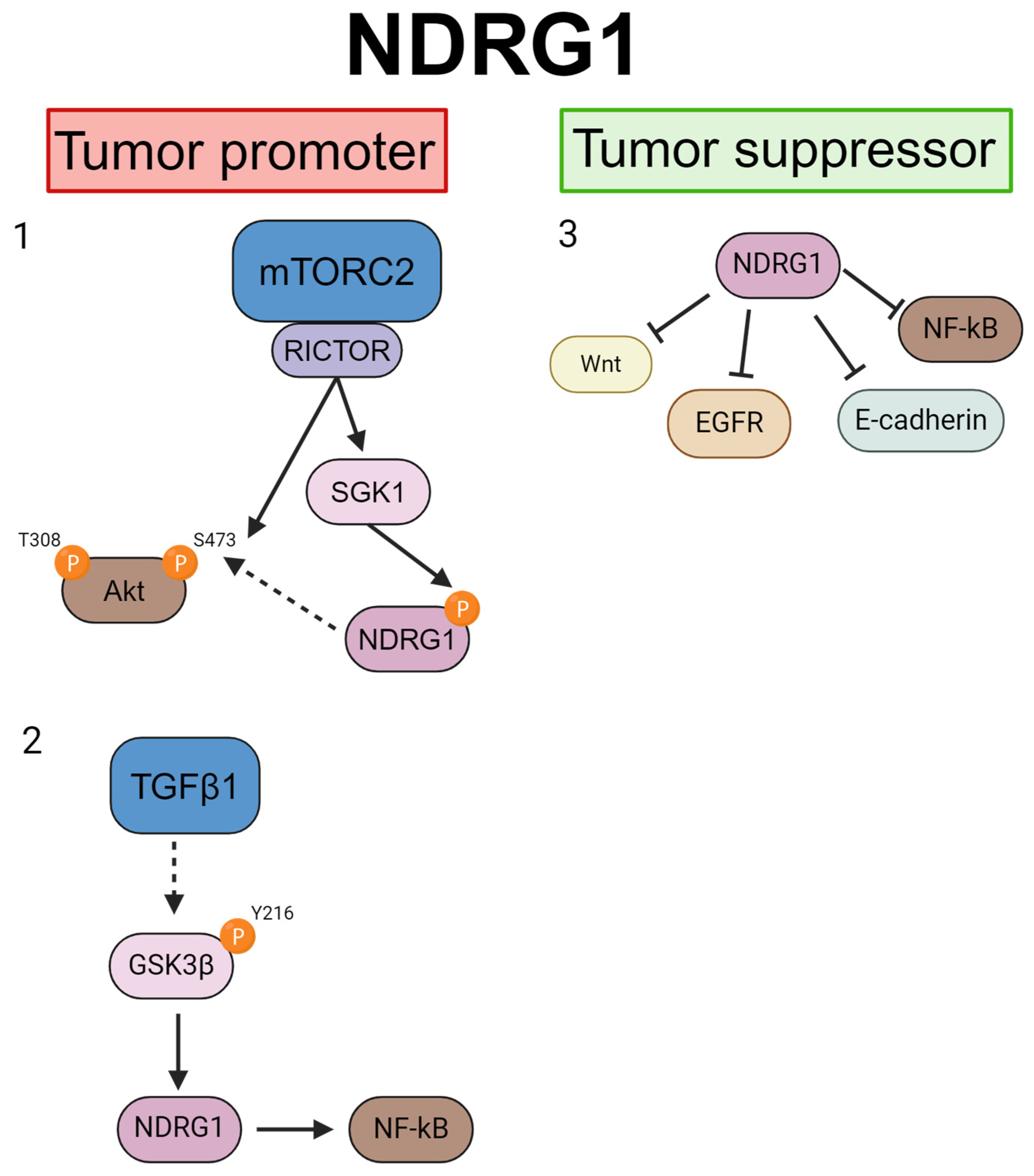
4. NDRG Signaling Pathways
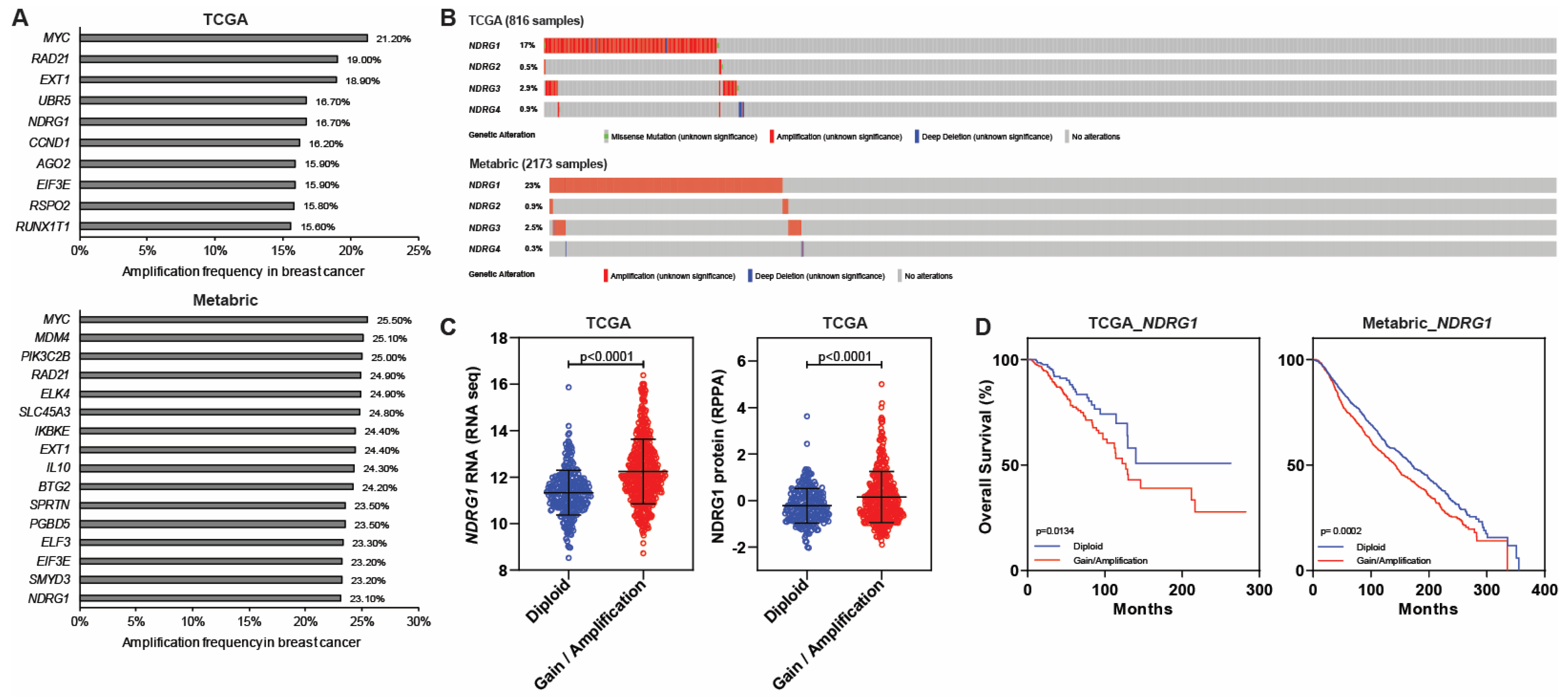
5. NDRGs and Amplification in Breast Cancer
6. NDRGs and Outcomes in Breast Cancer
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Davies, K.R.; Cantor, S.B.; Brewster, A.M. Better contralateral breast cancer risk estimation and alternative options to contralateral prophylactic mastectomy. Int. J. Womens Health 2015, 7, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Gnant, M. Breast cancer. Lancet 2017, 389, 1134–1150. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Perou, C.M. Deconstructing the molecular portraits of breast cancer. Mol. Oncol. 2011, 5, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Vuong, D.; Simpson, P.T.; Green, B.; Cummings, M.C.; Lakhani, S.R. Molecular classification of breast cancer. Virchows Arch. 2014, 465, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Dewan, K.; Mandal, A. Surrogate molecular classification of breast carcinoma: A classification in need or a dilemma indeed. Oncol. J. India 2020, 4, 79–86. [Google Scholar] [CrossRef]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Primers 2019, 5, 66. [Google Scholar] [CrossRef]
- Nascimento, R.G.d.; Otoni, K.M. Histological and molecular classification of breast cancer: What do we know? Mastology 2020, 30, 1–8. [Google Scholar] [CrossRef]
- Alluri, P.; Newman, L.A. Basal-like and triple-negative breast cancers: Searching for positives among many negatives. Surg. Oncol. Clin. N. Am. 2014, 23, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Badve, S.; Dabbs, D.J.; Schnitt, S.J.; Baehner, F.L.; Decker, T.; Eusebi, V.; Fox, S.B.; Ichihara, S.; Jacquemier, J.; Lakhani, S.R.; et al. Basal-like and triple-negative breast cancers: A critical review with an emphasis on the implications for pathologists and oncologists. Mod. Pathol. 2011, 24, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Bertucci, F.; Finetti, P.; Cervera, N.; Esterni, B.; Hermitte, F.; Viens, P.; Birnbaum, D. How basal are triple-negative breast cancers? Int. J. Cancer 2008, 123, 236–240. [Google Scholar] [CrossRef]
- Slamon, D.; Eiermann, W.; Robert, N.; Pienkowski, T.; Martin, M.; Press, M.; Mackey, J.; Glaspy, J.; Chan, A.; Pawlicki, M.; et al. Adjuvant trastuzumab in HER2-positive breast cancer. N. Engl. J. Med. 2011, 365, 1273–1283. [Google Scholar] [CrossRef]
- Romond, E.H.; Perez, E.A.; Bryant, J.; Suman, V.J.; Geyer, C.E., Jr.; Davidson, N.E.; Tan-Chiu, E.; Martino, S.; Paik, S.; Kaufman, P.A.; et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N. Engl. J. Med. 2005, 353, 1673–1684. [Google Scholar] [CrossRef] [PubMed]
- Piccart-Gebhart, M.J.; Procter, M.; Leyland-Jones, B.; Goldhirsch, A.; Untch, M.; Smith, I.; Gianni, L.; Baselga, J.; Bell, R.; Jackisch, C.; et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N. Engl. J. Med. 2005, 353, 1659–1672. [Google Scholar] [CrossRef]
- Chen, X.; Li, Y.; Lin, M.; Lu, Y. Efficacy and safety of treatment with or without pertuzumab for HER2-positive breast cancer: A meta-analysis. Medicine 2023, 102, e33925. [Google Scholar] [CrossRef]
- Masuda, H.; Zhang, D.; Bartholomeusz, C.; Doihara, H.; Hortobagyi, G.N.; Ueno, N.T. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res. Treat. 2012, 136, 331–345. [Google Scholar] [CrossRef]
- Kristensen, T.B.; Knutsson, M.L.; Wehland, M.; Laursen, B.E.; Grimm, D.; Warnke, E.; Magnusson, N.E. Anti-vascular endothelial growth factor therapy in breast cancer. Int. J. Mol. Sci. 2014, 15, 23024–23041. [Google Scholar] [CrossRef]
- Corkery, B.; Crown, J.; Clynes, M.; O’Donovan, N. Epidermal growth factor receptor as a potential therapeutic target in triple-negative breast cancer. Ann. Oncol. 2009, 20, 862–867. [Google Scholar] [CrossRef]
- Vaes, N.; Schonkeren, S.L.; Brosens, E.; Koch, A.; McCann, C.J.; Thapar, N.; Hofstra, R.M.W.; van Engeland, M.; Melotte, V. A combined literature and in silico analysis enlightens the role of the NDRG family in the gut. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2140–2151. [Google Scholar] [CrossRef] [PubMed]
- Schonkeren, S.L.; Massen, M.; van der Horst, R.; Koch, A.; Vaes, N.; Melotte, V. Nervous NDRGs: The N-myc downstream-regulated gene family in the central and peripheral nervous system. Neurogenetics 2019, 20, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Melotte, V.; Qu, X.; Ongenaert, M.; van Criekinge, W.; de Bruine, A.P.; Baldwin, H.S.; van Engeland, M. The N-myc downstream regulated gene (NDRG) family: Diverse functions, multiple applications. FASEB J. 2010, 24, 4153–4166. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.Y.; Chun, Y.S.; Shin, H.W.; Park, J.W. Potential role of the N-MYC downstream-regulated gene family in reprogramming cancer metabolism under hypoxia. Oncotarget 2016, 7, 57442–57451. [Google Scholar] [CrossRef] [PubMed]
- Stein, S.; Thomas, E.K.; Herzog, B.; Westfall, M.D.; Rocheleau, J.V.; Jackson, R.S., 2nd; Wang, M.; Liang, P. NDRG1 is necessary for p53-dependent apoptosis. J. Biol. Chem. 2004, 279, 48930–48940. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Wang, L.; Liu, X.; Yang, Q.; Zhang, J.; Zhang, W.; Wu, Y.; Shen, L.; Zhang, Y.; Yang, A.; et al. Promoter methylation, mutation, and genomic deletion are involved in the decreased NDRG2 expression levels in several cancer cell lines. Biochem. Biophys. Res. Commun. 2007, 358, 164–169. [Google Scholar] [CrossRef]
- Croessmann, S.; Wong, H.Y.; Zabransky, D.J.; Chu, D.; Mendonca, J.; Sharma, A.; Mohseni, M.; Rosen, D.M.; Scharpf, R.B.; Cidado, J.; et al. NDRG1 links p53 with proliferation-mediated centrosome homeostasis and genome stability. Proc. Natl. Acad. Sci. USA 2015, 112, 11583–11588. [Google Scholar] [CrossRef]
- Qu, X.; Zhai, Y.; Wei, H.; Zhang, C.; Xing, G.; Yu, Y.; He, F. Characterization and expression of three novel differentiation-related genes belong to the human NDRG gene family. Mol. Cell. Biochem. 2002, 229, 35–44. [Google Scholar] [CrossRef]
- Ulrix, W.; Swinnen, J.V.; Heyns, W.; Verhoeven, G. The differentiation-related gene 1, Drg1, is markedly upregulated by androgens in LNCaP prostatic adenocarcinoma cells. FEBS Lett. 1999, 455, 23–26. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- Ma, J.; Liu, W.; Guo, H.; Li, S.; Cao, W.; Du, X.; Lei, S.; Hou, W.; Xiong, L.; Yao, L.; et al. N-myc downstream-regulated gene 2 expression is associated with glucose transport and correlated with prognosis in breast carcinoma. Breast Cancer Res. 2014, 16, R27. [Google Scholar] [CrossRef]
- Zhao, W.; Tang, R.; Huang, Y.; Wang, W.; Zhou, Z.; Gu, S.; Dai, J.; Ying, K.; Xie, Y.; Mao, Y. Cloning and expression pattern of the human NDRG3 gene. Biochim. Biophys. Acta 2001, 1519, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; She, J.; Yang, J.; Bu, X.; Ji, G.; Zhu, S.; He, S.; Chu, D. NDRG4 in gastric cancer determines tumor cell proliferation and clinical outcome. Mol. Carcinog. 2018, 57, 762–771. [Google Scholar] [CrossRef]
- Chu, D.; Zhang, Z.; Zhou, Y.; Li, Y.; Zhu, S.; Zhang, J.; Zhao, Q.; Ji, G.; Wang, W.; Zheng, J. NDRG4, a novel candidate tumor suppressor, is a predictor of overall survival of colorectal cancer patients. Oncotarget 2015, 6, 7584–7596. [Google Scholar] [CrossRef]
- Ghalayini, M.K.; Dong, Q.; Richardson, D.R.; Assinder, S.J. Proteolytic cleavage and truncation of NDRG1 in human prostate cancer cells, but not normal prostate epithelial cells. Biosci. Rep. 2013, 33, e00042. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Kim, J.H. SUMO modification regulates the protein stability of NDRG1. Biochem. Biophys. Res. Commun. 2015, 459, 161–165. [Google Scholar] [CrossRef]
- Bandyopadhyay, S.; Pai, S.K.; Hirota, S.; Hosobe, S.; Tsukada, T.; Miura, K.; Takano, Y.; Saito, K.; Commes, T.; Piquemal, D.; et al. PTEN up-regulates the tumor metastasis suppressor gene Drg-1 in prostate and breast cancer. Cancer Res. 2004, 64, 7655–7660. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Pai, S.K.; Hirota, S.; Hosobe, S.; Takano, Y.; Saito, K.; Piquemal, D.; Commes, T.; Watabe, M.; Gross, S.C.; et al. Role of the putative tumor metastasis suppressor gene Drg-1 in breast cancer progression. Oncogene 2004, 23, 5675–5681. [Google Scholar] [CrossRef] [PubMed]
- Fotovati, A.; Fujii, T.; Yamaguchi, M.; Kage, M.; Shirouzu, K.; Oie, S.; Basaki, Y.; Ono, M.; Yamana, H.; Kuwano, M. 17Beta-estradiol induces down-regulation of Cap43/NDRG1/Drg-1, a putative differentiation-related and metastasis suppressor gene, in human breast cancer cells. Clin. Cancer Res. 2006, 12, 3010–3018. [Google Scholar] [CrossRef]
- Tian, S.; Wang, X.; Proud, C.G. Oncogenic MNK signalling regulates the metastasis suppressor NDRG1. Oncotarget 2017, 8, 46121–46135. [Google Scholar] [CrossRef]
- Salis, O.; Okuyucu, A.; Bedir, A.; Gor, U.; Kulcu, C.; Yenen, E.; Kilic, N. Antimetastatic effect of fluvastatin on breast and hepatocellular carcinoma cells in relation to SGK1 and NDRG1 genes. Tumour Biol. 2016, 37, 3017–3024. [Google Scholar] [CrossRef] [PubMed]
- Redmond, K.L.; Crawford, N.T.; Farmer, H.; D’Costa, Z.C.; O’Brien, G.J.; Buckley, N.E.; Kennedy, R.D.; Johnston, P.G.; Harkin, D.P.; Mullan, P.B. T-box 2 represses NDRG1 through an EGR1-dependent mechanism to drive the proliferation of breast cancer cells. Oncogene 2010, 29, 3252–3262. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.C.; Su, Y.Y.; Chen, K.C.; Tsai, M.H.; Sher, Y.P.; Lu, T.P.; Lee, C.Y.; Chuang, E.Y. Down-regulation of NDRG1 promotes migration of cancer cells during reoxygenation. PLoS ONE 2011, 6, e24375. [Google Scholar] [CrossRef] [PubMed]
- Han, L.L.; Hou, L.; Zhou, M.J.; Ma, Z.L.; Lin, D.L.; Wu, L.; Ge, Y.L. Aberrant NDRG1 methylation associated with its decreased expression and clinicopathological significance in breast cancer. J. Biomed. Sci. 2013, 20, 52. [Google Scholar] [CrossRef] [PubMed]
- Godbole, M.; Togar, T.; Patel, K.; Dharavath, B.; Yadav, N.; Janjuha, S.; Gardi, N.; Tiwary, K.; Terwadkar, P.; Desai, S.; et al. Up-regulation of the kinase gene SGK1 by progesterone activates the AP-1-NDRG1 axis in both PR-positive and -negative breast cancer cells. J. Biol. Chem. 2018, 293, 19263–19276. [Google Scholar] [CrossRef] [PubMed]
- Chiang, K.C.; Yeh, C.N.; Chung, L.C.; Feng, T.H.; Sun, C.C.; Chen, M.F.; Jan, Y.Y.; Yeh, T.S.; Chen, S.C.; Juang, H.H. WNT-1 inducible signaling pathway protein-1 enhances growth and tumorigenesis in human breast cancer. Sci. Rep. 2015, 5, 8686. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.T.; Campbell, D.G.; Morrice, N.; Auld, G.C.; Shpiro, N.; Marquez, R.; Peggie, M.; Bain, J.; Bloomberg, G.B.; Grahammer, F.; et al. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem. J. 2004, 384, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xing, F.; Iiizumi-Gairani, M.; Okuda, H.; Watabe, M.; Pai, S.K.; Pandey, P.R.; Hirota, S.; Kobayashi, A.; Mo, Y.Y.; et al. N-myc downstream regulated gene 1 modulates Wnt-beta-catenin signalling and pleiotropically suppresses metastasis. EMBO Mol. Med. 2012, 4, 93–108. [Google Scholar] [CrossRef] [PubMed]
- Abascal, M.F.; Elia, A.; Alvarez, M.; Pataccini, G.; Sequeira, G.; Riggio, M.; Figueroa, V.; Lamb, C.A.; Rojas, P.A.; Spengler, E.; et al. Progesterone receptor isoform ratio dictates antiprogestin/progestin effects on breast cancer growth and metastases: A role for NDRG1. Int. J. Cancer 2022, 150, 1481–1496. [Google Scholar] [CrossRef]
- Fotovati, A.; Abu-Ali, S.; Kage, M.; Shirouzu, K.; Yamana, H.; Kuwano, M. N-myc downstream-regulated gene 1 (NDRG1) a differentiation marker of human breast cancer. Pathol. Oncol. Res. 2011, 17, 525–533. [Google Scholar] [CrossRef]
- Villodre, E.S.; Hu, X.; Eckhardt, B.L.; Larson, R.; Huo, L.; Yoon, E.C.; Gong, Y.; Song, J.; Liu, S.; Ueno, N.T.; et al. NDRG1 in Aggressive Breast Cancer Progression and Brain Metastasis. J. Natl. Cancer Inst. 2022, 114, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Villodre, E.S.; Gong, Y.; Hu, X.; Huo, L.; Yoon, E.C.; Ueno, N.T.; Woodward, W.A.; Tripathy, D.; Song, J.; Debeb, B.G. NDRG1 Expression Is an Independent Prognostic Factor in Inflammatory Breast Cancer. Cancers 2020, 12, 3711. [Google Scholar] [CrossRef] [PubMed]
- Sommer, E.M.; Dry, H.; Cross, D.; Guichard, S.; Davies, B.R.; Alessi, D.R. Elevated SGK1 predicts resistance of breast cancer cells to Akt inhibitors. Biochem. J. 2013, 452, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Sevinsky, C.J.; Khan, F.; Kokabee, L.; Darehshouri, A.; Maddipati, K.R.; Conklin, D.S. NDRG1 regulates neutral lipid metabolism in breast cancer cells. Breast Cancer Res. 2018, 20, 55. [Google Scholar] [CrossRef] [PubMed]
- Parris, T.Z.; Kovacs, A.; Hajizadeh, S.; Nemes, S.; Semaan, M.; Levin, M.; Karlsson, P.; Helou, K. Frequent MYC coamplification and DNA hypomethylation of multiple genes on 8q in 8p11-p12-amplified breast carcinomas. Oncogenesis 2014, 3, e95. [Google Scholar] [CrossRef] [PubMed]
- Nagai, M.A.; Gerhard, R.; Fregnani, J.H.; Nonogaki, S.; Rierger, R.B.; Netto, M.M.; Soares, F.A. Prognostic value of NDRG1 and SPARC protein expression in breast cancer patients. Breast Cancer Res. Treat. 2011, 126, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Bonello, M.; Byron, A.; Langdon, S.P.; Sims, A.H. Evaluation of Gene Expression Data From Cybrids and Tumours Highlights Elevated NDRG1-Driven Proliferation in Triple-Negative Breast Cancer. Breast Cancer 2020, 14, 1178223420934447. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.Y.; Fan, C.F.; Wei, J.; Liu, C.; Zheng, H.C.; Yao, F.; Jin, F. Increased N-myc downstream-regulated gene 1 expression is associated with breast atypia-to-carcinoma progression. Tumour Biol. 2011, 32, 1271–1276. [Google Scholar] [CrossRef] [PubMed]
- Li, E.Y.; Huang, W.Y.; Chang, Y.C.; Tsai, M.H.; Chuang, E.Y.; Kuok, Q.Y.; Bai, S.T.; Chao, L.Y.; Sher, Y.P.; Lai, L.C. Aryl Hydrocarbon Receptor Activates NDRG1 Transcription under Hypoxia in Breast Cancer Cells. Sci. Rep. 2016, 6, 20808. [Google Scholar] [CrossRef]
- de Nonneville, A.; Finetti, P.; Mamessier, E.; Bertucci, F. RE: NDRG1 in Aggressive Breast Cancer Progression and Brain Metastasis. J. Natl. Cancer Inst. 2022, 114, 1046–1047. [Google Scholar] [CrossRef]
- Berghoff, A.S.; Liao, Y.; Karreman, M.A.; Ilhan-Mutlu, A.; Gunkel, K.; Sprick, M.R.; Eisen, C.; Kessler, T.; Osswald, M.; Wunsche, S.; et al. Identification and Characterization of Cancer Cells That Initiate Metastases to the Brain and Other Organs. Mol. Cancer Res. 2021, 19, 688–701. [Google Scholar] [CrossRef] [PubMed]
- Krug, K.; Jaehnig, E.J.; Satpathy, S.; Blumenberg, L.; Karpova, A.; Anurag, M.; Miles, G.; Mertins, P.; Geffen, Y.; Tang, L.C.; et al. Proteogenomic Landscape of Breast Cancer Tumorigenesis and Targeted Therapy. Cell 2020, 183, 1436–1456.e31. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Tejada, A.; Grinan-Lison, C.; Gonzalez-Gonzalez, A.; Cara, F.E.; Luque, R.J.; Rosa-Garrido, C.; Blaya-Canovas, J.L.; Navarro-Ocon, A.; Valenzuela-Torres, M.; Parra-Lopez, M.; et al. TGFbeta Governs the Pleiotropic Activity of NDRG1 in Triple-Negative Breast Cancer Progression. Int. J. Biol. Sci. 2023, 19, 204–224. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.; Mu, T.; Zhao, L.; Li, Y.; Zhu, D.; Pan, Y. MiR-181a-5p facilitates proliferation, invasion, and glycolysis of breast cancer through NDRG2-mediated activation of PTEN/AKT pathway. Bioengineered 2022, 13, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Yu, S.; Zhang, Y.; Zhang, Y.; Zhao, H.; Xiao, Z.; Yao, L.; Chen, S.; Zhang, J. NDRG2 promotes adriamycin sensitivity through a Bad/p53 complex at the mitochondria in breast cancer. Oncotarget 2017, 8, 29038–29047. [Google Scholar] [CrossRef] [PubMed]
- Shon, S.K.; Kim, A.; Kim, J.Y.; Kim, K.I.; Yang, Y.; Lim, J.S. Bone morphogenetic protein-4 induced by NDRG2 expression inhibits MMP-9 activity in breast cancer cells. Biochem. Biophys. Res. Commun. 2009, 385, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Shon, S.K.; Kim, A.; Kim, K.I.; Yang, Y.; Cho, D.H.; Lee, M.S.; Lim, J.S. SOCS1 induced by NDRG2 expression negatively regulates STAT3 activation in breast cancer cells. Biochem. Biophys. Res. Commun. 2007, 363, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.S.; Kim, D.; Kim, D.H.; Chang, H.H.; Sohn, K.C.; Kim, K.H.; Jung, S.H.; Lee, B.K.; Kim, J.H.; Kim, K.D. NDRG2 correlated with favorable recurrence-free survival inhibits metastasis of mouse breast cancer cells via attenuation of active TGF-beta production. Carcinogenesis 2012, 33, 1882–1888. [Google Scholar] [CrossRef] [PubMed]
- Lorentzen, A.; Lewinsky, R.H.; Bornholdt, J.; Vogel, L.K.; Mitchelmore, C. Expression profile of the N-myc Downstream Regulated Gene 2 (NDRG2) in human cancers with focus on breast cancer. BMC Cancer 2011, 11, 14. [Google Scholar] [CrossRef]
- Lee, A.; Lim, S.; Oh, J.; Lim, J.; Yang, Y.; Lee, M.S.; Lim, J.S. NDRG2 Expression in Breast Cancer Cells Downregulates PD-L1 Expression and Restores T Cell Proliferation in Tumor-Coculture. Cancers 2021, 13, 6112. [Google Scholar] [CrossRef]
- Kim, M.J.; Lim, J.; Yang, Y.; Lee, M.S.; Lim, J.S. N-myc downstream-regulated gene 2 (NDRG2) suppresses the epithelial-mesenchymal transition (EMT) in breast cancer cells via STAT3/Snail signaling. Cancer Lett. 2014, 354, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Kim, H.S.; Lee, S.H.; Yang, Y.; Lee, M.S.; Lim, J.S. NDRG2 controls COX-2/PGE(2)-mediated breast cancer cell migration and invasion. Mol. Cells 2014, 37, 759–765. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, M.J.; Lim, J.; Yang, Y.; Lee, M.S.; Lim, J.S. NDRG2 overexpression enhances glucose deprivation-mediated apoptosis in breast cancer cells via inhibition of the LKB1-AMPK pathway. Genes Cancer 2014, 5, 175–185. [Google Scholar] [CrossRef]
- Kim, B.; Nam, S.; Lim, J.H.; Lim, J.S. NDRG2 Expression Decreases Tumor-Induced Osteoclast Differentiation by Down-regulating ICAM1 in Breast Cancer Cells. Biomol. Ther. 2016, 24, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Kloten, V.; Schlensog, M.; Eschenbruch, J.; Gasthaus, J.; Tiedemann, J.; Mijnes, J.; Heide, T.; Braunschweig, T.; Knuchel, R.; Dahl, E. Abundant NDRG2 Expression Is Associated with Aggressiveness and Unfavorable Patients’ Outcome in Basal-Like Breast Cancer. PLoS ONE 2016, 11, e0159073. [Google Scholar] [CrossRef]
- Ma, J.; Liu, W.; Yan, X.; Wang, Q.; Zhao, Q.; Xue, Y.; Ren, H.; Wu, L.; Cheng, Y.; Li, S.; et al. Inhibition of endothelial cell proliferation and tumor angiogenesis by up-regulating NDRG2 expression in breast cancer cells. PLoS ONE 2012, 7, e32368. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Liu, Q.; Li, Y.; Yang, J.; Ma, J.; Yu, F.; Shi, H.; Ren, Q.; Zhang, R.; Zhang, J.; et al. NDRG2 expression regulates CD24 and metastatic potential of breast cancer cells. Asian Pac. J. Cancer Prev. 2010, 11, 1817–1821. [Google Scholar]
- Kim, M.C.; Park, M.H.; Kang, S.H.; Bae, Y.K. NDRG3 protein expression is associated with aggressive biologic phenotype and unfavorable outcome in patients with invasive breast cancer. Int. J. Clin. Exp. Pathol. 2019, 12, 3886–3893. [Google Scholar] [PubMed]
- Estiar, M.A.; Zare, A.A.; Esmaeili, R.; Farahmand, L.; Fazilaty, H.; Jafari, D.; Samadi, T.; Majidzadeh, A.K. Clinical significance of NDRG3 in patients with breast cancer. Future Oncol. 2017, 13, 961–969. [Google Scholar] [CrossRef]
- Jandrey, E.H.F.; Moura, R.P.; Andrade, L.N.S.; Machado, C.L.; Campesato, L.F.; Leite, K.R.M.; Inoue, L.T.; Asprino, P.F.; da Silva, A.P.M.; de Barros, A.; et al. NDRG4 promoter hypermethylation is a mechanistic biomarker associated with metastatic progression in breast cancer patients. NPJ Breast Cancer 2019, 5, 11. [Google Scholar] [CrossRef]
- Menezes, S.V.; Sahni, S.; Kovacevic, Z.; Richardson, D.R. Interplay of the iron-regulated metastasis suppressor NDRG1 with epidermal growth factor receptor (EGFR) and oncogenic signaling. J. Biol. Chem. 2017, 292, 12772–12782. [Google Scholar] [CrossRef]
- Bae, D.H.; Jansson, P.J.; Huang, M.L.; Kovacevic, Z.; Kalinowski, D.; Lee, C.S.; Sahni, S.; Richardson, D.R. The role of NDRG1 in the pathology and potential treatment of human cancers. J. Clin. Pathol. 2013, 66, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhang, D.; Yue, F.; Zheng, M.; Kovacevic, Z.; Richardson, D.R. The iron chelators Dp44mT and DFO inhibit TGF-beta-induced epithelial-mesenchymal transition via up-regulation of N-Myc downstream-regulated gene 1 (NDRG1). J. Biol. Chem. 2012, 287, 17016–17028. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yin, D.; Xie, C.; Zheng, T.; Liang, Y.; Hong, X.; Lu, Z.; Song, X.; Song, R.; Yang, H.; et al. The iron chelator Dp44mT inhibits hepatocellular carcinoma metastasis via N-Myc downstream-regulated gene 2 (NDRG2)/gp130/STAT3 pathway. Oncotarget 2014, 5, 8478–8491. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.; Kim, M.J.; Yang, Y.; Kim, J.W.; Yeom, Y.I.; Lim, J.S. Suppression of NF-kappaB activity by NDRG2 expression attenuates the invasive potential of highly malignant tumor cells. Carcinogenesis 2009, 30, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Matsui, A.; Ihara, T.; Suda, H.; Mikami, H.; Semba, K. Gene amplification: Mechanisms and involvement in cancer. Biomol. Concepts 2013, 4, 567–582. [Google Scholar] [CrossRef] [PubMed]
- Albertson, D.G. Gene amplification in cancer. Trends Genet. 2006, 22, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Kadota, M.; Sato, M.; Duncan, B.; Ooshima, A.; Yang, H.H.; Diaz-Meyer, N.; Gere, S.; Kageyama, S.; Fukuoka, J.; Nagata, T.; et al. Identification of novel gene amplifications in breast cancer and coexistence of gene amplification with an activating mutation of PIK3CA. Cancer Res. 2009, 69, 7357–7365. [Google Scholar] [CrossRef] [PubMed]
- Bizari, L.; Borim, A.A.; Leite, K.R.; Goncalves Fde, T.; Cury, P.M.; Tajara, E.H.; Silva, A.E. Alterations of the CCND1 and HER-2/neu (ERBB2) proteins in esophageal and gastric cancers. Cancer Genet. Cytogenet. 2006, 165, 41–50. [Google Scholar] [CrossRef]
- Cuny, M.; Kramar, A.; Courjal, F.; Johannsdottir, V.; Iacopetta, B.; Fontaine, H.; Grenier, J.; Culine, S.; Theillet, C. Relating genotype and phenotype in breast cancer: An analysis of the prognostic significance of amplification at eight different genes or loci and of p53 mutations. Cancer Res. 2000, 60, 1077–1083. [Google Scholar]
- Al-Kuraya, K.; Schraml, P.; Torhorst, J.; Tapia, C.; Zaharieva, B.; Novotny, H.; Spichtin, H.; Maurer, R.; Mirlacher, M.; Kochli, O.; et al. Prognostic relevance of gene amplifications and coamplifications in breast cancer. Cancer Res. 2004, 64, 8534–8540. [Google Scholar] [CrossRef]
- Ciriello, G.; Gatza, M.L.; Beck, A.H.; Wilkerson, M.D.; Rhie, S.K.; Pastore, A.; Zhang, H.; McLellan, M.; Yau, C.; Kandoth, C.; et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015, 163, 506–519. [Google Scholar] [CrossRef] [PubMed]
- Rueda, O.M.; Sammut, S.J.; Seoane, J.A.; Chin, S.F.; Caswell-Jin, J.L.; Callari, M.; Batra, R.; Pereira, B.; Bruna, A.; Ali, H.R.; et al. Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups. Nature 2019, 567, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Pereira, B.; Chin, S.F.; Rueda, O.M.; Vollan, H.K.; Provenzano, E.; Bardwell, H.A.; Pugh, M.; Jones, L.; Russell, R.; Sammut, S.J.; et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 2016, 7, 11479. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Mena, O.; Medina-Martinez, I.; Juarez-Torres, E.; Barron, V.; Espinosa, A.; Villegas-Sepulveda, N.; Gomez-Laguna, L.; Nieto-Martinez, K.; Orozco, L.; Roman-Basaure, E.; et al. Amplified genes may be overexpressed, unchanged, or downregulated in cervical cancer cell lines. PLoS ONE 2012, 7, e32667. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Chen, L.; Jia, Q.; Dou, X.; Xu, N.; Liao, D.J. The well-accepted notion that gene amplification contributes to increased expression still remains, after all these years, a reasonable but unproven assumption. J. Carcinog. 2016, 15, 3. [Google Scholar] [CrossRef] [PubMed]
- Press, M.F.; Slamon, D.J.; Flom, K.J.; Park, J.; Zhou, J.Y.; Bernstein, L. Evaluation of HER-2/neu gene amplification and overexpression: Comparison of frequently used assay methods in a molecularly characterized cohort of breast cancer specimens. J. Clin. Oncol. 2002, 20, 3095–3105. [Google Scholar] [CrossRef] [PubMed]
- Hoang, M.P.; Sahin, A.A.; Ordonez, N.G.; Sneige, N. HER-2/neu gene amplification compared with HER-2/neu protein overexpression and interobserver reproducibility in invasive breast carcinoma. Am. J. Clin. Pathol. 2000, 113, 852–859. [Google Scholar] [CrossRef]
- Luoh, S.W.; Ramsey, B.; Hanlon Newell, A.; Troxell, M.; Hu, Z.; Chin, K.; Spellman, P.; Olson, S.; Keenan, E. HER-2 gene amplification in human breast cancer without concurrent HER-2 over-expression. Springerplus 2013, 2, 386. [Google Scholar] [CrossRef]
- Kao, K.J.; Chang, K.M.; Hsu, H.C.; Huang, A.T. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: Implications for treatment optimization. BMC Cancer 2011, 11, 143. [Google Scholar] [CrossRef] [PubMed]
- Esserman, L.J.; Moore, D.H.; Tsing, P.J.; Chu, P.W.; Yau, C.; Ozanne, E.; Chung, R.E.; Tandon, V.J.; Park, J.W.; Baehner, F.L.; et al. Biologic markers determine both the risk and the timing of recurrence in breast cancer. Breast Cancer Res. Treat. 2011, 129, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Woodward, W.A.; Krishnamurthy, S.; Yamauchi, H.; El-Zein, R.; Ogura, D.; Kitadai, E.; Niwa, S.; Cristofanilli, M.; Vermeulen, P.; Dirix, L.; et al. Genomic and expression analysis of microdissected inflammatory breast cancer. Breast Cancer Res. Treat. 2013, 138, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Hatzis, C.; Pusztai, L.; Valero, V.; Booser, D.J.; Esserman, L.; Lluch, A.; Vidaurre, T.; Holmes, F.; Souchon, E.; Wang, H.; et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA 2011, 305, 1873–1881. [Google Scholar] [CrossRef]
- Desmedt, C.; Piette, F.; Loi, S.; Wang, Y.; Lallemand, F.; Haibe-Kains, B.; Viale, G.; Delorenzi, M.; Zhang, Y.; d’Assignies, M.S.; et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin. Cancer Res. 2007, 13, 3207–3214. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villodre, E.S.; Nguyen, A.P.N.; Debeb, B.G. NDRGs in Breast Cancer: A Review and In Silico Analysis. Cancers 2024, 16, 1342. https://doi.org/10.3390/cancers16071342
Villodre ES, Nguyen APN, Debeb BG. NDRGs in Breast Cancer: A Review and In Silico Analysis. Cancers. 2024; 16(7):1342. https://doi.org/10.3390/cancers16071342
Chicago/Turabian StyleVillodre, Emilly S., Anh P. N. Nguyen, and Bisrat G. Debeb. 2024. "NDRGs in Breast Cancer: A Review and In Silico Analysis" Cancers 16, no. 7: 1342. https://doi.org/10.3390/cancers16071342
APA StyleVillodre, E. S., Nguyen, A. P. N., & Debeb, B. G. (2024). NDRGs in Breast Cancer: A Review and In Silico Analysis. Cancers, 16(7), 1342. https://doi.org/10.3390/cancers16071342







