Preclinical Models for Functional Precision Lung Cancer Research
Simple Summary
Abstract
1. Introduction
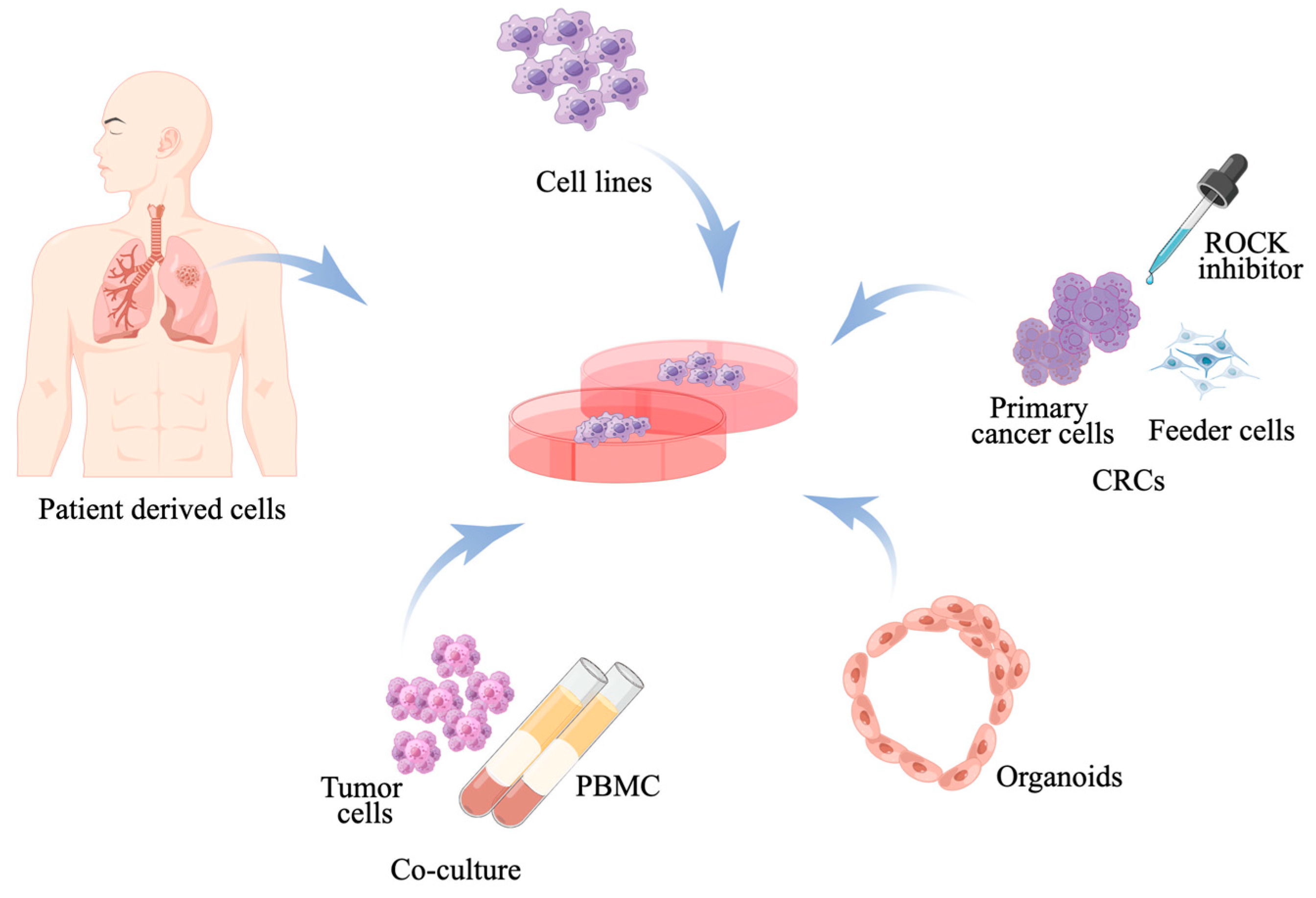
2. In Vitro Model
2.1. Human Lung Cancer Cell Line Models
2.2. Primary Cell Cultures
2.3. Conditionally Reprogrammed Cell
2.4. Cancer Spheroids and Organoids
2.5. Co-Culture System of Patient-Derived Immune Cells and Patient-Derived Tumors
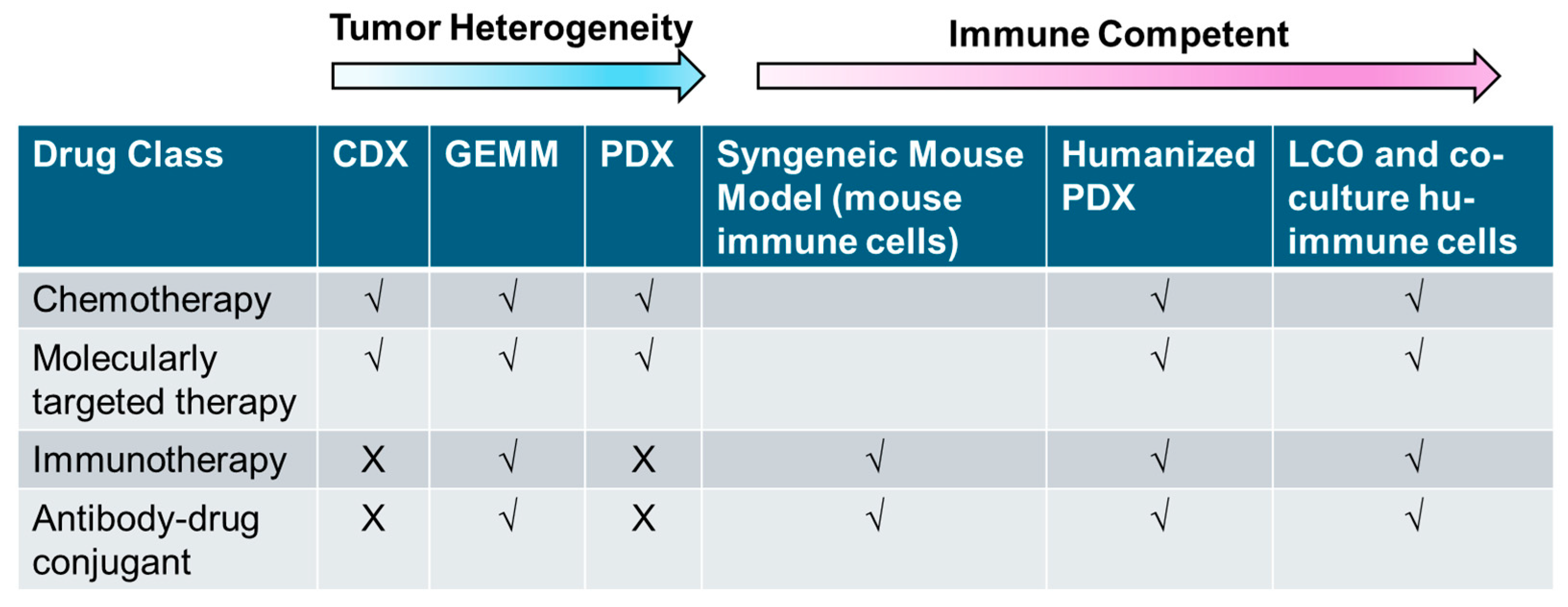
3. In Vivo Models
3.1. Carcinogen-Induced Mouse Model
3.2. Syngeneic Mouse Models
3.3. Transgenic/Genetically Engineered Mouse Models
3.4. Cell Line-Derived Xenografts (CDXs)
3.5. Patient-Derived Xenografts (PDXs)
3.6. Humanized Mouse Models
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ALK | anaplastic lymphoma kinase |
| BaP | polyaromatic hydrocarbons as benzo(a)pyrene |
| BRAF | v-raf murine sarcoma viral oncogene homolog B |
| CDX | cell line-derived xenograft |
| CR | conditional reprogramming |
| CRCs | conditional reprogramming cultures |
| CR-PDX | conditionally reprogrammed cell lines |
| CTLA-4 | cytotoxic T-lymphocyte-associated protein 4 |
| EGFR | epidermal growth factor receptor |
| ErbB2 | erb-b2 receptor tyrosine kinase 2 |
| FEN1 | flap endonuclease 1 |
| GEM | genetically engineered model |
| GEMMs | genetically engineered mouse models |
| GLI1 | glioma-associated oncogene homolog 1 |
| GVHD | graft-versus-host disease |
| GM-CSF | granulocyte–macrophage colony-stimulating factor |
| HER2 | human epidermal growth factor 2 |
| HLA | human leukocyte antigen |
| HPSC | hematopoietic stem and progenitor cell |
| Hu-HPSCs | human pluripotent stem cells |
| Hu-PBMCs | human peripheral blood mononuclear cells |
| ICIs | immune checkpoint inhibitors |
| IF | interfemoral |
| IP | intraperitoneal |
| iPSC | induced pluripotent stem cells |
| IV | intravenous |
| KRAS | Kirsten rat sarcoma |
| LCOs | lung cancer organoids |
| LUAD | lung adenocarcinoma |
| LUSC | lung squamous cell carcinoma |
| MCA | diethylnitrosamine and 2-methycholanthrene |
| MDSCs | myeloid-derived suppressor cells |
| MET | mesenchymal epithelial transition |
| NK | natural killer |
| NNK | 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone |
| NRAS | neuroblastoma RAS viral |
| NSCLC | non-small cell lung cancer |
| NSG | nod-scid il2rgammanull |
| NTCU | n-nitroso-tris-chloroethylurea |
| PD | pharmacodynamic |
| PD-1 | programmed cell death protein 1 |
| PDOs | patient-derived organoids |
| PDXs | patient-derived xenografts |
| PI3K | phosphatidylinositol 3-kinase |
| PK | pharmacokinetic |
| PMBC | peripheral blood mononuclear cell |
| RET | rearranged during transfection |
| ROCK | rho kinase inhibitor |
| ROS1 | c-ros oncogene 1 |
| SCLC | small cell lung cancer |
| scRNA | single-cell RNA |
| TKIs | tyrosine kinase inhibitors |
| TME | tumor microenvironment |
| Tregs | regulatory T cells |
| UCB | umbilical cord blood |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; de Camargo Correia, G.S.; Wang, J.; Manochakian, R.; Zhao, Y.; Lou, Y. Emerging Targeted Therapies in Advanced Non-Small-Cell Lung Cancer. Cancers 2023, 15, 2899. [Google Scholar] [CrossRef]
- Reyes, A.; Pharaon, R.; Mohanty, A.; Massarelli, E. Arising Novel Agents in Lung Cancer: Are Bispecifics and ADCs the New Paradigm? Cancers 2023, 15, 3162. [Google Scholar] [CrossRef]
- Patel, S.R.; Das, M. Small Cell Lung Cancer: Emerging Targets and Strategies for Precision Therapy. Cancers 2023, 15, 4016. [Google Scholar] [CrossRef] [PubMed]
- Mulvey, C.K. Emerging Precision Medicine Approaches for Lung Neuroendocrine Tumors. Cancers 2023, 15, 5575. [Google Scholar] [CrossRef]
- Yun, K.M.; Bazhenova, L. Emerging New Targets in Systemic Therapy for Malignant Pleural Mesothelioma. Cancers 2024, 16, 1252. [Google Scholar] [CrossRef]
- Li, T.; Ma, W.; Al-Obeidi, E. Evolving Precision First-Line Systemic Treatment for Patients with Unresectable Non-Small Cell Lung Cancer. Cancers 2024, 16, 2350. [Google Scholar] [CrossRef]
- Ettinger, D.S.; Wood, D.E.; Aisner, D.L.; Akerley, W.; Bauman, J.R.; Bharat, A.; Bruno, D.S.; Chang, J.Y.; Chirieac, L.R.; D’Amico, T.A.; et al. Non-Small Cell Lung Cancer, Version 3.2022, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2022, 20, 497–530. [Google Scholar] [CrossRef] [PubMed]
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment with Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J. Mol. Diagn. 2018, 20, 129–159. [Google Scholar]
- Xie, T.; Qiu, B.M.; Luo, J.; Diao, Y.F.; Hu, L.W.; Liu, X.L.; Shen, Y. Distant metastasis patterns among lung cancer subtypes and impact of primary tumor resection on survival in metastatic lung cancer using SEER database. Sci. Rep. 2024, 14, 22445. [Google Scholar] [CrossRef]
- Chen, Z.; Fillmore, C.M.; Hammerman, P.S.; Kim, C.F.; Wong, K.K. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat. Rev. Cancer 2014, 14, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Ferone, G.; Lee, M.C.; Sage, J.; Berns, A. Cells of origin of lung cancers: Lessons from mouse studies. Genes Dev. 2020, 34, 1017–1032. [Google Scholar] [CrossRef] [PubMed]
- Tata, P.R.; Rajagopal, J. Plasticity in the lung: Making and breaking cell identity. Development 2017, 144, 755–766. [Google Scholar] [CrossRef]
- Collins, F.S.; Varmus, H. A new initiative on precision medicine. N. Engl. J. Med. 2015, 372, 793–795. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Gandhi, J.; Zhang, J.; Xie, Y.; Soh, J.; Shigematsu, H.; Zhang, W.; Yamamoto, H.; Peyton, M.; Girard, L.; Lockwood, W.W.; et al. Alterations in genes of the EGFR signaling pathway and their relationship to EGFR tyrosine kinase inhibitor sensitivity in lung cancer cell lines. PLoS ONE 2009, 4, e4576. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Greninger, P.; Rhodes, D.; Koopman, L.; Violette, S.; Bardeesy, N.; Settleman, J. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 2009, 15, 489–500. [Google Scholar] [CrossRef]
- Soh, J.; Okumura, N.; Lockwood, W.W.; Yamamoto, H.; Shigematsu, H.; Zhang, W.; Chari, R.; Shames, D.S.; Tang, X.; MacAulay, C.; et al. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS ONE 2009, 4, e7464. [Google Scholar] [CrossRef] [PubMed]
- Harbour, J.W.; Lai, S.L.; Whang-Peng, J.; Gazdar, A.F.; Minna, J.D.; Kaye, F.J. Abnormalities in structure and expression of the human retinoblastoma gene in SCLC. Science 1988, 241, 353–357. [Google Scholar] [CrossRef]
- Otterson, G.A.; Kratzke, R.A.; Coxon, A.; Kim, Y.W.; Kaye, F.J. Absence of p16INK4 protein is restricted to the subset of lung cancer lines that retains wildtype RB. Oncogene 1994, 9, 3375–3378. [Google Scholar]
- FJ, K. B and cyclin dependent kinase pathways: Defining a distinction between RB and p16 loss in lung cancer. Oncogene 2002, 2145, 6908–6914. [Google Scholar]
- Rudin, C.M.; Poirier, J.T.; Byers, L.A.; Dive, C.; Dowlati, A.; George, J.; Heymach, J.V.; Johnson, J.E.; Lehman, J.M.; MacPherson, D.; et al. Author Correction: Molecular subtypes of small cell lung cancer: A synthesis of human and mouse model data. Nat. Rev. Cancer 2019, 19, 415. [Google Scholar] [CrossRef] [PubMed]
- Saunders, L.R.; Bankovich, A.J.; Anderson, W.C.; Aujay, M.A.; Bheddah, S.; Black, K.; Desai, R.; Escarpe, P.A.; Hampl, J.; Laysang, A.; et al. A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci. Transl. Med. 2015, 7, 302ra136. [Google Scholar] [CrossRef]
- Mok, T.S.; Wu, Y.L.; Thongprasert, S.; Yang, C.H.; Chu, D.T.; Saijo, N.; Sunpaweravong, P.; Han, B.; Margono, B.; Ichinose, Y.; et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 2009, 361, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Druker, B.J.; Guilhot, F.; O’Brien, S.G.; Gathmann, I.; Kantarjian, H.; Gattermann, N.; Deininger, M.W.; Silver, R.T.; Goldman, J.M.; Stone, R.M.; et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N. Engl. J. Med. 2006, 355, 2408–2417. [Google Scholar] [CrossRef]
- Shepherd, F.A.; Rodrigues Pereira, J.; Ciuleanu, T.; Tan, E.H.; Hirsh, V.; Thongprasert, S.; Campos, D.; Maoleekoonpiroj, S.; Smylie, M.; Martins, R.; et al. Erlotinib in previously treated non-small-cell lung cancer. N. Engl. J. Med. 2005, 353, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Demetri, G.D.; von Mehren, M.; Blanke, C.D.; Van den Abbeele, A.D.; Eisenberg, B.; Roberts, P.J.; Heinrich, M.C.; Tuveson, D.A.; Singer, S.; Janicek, M.; et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N. Engl. J. Med. 2002, 347, 472–480. [Google Scholar] [CrossRef] [PubMed]
- Coe, B.P.; Lockwood, W.W.; Girard, L.; Chari, R.; Macaulay, C.; Lam, S.; Gazdar, A.F.; Minna, J.D.; Lam, W.L. Differential disruption of cell cycle pathways in small cell and non-small cell lung cancer. Br. J. Cancer 2006, 94, 1927–1935. [Google Scholar] [CrossRef] [PubMed]
- Lockwood, W.W.; Chari, R.; Coe, B.P.; Girard, L.; Macaulay, C.; Lam, S.; Gazdar, A.F.; Minna, J.D.; Lam, W.L. DNA amplification is a ubiquitous mechanism of oncogene activation in lung and other cancers. Oncogene 2008, 27, 4615–4624. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kwei, K.A.; Kim, Y.H.; Girard, L.; Kao, J.; Pacyna-Gengelbach, M.; Salari, K.; Lee, J.; Choi, Y.L.; Sato, M.; Wang, P.; et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene 2008, 27, 3635–3640. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Yanagisawa, K.; Shinjo, K.; Taguchi, A.; Maeno, K.; Tomida, S.; Shimada, Y.; Osada, H.; Kosaka, T.; Matsubara, H.; et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007, 67, 6007–6011. [Google Scholar] [CrossRef] [PubMed]
- Paez, J.G.; Janne, P.A.; Lee, J.C.; Tracy, S.; Greulich, H.; Gabriel, S.; Herman, P.; Kaye, F.J.; Lindeman, N.; Boggon, T.J.; et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 2004, 304, 1497–1500. [Google Scholar] [CrossRef]
- Engelman, J.A.; Janne, P.A. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin. Cancer Res. 2008, 14, 2895–2899. [Google Scholar] [CrossRef] [PubMed]
- Gazdar, A.F. Activating and resistance mutations of EGFR in non-small-cell lung cancer: Role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 2009, 28 (Suppl. S1), S24–S31. [Google Scholar] [CrossRef]
- Geraghty, R.J.; Capes-Davis, A.; Davis, J.M.; Downward, J.; Freshney, R.I.; Knezevic, I.; Lovell-Badge, R.; Masters, J.R.; Meredith, J.; Stacey, G.N.; et al. Guidelines for the use of cell lines in biomedical research. Br. J. Cancer 2014, 111, 1021–1046. [Google Scholar] [CrossRef] [PubMed]
- Wilding, J.L.; Bodmer, W.F. Cancer cell lines for drug discovery and development. Cancer Res. 2014, 74, 2377–2384. [Google Scholar] [CrossRef] [PubMed]
- Gazdar, A.F.; Hirsch, F.R.; Minna, J.D. Correction: “From Mice to Men and Back: An Assessment of Preclinical Model Systems for the Study of Lung Cancers”. J. Thorac. Oncol. 2016, 11, e88–e89. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gazdar, A.F.; Girard, L.; Lockwood, W.W.; Lam, W.L.; Minna, J.D. Lung cancer cell lines as tools for biomedical discovery and research. J. Natl. Cancer Inst. 2016, 102, 1310–1320. [Google Scholar] [CrossRef] [PubMed]
- Cross, D.A.; Ashton, S.E.; Ghiorghiu, S.; Eberlein, C.; Nebhan, C.A.; Spitzler, P.J.; Orme, J.P.; Finlay, M.R.; Ward, R.A.; Mellor, M.J.; et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014, 4, 1046–1061. [Google Scholar] [CrossRef] [PubMed]
- Wistuba, I.I.; Bryant, D.; Behrens, C.; Milchgrub, S.; Virmani, A.K.; Ashfaq, R.; Minna, J.D.; Gazdar, A.F. Comparison of features of human lung cancer cell lines and their corresponding tumors. Clin. Cancer Res. 1999, 5, 991–1000. [Google Scholar]
- Li, T.; Ling, Y.H.; Goldman, I.D.; Perez-Soler, R. Schedule-dependent cytotoxic synergism of pemetrexed and erlotinib in human non-small cell lung cancer cells. Clin. Cancer Res. 2007, 13, 3413–3422. [Google Scholar] [CrossRef]
- Osude, C.; Lin, L.; Patel, M.; Eckburg, A.; Berei, J.; Kuckovic, A.; Dube, N.; Rastogi, A.; Gautam, S.; Smith, T.J.; et al. Mediating EGFR-TKI Resistance by VEGF/VEGFR Autocrine Pathway in Non-Small Cell Lung Cancer. Cells 2022, 11, 1694. [Google Scholar] [CrossRef]
- Cragg, M.S.; Kuroda, J.; Puthalakath, H.; Huang, D.C.; Strasser, A. Gefitinib-induced killing of NSCLC cell lines expressing mutant EGFR requires BIM and can be enhanced by BH3 mimetics. PLoS Med. 2007, 4, 1681–1689; discussion 1690. [Google Scholar] [CrossRef]
- Chava, S.; Bugide, S.; Zhang, X.; Gupta, R.; Wajapeyee, N. Betacellulin promotes tumor development and EGFR mutant lung cancer growth by stimulating the EGFR pathway and suppressing apoptosis. iScience 2022, 25, 104211. [Google Scholar] [CrossRef]
- Forcella, M.; Oldani, M.; Epistolio, S.; Freguia, S.; Monti, E.; Fusi, P.; Frattini, M. Non-small cell lung cancer (NSCLC), EGFR downstream pathway activation and TKI targeted therapies sensitivity: Effect of the plasma membrane-associated NEU3. PLoS ONE 2017, 12, e0187289. [Google Scholar] [CrossRef]
- Wu, L.; Yu, Y.; Xu, L.; Wang, X.; Zhou, J.; Wang, Y. TROY Modulates Cancer Stem-Like Cell Properties and Gefitinib Resistance Through EMT Signaling in Non-Small Cell Lung Cancer. Front. Genet. 2022, 13, 881875. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.A.; Ma, T.; Zhang, X.Y.; Cheng, X.S.; Olajuyin, A.M.; Sun, Z.F.; Zhang, X.J. Apatinib preferentially inhibits PC9 gefitinib-resistant cancer cells by inducing cell cycle arrest and inhibiting VEGFR signaling pathway. Cancer Cell Int. 2019, 19, 117. [Google Scholar] [CrossRef] [PubMed]
- Guerra, S.L.; Maertens, O.; Kuzmickas, R.; De Raedt, T.; Adeyemi, R.O.; Guild, C.J.; Guillemette, S.; Redig, A.J.; Chambers, E.S.; Xu, M.; et al. A Deregulated HOX Gene Axis Confers an Epigenetic Vulnerability in KRAS-Mutant Lung Cancers. Cancer Cell 2020, 37, 705–719.e6. [Google Scholar] [CrossRef] [PubMed]
- Sunaga, N.; Shames, D.S.; Girard, L.; Peyton, M.; Larsen, J.E.; Imai, H.; Soh, J.; Sato, M.; Yanagitani, N.; Kaira, K.; et al. Knockdown of oncogenic KRAS in non-small cell lung cancers suppresses tumor growth and sensitizes tumor cells to targeted therapy. Mol. Cancer Ther. 2011, 10, 336–346. [Google Scholar] [CrossRef]
- Blanco, R.; Iwakawa, R.; Tang, M.; Kohno, T.; Angulo, B.; Pio, R.; Montuenga, L.M.; Minna, J.D.; Yokota, J.; Sanchez-Cespedes, M. A gene-alteration profile of human lung cancer cell lines. Hum. Mutat. 2009, 30, 1199–1206. [Google Scholar] [CrossRef] [PubMed]
- Fawdar, S.; Trotter, E.W.; Li, Y.; Stephenson, N.L.; Hanke, F.; Marusiak, A.A.; Edwards, Z.C.; Ientile, S.; Waszkowycz, B.; Miller, C.J.; et al. Targeted genetic dependency screen facilitates identification of actionable mutations in FGFR4, MAP3K9, and PAK5 in lung cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 12426–12431. [Google Scholar] [CrossRef]
- Song, Z.; Liu, F.; Zhang, J. Targeting NRAS(Q61K) mutant delays tumor growth and angiogenesis in non-small cell lung cancer. Am. J. Cancer Res. 2017, 7, 831–844. [Google Scholar]
- Lee, G.D.; Lee, S.E.; Oh, D.Y.; Yu, D.B.; Jeong, H.M.; Kim, J.; Hong, S.; Jung, H.S.; Oh, E.; Song, J.Y.; et al. MET Exon 14 Skipping Mutations in Lung Adenocarcinoma: Clinicopathologic Implications and Prognostic Values. J. Thorac. Oncol. 2017, 12, 1233–1246. [Google Scholar] [CrossRef]
- Koivunen, J.P.; Mermel, C.; Zejnullahu, K.; Murphy, C.; Lifshits, E.; Holmes, A.J.; Choi, H.G.; Kim, J.; Chiang, D.; Thomas, R.; et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin. Cancer Res. 2008, 14, 4275–4283. [Google Scholar] [CrossRef] [PubMed]
- Giannopoulou, E.; Nikolakopoulos, A.; Kotsirilou, D.; Lampropoulou, A.; Raftopoulou, S.; Papadimitriou, E.; Theocharis, A.D.; Makatsoris, T.; Fasseas, K.; Kalofonos, H.P. Epidermal growth factor receptor status and Notch inhibition in non-small cell lung cancer cells. J. Biomed. Sci. 2015, 22, 98. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bamford, S.; Dawson, E.; Forbes, S.; Clements, J.; Pettett, R.; Dogan, A.; Flanagan, A.; Teague, J.; Futreal, P.A.; Stratton, M.R.; et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer 2004, 91, 355–358. [Google Scholar] [CrossRef]
- Palanikumar, L.; Karpauskaite, L.; Al-Sayegh, M.; Chehade, I.; Alam, M.; Hassan, S.; Maity, D.; Ali, L.; Kalmouni, M.; Hunashal, Y.; et al. Protein mimetic amyloid inhibitor potently abrogates cancer-associated mutant p53 aggregation and restores tumor suppressor function. Nat. Commun. 2021, 12, 3962. [Google Scholar] [CrossRef] [PubMed]
- Ogoshi, Y.; Shien, K.; Yoshioka, T.; Torigoe, H.; Sato, H.; Sakaguchi, M.; Tomida, S.; Namba, K.; Kurihara, E.; Takahashi, Y.; et al. Anti-tumor effect of neratinib against lung cancer cells harboring HER2 oncogene alterations. Oncol. Lett. 2019, 17, 2729–2736. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.P.; Pinkoski, M.J.; Bhardwaj, G.; Deeley, R.G. Elevated expression of annexin II (lipocortin II, p36) in a multidrug resistant small cell lung cancer cell line. Br. J. Cancer 1992, 65, 498–502. [Google Scholar] [CrossRef][Green Version]
- Pettengill, O.S.; Sorenson, G.D.; Wurster-Hill, D.H.; Curphey, T.J.; Noll, W.W.; Cate, C.C.; Maurer, L.H. Isolation and growth characteristics of continuous cell lines from small-cell carcinoma of the lung. Cancer 1980, 45, 906–918. [Google Scholar] [CrossRef]
- Dutil, J.; Chen, Z.; Monteiro, A.N.; Teer, J.K.; Eschrich, S.A. An Interactive Resource to Probe Genetic Diversity and Estimated Ancestry in Cancer Cell Lines. Cancer Res. 2019, 79, 1263–1273. [Google Scholar] [CrossRef]
- Twentyman, P.R.; Wright, K.A.; Mistry, P.; Kelland, L.R.; Murrer, B.A. Sensitivity to novel platinum compounds of panels of human lung cancer cell lines with acquired and inherent resistance to cisplatin. Cancer Res. 1992, 52, 5674–5680. [Google Scholar] [PubMed]
- Fisher, E.R.; Paulson, J.D. A new in vitro cell line established from human large cell variant of oat cell lung cancer. Cancer Res. 1978, 38, 3830–3835. [Google Scholar]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef]
- Huo, K.G.; D’Arcangelo, E.; Tsao, M.S. Patient-derived cell line, xenograft and organoid models in lung cancer therapy. Transl. Lung Cancer Res. 2020, 9, 2214–2232. [Google Scholar] [CrossRef] [PubMed]
- Saforo, D.; Omer, L.; Smolenkov, A.; Barve, A.; Casson, L.; Boyd, N.; Clark, G.; Siskind, L.; Beverly, L. Primary lung cancer samples cultured under microenvironment-mimetic conditions enrich for mesenchymal stem-like cells that promote metastasis. Sci. Rep. 2019, 9, 4177. [Google Scholar] [CrossRef] [PubMed]
- Kodack, D.P.; Farago, A.F.; Dastur, A.; Held, M.A.; Dardaei, L.; Friboulet, L.; von Flotow, F.; Damon, L.J.; Lee, D.; Parks, M.; et al. Primary Patient-Derived Cancer Cells and Their Potential for Personalized Cancer Patient Care. Cell Rep. 2017, 21, 3298–3309. [Google Scholar] [CrossRef] [PubMed]
- Tiran, V.; Lindenmann, J.; Brcic, L.; Heitzer, E.; Stanzer, S.; Tabrizi-Wizsy, N.G.; Stacher, E.; Stoeger, H.; Popper, H.H.; Balic, M.; et al. Primary patient-derived lung adenocarcinoma cell culture challenges the association of cancer stem cells with epithelial-to-mesenchymal transition. Sci. Rep. 2017, 7, 10040. [Google Scholar] [CrossRef] [PubMed]
- Odintsov, I.; Mattar, M.S.; Lui, A.J.; Offin, M.; Kurzatkowski, C.; Delasos, L.; Khodos, I.; Asher, M.; Daly, R.M.; Rekhtman, N.; et al. Novel Preclinical Patient-Derived Lung Cancer Models Reveal Inhibition of HER3 and MTOR Signaling as Therapeutic Strategies for NRG1 Fusion-Positive Cancers. J. Thorac. Oncol. 2021, 16, 1149–1165. [Google Scholar] [CrossRef] [PubMed]
- Crystal, A.S.; Shaw, A.T.; Sequist, L.V.; Friboulet, L.; Niederst, M.J.; Lockerman, E.L.; Frias, R.L.; Gainor, J.F.; Amzallag, A.; Greninger, P.; et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science 2014, 346, 1480–1486. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ory, V.; Chapman, S.; Yuan, H.; Albanese, C.; Kallakury, B.; Timofeeva, O.A.; Nealon, C.; Dakic, A.; Simic, V.; et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 2012, 180, 599–607. [Google Scholar] [CrossRef]
- Liu, X.; Krawczyk, E.; Suprynowicz, F.A.; Palechor-Ceron, N.; Yuan, H.; Dakic, A.; Simic, V.; Zheng, Y.L.; Sripadhan, P.; Chen, C.; et al. Conditional reprogramming and long-term expansion of normal and tumor cells from human biospecimens. Nat. Protoc. 2017, 12, 439–451. [Google Scholar] [CrossRef]
- Talwelkar, S.S.; Nagaraj, A.S.; Devlin, J.R.; Hemmes, A.; Potdar, S.; Kiss, E.A.; Saharinen, P.; Salmenkivi, K.; Mayranpaa, M.I.; Wennerberg, K.; et al. Receptor Tyrosine Kinase Signaling Networks Define Sensitivity to ERBB Inhibition and Stratify Kras-Mutant Lung Cancers. Mol. Cancer Ther. 2019, 18, 1863–1874. [Google Scholar] [CrossRef]
- Borodovsky, A.; McQuiston, T.J.; Stetson, D.; Ahmed, A.; Whitston, D.; Zhang, J.; Grondine, M.; Lawson, D.; Challberg, S.S.; Zinda, M.; et al. Generation of stable PDX derived cell lines using conditional reprogramming. Mol. Cancer 2017, 16, 177. [Google Scholar] [CrossRef]
- Yuan, H.; Myers, S.; Wang, J.; Zhou, D.; Woo, J.A.; Kallakury, B.; Ju, A.; Bazylewicz, M.; Carter, Y.M.; Albanese, C.; et al. Use of reprogrammed cells to identify therapy for respiratory papillomatosis. N. Engl. J. Med. 2012, 367, 1220–1227. [Google Scholar] [CrossRef] [PubMed]
- Alamri, A.M.; Liu, X.; Blancato, J.K.; Haddad, B.R.; Wang, W.; Zhong, X.; Choudhary, S.; Krawczyk, E.; Kallakury, B.V.; Davidson, B.J.; et al. Expanding primary cells from mucoepidermoid and other salivary gland neoplasms for genetic and chemosensitivity testing. Dis. Model. Mech. 2018, 11, dmm031716. [Google Scholar] [CrossRef] [PubMed]
- Zhong, M.; Fu, L. Culture and application of conditionally reprogrammed primary tumor cells. Gastroenterol. Rep. 2020, 8, 224–233. [Google Scholar] [CrossRef]
- Jiang, S.; Wang, J.; Yang, C.; Tan, R.; Hou, J.; Shi, Y.; Zhang, H.; Ma, S.; Wang, J.; Zhang, M.; et al. Continuous culture of urine-derived bladder cancer cells for precision medicine. Protein Cell 2019, 10, 902–907. [Google Scholar] [CrossRef]
- Hirano, T.; Yasuda, H.; Tani, T.; Hamamoto, J.; Oashi, A.; Ishioka, K.; Arai, D.; Nukaga, S.; Miyawaki, M.; Kawada, I.; et al. In Vitro modeling to determine mutation specificity of EGFR tyrosine kinase inhibitors against clinically relevant EGFR mutants in non-small-cell lung cancer. Oncotarget 2015, 6, 38789–38803. [Google Scholar] [CrossRef]
- Eramo, A.; Lotti, F.; Sette, G.; Pilozzi, E.; Biffoni, M.; Di Virgilio, A.; Conticello, C.; Ruco, L.; Peschle, C.; De Maria, R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008, 15, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Camp, J.G.; Sekine, K.; Gerber, T.; Loeffler-Wirth, H.; Binder, H.; Gac, M.; Kanton, S.; Kageyama, J.; Damm, G.; Seehofer, D.; et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 2017, 546, 533–538. [Google Scholar] [CrossRef]
- Orienti, I.; Francescangeli, F.; de Angelis, M.L.; Fecchi, K.; Bongiorno-Borbone, L.; Signore, M.; Peschiaroli, A.; Boe, A.; Bruselles, A.; Costantino, A.; et al. A new bioavailable fenretinide formulation with antiproliferative, antimetabolic, and cytotoxic effects on solid tumors. Cell Death Dis. 2019, 10, 529. [Google Scholar] [CrossRef] [PubMed]
- Orienti, I.; Salvati, V.; Sette, G.; Zucchetti, M.; Bongiorno-Borbone, L.; Peschiaroli, A.; Zolla, L.; Francescangeli, F.; Ferrari, M.; Matteo, C.; et al. A novel oral micellar fenretinide formulation with enhanced bioavailability and antitumour activity against multiple tumours from cancer stem cells. J. Exp. Clin. Cancer Res. 2019, 38, 373. [Google Scholar] [CrossRef] [PubMed]
- Zeuner, A.; Francescangeli, F.; Contavalli, P.; Zapparelli, G.; Apuzzo, T.; Eramo, A.; Baiocchi, M.; De Angelis, M.L.; Biffoni, M.; Sette, G.; et al. Elimination of quiescent/slow-proliferating cancer stem cells by Bcl-XL inhibition in non-small cell lung cancer. Cell Death Differ. 2014, 21, 1877–1888. [Google Scholar] [CrossRef] [PubMed]
- Kamer, I.; Bab-Dinitz, E.; Zadok, O.; Ofek, E.; Gottfried, T.; Daniel-Meshulam, I.; Hout-Siloni, G.; Ben Nun, A.; Barshack, I.; Onn, A.; et al. Immunotherapy response modeling by ex-vivo organ culture for lung cancer. Cancer Immunol. Immunother. 2021, 70, 2223–2234. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, E.; Kuraguchi, M.; Xu, M.; Portell, A.J.; Taus, L.; Diala, I.; Lalani, A.S.; Choi, J.; Chambers, E.S.; Li, S.; et al. Use of Ex Vivo Patient-Derived Tumor Organotypic Spheroids to Identify Combination Therapies for HER2 Mutant Non-Small Cell Lung Cancer. Clin. Cancer Res. 2020, 26, 2393–2403. [Google Scholar] [CrossRef]
- Endo, H.; Okami, J.; Okuyama, H.; Kumagai, T.; Uchida, J.; Kondo, J.; Takehara, T.; Nishizawa, Y.; Imamura, F.; Higashiyama, M.; et al. Spheroid culture of primary lung cancer cells with neuregulin 1/HER3 pathway activation. J. Thorac. Oncol. 2013, 8, 131–139. [Google Scholar] [CrossRef]
- Benali, R.; Tournier, J.M.; Chevillard, M.; Zahm, J.M.; Klossek, J.M.; Hinnrasky, J.; Gaillard, D.; Maquart, F.X.; Puchelle, E. Tubule formation by human surface respiratory epithelial cells cultured in a three-dimensional collagen lattice. Am. J. Physiol. 1993, 264, L183–L192. [Google Scholar] [CrossRef]
- Chen, Y.W.; Huang, S.X.; de Carvalho, A.; Ho, S.H.; Islam, M.N.; Volpi, S.; Notarangelo, L.D.; Ciancanelli, M.; Casanova, J.L.; Bhattacharya, J.; et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 2017, 19, 542–549. [Google Scholar] [CrossRef]
- Dye, B.R.; Hill, D.R.; Ferguson, M.A.; Tsai, Y.H.; Nagy, M.S.; Dyal, R.; Wells, J.M.; Mayhew, C.N.; Nattiv, R.; Klein, O.D.; et al. In Vitro generation of human pluripotent stem cell derived lung organoids. eLife 2015, 4, e05098. [Google Scholar] [CrossRef]
- McCauley, K.B.; Hawkins, F.; Serra, M.; Thomas, D.C.; Jacob, A.; Kotton, D.N. Efficient Derivation of Functional Human Airway Epithelium from Pluripotent Stem Cells via Temporal Regulation of Wnt Signaling. Cell Stem Cell 2017, 20, 844–857.e6. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; Choi, K.M.; Sicard, D.; Tschumperlin, D.J. Human airway organoid engineering as a step toward lung regeneration and disease modeling. Biomaterials 2017, 113, 118–132. [Google Scholar] [CrossRef]
- Nikolic, M.Z.; Caritg, O.; Jeng, Q.; Johnson, J.A.; Sun, D.; Howell, K.J.; Brady, J.L.; Laresgoiti, U.; Allen, G.; Butler, R.; et al. Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids. eLife 2017, 6, e26575. [Google Scholar] [CrossRef] [PubMed]
- Sachs, N.; Papaspyropoulos, A.; Zomer-van Ommen, D.D.; Heo, I.; Bottinger, L.; Klay, D.; Weeber, F.; Huelsz-Prince, G.; Iakobachvili, N.; Amatngalim, G.D.; et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019, 38, e100300. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Sui, X.; Song, F.; Li, Y.; Li, K.; Chen, Z.; Yang, F.; Chen, X.; Zhang, Y.; Wang, X.; et al. Lung cancer organoids analyzed on microwell arrays predict drug responses of patients within a week. Nat. Commun. 2021, 12, 2581. [Google Scholar] [CrossRef]
- Dijkstra, K.K.; Monkhorst, K.; Schipper, L.J.; Hartemink, K.J.; Smit, E.F.; Kaing, S.; de Groot, R.; Wolkers, M.C.; Clevers, H.; Cuppen, E.; et al. Challenges in Establishing Pure Lung Cancer Organoids Limit Their Utility for Personalized Medicine. Util. Pers. Med. 2020, 31, 107588. [Google Scholar] [CrossRef]
- Li, Z.; Yu, L.; Chen, D.; Meng, Z.; Chen, W.; Huang, W. Protocol for generation of lung adenocarcinoma organoids from clinical samples. STAR Protoc. 2021, 2, 100239. [Google Scholar] [CrossRef] [PubMed]
- Finnberg, N.K.; Gokare, P.; Lev, A.; Grivennikov, S.I.; MacFarlane, A.W.t.; Campbell, K.S.; Winters, R.M.; Kaputa, K.; Farma, J.M.; Abbas, A.E.; et al. Application of 3D tumoroid systems to define immune and cytotoxic therapeutic responses based on tumoroid and tissue slice culture molecular signatures. Oncotarget 2017, 8, 66747–66757. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, S.M.; Lim, S.; Lee, J.Y.; Choi, S.J.; Yang, S.D.; Yun, M.R.; Kim, C.G.; Gu, S.R.; Park, C.; et al. Modeling Clinical Responses to Targeted Therapies by Patient-Derived Organoids of Advanced Lung Adenocarcinoma. Clin. Cancer Res. 2021, 27, 4397–4409. [Google Scholar] [CrossRef] [PubMed]
- Yokota, E.; Iwai, M.; Yukawa, T.; Yoshida, M.; Naomoto, Y.; Haisa, M.; Monobe, Y.; Takigawa, N.; Guo, M.; Maeda, Y.; et al. Clinical application of a lung cancer organoid (tumoroid) culture system. NPJ Precis. Oncol. 2021, 5, 29. [Google Scholar] [CrossRef]
- Mazzocchi, A.; Dominijanni, A.; Soker, S. Pleural Effusion Aspirate for Use in 3D Lung Cancer Modeling and Chemotherapy Screening. Methods Mol. Biol. 2022, 2394, 471–483. [Google Scholar] [PubMed]
- Ma, X.; Yang, S.; Jiang, H.; Wang, Y.; Xiang, Z. Transcriptomic analysis of tumor tissues and organoids reveals the crucial genes regulating the proliferation of lung adenocarcinoma. J. Transl. Med. 2021, 19, 368. [Google Scholar] [CrossRef]
- Lo, Y.H.; Karlsson, K.; Kuo, C.J. Applications of Organoids for Cancer Biology and Precision Medicine. Nat. Cancer 2020, 1, 761–773. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, M.L.; Francescangeli, F.; Nicolazzo, C.; Signore, M.; Giuliani, A.; Colace, L.; Boe, A.; Magri, V.; Baiocchi, M.; Ciardi, A.; et al. An organoid model of colorectal circulating tumor cells with stem cell features, hybrid EMT state and distinctive therapy response profile. J. Exp. Clin. Cancer Res. 2022, 41, 86. [Google Scholar] [CrossRef]
- Bolhaqueiro, A.C.F.; Ponsioen, B.; Bakker, B.; Klaasen, S.J.; Kucukkose, E.; van Jaarsveld, R.H.; Vivie, J.; Verlaan-Klink, I.; Hami, N.; Spierings, D.C.J.; et al. Ongoing chromosomal instability and karyotype evolution in human colorectal cancer organoids. Nat. Genet. 2019, 51, 824–834. [Google Scholar] [CrossRef]
- Drost, J.; van Boxtel, R.; Blokzijl, F.; Mizutani, T.; Sasaki, N.; Sasselli, V.; de Ligt, J.; Behjati, S.; Grolleman, J.E.; van Wezel, T.; et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 2017, 358, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; van Jaarsveld, R.H.; Ponsioen, B.; Zimberlin, C.; van Boxtel, R.; Buijs, A.; Sachs, N.; Overmeer, R.M.; Offerhaus, G.J.; Begthel, H.; et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015, 521, 43–47. [Google Scholar] [CrossRef]
- Li, X.; Nadauld, L.; Ootani, A.; Corney, D.C.; Pai, R.K.; Gevaert, O.; Cantrell, M.A.; Rack, P.G.; Neal, J.T.; Chan, C.W.; et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 2014, 20, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Radulovich, N.; Ng, C.; Liu, N.; Notsuda, H.; Cabanero, M.; Martins-Filho, S.N.; Raghavan, V.; Li, Q.; Mer, A.S.; et al. Organoid Cultures as Preclinical Models of Non-Small Cell Lung Cancer. Clin. Cancer Res. 2021, 26, 1162–1174. [Google Scholar] [CrossRef]
- Dost, A.F.M.; Moye, A.L.; Vedaie, M.; Tran, L.M.; Fung, E.; Heinze, D.; Villacorta-Martin, C.; Huang, J.; Hekman, R.; Kwan, J.H.; et al. Organoids Model Transcriptional Hallmarks of Oncogenic KRAS Activation in Lung Epithelial Progenitor Cells. Cell Stem Cell 2020, 27, 663–678 e668. [Google Scholar] [CrossRef]
- Semba, T.; Sato, R.; Kasuga, A.; Suina, K.; Shibata, T.; Kohno, T.; Suzuki, M.; Saya, H.; Arima, Y. Lung Adenocarcinoma Mouse Models Based on Orthotopic Transplantation of Syngeneic Tumor-Initiating Cells Expressing EpCAM, SCA-1, and Ly6d. Cancers 2020, 12, 3805. [Google Scholar] [CrossRef] [PubMed]
- Na, F.; Pan, X.; Chen, J.; Chen, X.; Wang, M.; Chi, P.; You, L.; Zhang, L.; Zhong, A.; Zhao, L.; et al. KMT2C deficiency promotes small cell lung cancer metastasis through DNMT3A-mediated epigenetic reprogramming. Nat. Cancer 2023, 3, 753–767. [Google Scholar] [CrossRef] [PubMed]
- Nacarino-Palma, A.; Rejano-Gordillo, C.M.; Gonzalez-Rico, F.J.; Ordiales-Talavero, A.; Roman, A.C.; Cuadrado, M.; Bustelo, X.R.; Merino, J.M.; Fernandez-Salguero, P.M. Loss of Aryl Hydrocarbon Receptor Favors K-Ras(G12D)-Driven Non-Small Cell Lung Cancer. Cancers 2021, 13, 4071. [Google Scholar] [CrossRef]
- Wu, M.; Liao, Y.; Tang, L. Non-small cell lung cancer organoids: Advances and challenges in current applications. Chin. J. Cancer Res. 2024, 36, 455–473. [Google Scholar] [CrossRef]
- Takahashi, N.; Hoshi, H.; Higa, A.; Hiyama, G.; Tamura, H.; Ogawa, M.; Takagi, K.; Goda, K.; Okabe, N.; Muto, S.; et al. An In Vitro System for Evaluating Molecular Targeted Drugs Using Lung Patient-Derived Tumor Organoids. Cells 2019, 8, 481. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.H.; Chu, X.P.; Zhang, J.T.; Nie, Q.; Tang, W.F.; Su, J.; Yan, H.H.; Zheng, H.P.; Chen, Z.X.; Chen, X.; et al. Genomic characteristics and drug screening among organoids derived from non-small cell lung cancer patients. Thorat. Cancer 2020, 11, 2279–2290. [Google Scholar] [CrossRef] [PubMed]
- Ooft, S.N.; Weeber, F.; Dijkstra, K.K.; McLean, C.M.; Kaing, S.; van Werkhoven, E.; Schipper, L.; Hoes, L.; Vis, D.J.; van de Haar, J.; et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 2019, 11, 2574. [Google Scholar] [CrossRef]
- Ooft, S.N.; Weeber, F.; Schipper, L.; Dijkstra, K.; McLean, C.; Kaing, S.; van de Haar, J.; Prevoo, W.; van Werkhoven, E.; Snaebjornsson, P.; et al. Prospective experimental treatment of colorectal cancer patients based on organoid drug responses. ESMO Open 2021, 6, 100103. [Google Scholar] [CrossRef]
- Li, Z.; Qian, Y.; Li, W.; Liu, L.; Yu, L.; Liu, X.; Wu, G.; Wang, Y.; Luo, W.; Fang, F.; et al. Human Lung Adenocarcinoma-Derived Organoid Models for Drug Screening. eScience 2020, 23, 101411. [Google Scholar] [CrossRef]
- Wang, H.M.; Zhang, C.Y.; Peng, K.C.; Chen, Z.X.; Su, J.W.; Li, Y.F.; Li, W.F.; Gao, Q.Y.; Zhang, S.L.; Chen, Y.Q.; et al. Using patient-derived organoids to predict locally advanced or metastatic lung cancer tumor response: A real-world study. Cell Rep. Med. 2023, 4, 100911. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, N.A.; Taube, J.M. PD-L1 and Other Immunological Diagnosis Tools. In Oncoimmunology: A Practical Guide for Cancer Immunotherapy; Springer: Cham, Switzerland, 2018; pp. 371–385. [Google Scholar]
- Giraldo, N.A.; Sanchez-Salas, R.; Peske, J.D.; Vano, Y.; Becht, E.; Petitprez, F.; Validire, P.; Ingels, A.; Cathelineau, X.; Fridman, W.H.; et al. The clinical role of the TME in solid cancer. Br. J. Cancer 2019, 120, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Toor, S.M.; Sasidharan Nair, V.; Decock, J.; Elkord, E. Immune checkpoints in the tumor microenvironment. Semin. Cancer Biol. 2020, 65, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hammers, H.J.; Plimack, E.R.; Infante, J.R.; Rini, B.I.; McDermott, D.F.; Lewis, L.D.; Voss, M.H.; Sharma, P.; Pal, S.K.; Razak, A.R.A.; et al. Safety and Efficacy of Nivolumab in Combination with Ipilimumab in Metastatic Renal Cell Carcinoma: The CheckMate 016 Study. J. Clin. Oncol. 2017, 35, 3851–3858. [Google Scholar] [CrossRef]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Cao, D.; Xu, H.; Xu, X.; Guo, T.; Ge, W. High tumor mutation burden predicts better efficacy of immunotherapy: A pooled analysis of 103078 cancer patients. Oncoimmunology 2019, 8, e1629258. [Google Scholar] [CrossRef]
- Menon, S.; Shin, S.; Dy, G. Advances in Cancer Immunotherapy in Solid Tumors. Cancers 2016, 8, 106. [Google Scholar] [CrossRef]
- Saraiva, D.P.; Matias, A.T.; Braga, S.; Jacinto, A.; Cabral, M.G. Establishment of a 3D Co-culture with MDA-MB-231 Breast Cancer Cell Line and Patient-Derived Immune Cells for Application in the Development of Immunotherapies. Front. Oncol. 2020, 10, 1543. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, K.K.; Cattaneo, C.M.; Weeber, F.; Chalabi, M.; van de Haar, J.; Fanchi, L.F.; Slagter, M.; van der Velden, D.L.; Kaing, S.; Kelderman, S.; et al. Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 2018, 174, 1586–1598.e12. [Google Scholar] [CrossRef]
- Ma, W.; Zeng, J.; Chen, S.; Lyu, Y.; Toomey, K.A.; Phan, C.T.; Yoneda, K.Y.; Li, T. Small molecule tyrosine kinase inhibitors modulated blood immune cell counts in patients with oncogene-driven NSCLC. Biomark. Res. 2021, 9, 69. [Google Scholar] [CrossRef]
- Lyman, G. Risk factors for cancer. Prim. Care 1992, 19, 465–479. [Google Scholar] [CrossRef]
- Kasala, E.R.; Bodduluru, L.N.; Barua, C.C.; Sriram, C.S.; Gogoi, R. Benzo(a)pyrene induced lung cancer: Role of dietary phytochemicals in chemoprevention. Pharmacol. Rep. 2015, 67, 996–1009. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Z.; Yan, Y.; Lemon, W.J.; LaRegina, M.; Morrison, C.; Lubet, R.; You, M. A chemically induced model for squamous cell carcinoma of the lung in mice: Histopathology and strain susceptibility. Cancer Res. 2004, 64, 1647–1654. [Google Scholar] [CrossRef] [PubMed]
- Nettesheim, P.; AS, H. nduction of squamous cell carcinoma in the respiratory tract of mice. J. Natl. Cancer Inst. 1971, 47, 697–701. [Google Scholar]
- Rajendran, P.; Ekambaram, G.; Sakthisekaran, D. Protective role of mangiferin against Benzo(a)pyrene induced lung carcinogenesis in experimental animals. Biol. Pharm. Bull. 2008, 31, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wang, Y.; Tan, Q.; Lubet, R.A.; You, M. Efficacy of deguelin and silibinin on benzo(a)pyrene-induced lung tumorigenesis in A/J mice. Neoplasia 2005, 7, 1053–1057. [Google Scholar] [CrossRef]
- Nolan, K.; Verzosa, G.; Cleaver, T.; Tippimanchai, D.; DePledge, L.N.; Wang, X.J.; Young, C.; Le, A.; Doebele, R.; Li, H.; et al. Development of syngeneic murine cell lines for use in immunocompetent orthotopic lung cancer models. Cancer Cell Int. 2020, 20, 417. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, L.M.; Urbiola, C.; Das, K.; Spiesschaert, B.; Kimpel, J.; Heinemann, F.; Stierstorfer, B.; Muller, P.; Petersson, M.; Erlmann, P.; et al. The lytic activity of VSV-GP treatment dominates the therapeutic effects in a syngeneic model of lung cancer. Br. J. Cancer 2019, 121, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Wang, L.; Zhang, R.; Pu, X.; Wu, S.; Yu, L.; Meraz, I.M.; Zhang, X.; Wang, J.F.; Gibbons, D.L.; et al. Overcoming resistance to anti-PD immunotherapy in a syngeneic mouse lung cancer model using locoregional virotherapy. Oncoimmunology 2017, 7, e1376156. [Google Scholar] [CrossRef] [PubMed]
- Ajona, D.; Ortiz-Espinosa, S.; Moreno, H.; Lozano, T.; Pajares, M.J.; Agorreta, J.; Bertolo, C.; Lasarte, J.J.; Vicent, S.; Hoehlig, K.; et al. A Combined PD-1/C5a Blockade Synergistically Protects against Lung Cancer Growth and Metastasis. Cancer Discov. 2017, 7, 694–703. [Google Scholar] [CrossRef]
- Meraz, I.M.; Majidi, M.; Cao, X.; Lin, H.; Li, L.; Wang, J.; Baladandayuthapani, V.; Rice, D.; Sepesi, B.; Ji, L.; et al. TUSC2 Immunogene Therapy Synergizes with Anti-PD-1 through Enhanced Proliferation and Infiltration of Natural Killer Cells in Syngeneic Kras-Mutant Mouse Lung Cancer Models. Immunol. Res. 2018, 6, 163–177. [Google Scholar] [CrossRef] [PubMed]
- Doetschman, T. GI GEMs: Genetically engineered mouse models of gastrointestinal disease. Gastroenterology 2011, 140, 380–385 e382. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Frese, K.K.; Tuveson, D.A. Maximizing mouse cancer models. Nat. Rev. Cancer 2007, 7, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, B.E.; Park, K.S.; Yiu, G.; Conklin, J.F.; Lin, C.; Burkhart, D.L.; Karnezis, A.N.; Sweet-Cordero, E.A.; Sage, J. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res. 2010, 70, 3877–3883. [Google Scholar] [CrossRef]
- Manchado, E.; Weissmueller, S.; Morris, J.P.; Chen, C.C.; Wullenkord, R.; Lujambio, A.; de Stanchina, E.; Poirier, J.T.; Gainor, J.F.; Corcoran, R.B.; et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature 2016, 534, 647–651. [Google Scholar] [CrossRef] [PubMed]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Carroll, D. Genome engineering with zinc-finger nucleases. Genetics 2011, 188, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Wyman, C.; Kanaar, R. DNA double-strand break repair: All’s well that ends well. Annu. Rev. Genet. 2006, 40, 363–383. [Google Scholar] [CrossRef]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F., 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Mashimo, T. Gene targeting technologies in rats: Zinc finger nucleases, transcription activator-like effector nucleases, and clustered regularly interspaced short palindromic repeats. Dev. Growth Differ. 2014, 56, 46–52. [Google Scholar] [CrossRef]
- Wright, A.V.; Nuñez, J.K.; Doudna, J.A. Biology and applications of CRISPR systems: Harnessing nature’s toolbox for genome engineering. Cell 2016, 164, 29–44. [Google Scholar] [CrossRef] [PubMed]
- Rakhit, C.P.; Trigg, R.M.; Le Quesne, J.; Kelly, M.; Shaw, J.A.; Pritchard, C.; Martins, L.M. Early detection of pre-malignant lesions in a KRASG12D-driven mouse lung cancer model by monitoring circulating free DNA. Dis. Models Mech. 2019, 12, dmm036863. [Google Scholar] [CrossRef]
- Politi, K.; Zakowski, M.F.; Fan, P.D.; Schonfeld, E.A.; Pao, W.; Varmus, H.E. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 2006, 20, 1496–1510. [Google Scholar] [CrossRef]
- Politi, K.; Fan, P.D.; Shen, R.; Zakowski, M.; Varmus, H. Erlotinib resistance in mouse models of epidermal growth factor receptor-induced lung adenocarcinoma. Dis. Models Mech. 2010, 3, 111–119. [Google Scholar] [CrossRef]
- Li, D.; Ambrogio, L.; Shimamura, T.; Kubo, S.; Takahashi, M.; Chirieac, L.R.; Padera, R.F.; Shapiro, G.I.; Baum, A.; Himmelsbach, F.; et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene 2008, 27, 4702–4711. [Google Scholar] [CrossRef]
- Mancini, M.; Thomas, Q.D.; Bourdel, S.; Papon, L.; Bousquet, E.; Jalta, P.; La Monica, S.; Travert, C.; Alfieri, R.; Quantin, X.; et al. Generation and Characterization of a New Preclinical Mouse Model of EGFR-Driven Lung Cancer with MET-Induced Osimertinib Resistance. Cancers 2021, 13, 3441. [Google Scholar] [CrossRef]
- He, L.; Luo, L.; Zhu, H.; Yang, H.; Zhang, Y.; Wu, H.; Sun, H.; Jiang, F.; Kathera, C.S.; Liu, L.; et al. FEN1 promotes tumor progression and confers cisplatin resistance in non-small-cell lung cancer. Mol. Oncol. 2017, 11, 1302–1303. [Google Scholar] [CrossRef] [PubMed]
- Kasiri, S.; Shao, C.; Chen, B.; Wilson, A.N.; Yenerall, P.; Timmons, B.C.; Girard, L.; Tian, H.; Behrens, C.; Wistuba, I.I.; et al. GLI1 Blockade Potentiates the Antitumor Activity of PI3K Antagonists in Lung Squamous Cell Carcinoma. Cancer Res. 2017, 77, 4448–4459. [Google Scholar] [CrossRef]
- Lai, Y.; Wei, X.; Lin, S.; Qin, L.; Cheng, L.; Li, P. Current status and perspectives of patient-derived xenograft models in cancer research. J. Hematol. Oncol. 2017, 10, 106. [Google Scholar] [CrossRef] [PubMed]
- Fichtner, I.; Rolff, J.; Soong, R.; Hoffmann, J.; Hammer, S.; Sommer, A.; Becker, M.; Merk, J. Establishment of patient-derived non-small cell lung cancer xenografts as models for the identification of predictive biomarkers. Clin. Cancer Res. 2008, 14, 6456–6468. [Google Scholar] [CrossRef] [PubMed]
- Merk, J.; Rolff, J.; Becker, M.; Leschber, G.; Fichtner, I. Patient-derived xenografts of non-small-cell lung cancer: A pre-clinical model to evaluate adjuvant chemotherapy? Eur. J. Cardiothorac. Surg. 2009, 36, 454–459. [Google Scholar] [CrossRef]
- Blake, J.A.; Baldarelli, R.; Kadin, J.A.; Richardson, J.E.; Smith, C.L.; Bult, C.J.; Mouse Genome Database, G. Mouse Genome Database (MGD): Knowledgebase for mouse-human comparative biology. Nucleic Acids Res. 2021, 49, D981–D987. [Google Scholar] [CrossRef] [PubMed]
- Baldarelli, R.M.; Smith, C.M.; Finger, J.H.; Hayamizu, T.F.; McCright, I.J.; Xu, J.; Shaw, D.R.; Beal, J.S.; Blodgett, O.; Campbell, J.; et al. The mouse Gene Expression Database (GXD): 2021 update. Nucleic Acids Res. 2021, 49, D924–D931. [Google Scholar] [CrossRef]
- Begley, D.A.; Krupke, D.M.; Neuhauser, S.; Sundberg, J.; Bult, C.J. Abstract 1190: MMHCdb: A knowledgebase for the evolving landscape of mouse models of human cancer. Cancer Res. 2022, 82, 1190. [Google Scholar] [CrossRef]
- Meehan, T.F.; Conte, N.; Goldstein, T.; Inghirami, G.; Murakami, M.A.; Brabetz, S.; Gu, Z.; Wiser, J.A.; Dunn, P.; Begley, D.A.; et al. PDX-MI: Minimal Information for Patient-Derived Tumor Xenograft Models. Cancer Res. 2017, 77, e62–e66. [Google Scholar] [CrossRef]
- Stewart, E.L.; Mascaux, C.; Pham, N.A.; Sakashita, S.; Sykes, J.; Kim, L.; Yanagawa, N.; Allo, G.; Ishizawa, K.; Wang, D.; et al. Clinical Utility of Patient-Derived Xenografts to Determine Biomarkers of Prognosis and Map Resistance Pathways in EGFR-Mutant Lung Adenocarcinoma. J. Clin. Oncol. 2015, 33, 2472–2480. [Google Scholar] [CrossRef] [PubMed]
- Woo, X.Y.; Srivastava, A.; Graber, J.H.; Yadav, V.; Sarsani, V.K.; Simons, A.; Beane, G.; Grubb, S.; Ananda, G.; Liu, R.; et al. Genomic data analysis workflows for tumors from patient-derived xenografts (PDXs): Challenges and guidelines. BMC Med. Genom. 2019, 12, 92. [Google Scholar] [CrossRef] [PubMed]
- Jo, H.; Yagishita, S.; Hayashi, Y.; Ryu, S.; Suzuki, M.; Kohsaka, S.; Ueno, T.; Matsumoto, Y.; Horinouchi, H.; Ohe, Y.; et al. Comparative Study on the Efficacy and Exposure of Molecular Target Agents in Non-small Cell Lung Cancer PDX Models with Driver Genetic Alterations. Mol. Cancer Ther. 2022, 21, 359–370. [Google Scholar] [CrossRef]
- Rydberg Millrud, C.; Deronic, A.; Gronberg, C.; Jaensson Gyllenback, E.; von Wachenfeldt, K.; Forsberg, G.; Liberg, D. Blockade of IL-1alpha and IL-1beta signaling by the anti-IL1RAP antibody nadunolimab (CAN04) mediates synergistic anti-tumor efficacy with chemotherapy. Cancer Immunol. Immunother. 2023, 72, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Guan, J.; English, J.C.; Flint, J.; Yee, J.; Evans, K.; Murray, N.; Macaulay, C.; Ng, R.T.; Gout, P.W.; et al. Patient-derived first generation xenografts of non-small cell lung cancers: Promising tools for predicting drug responses for personalized chemotherapy. Clin. Cancer Res. 2010, 16, 1442–1451. [Google Scholar] [CrossRef]
- Wang, J.; Anderson, M.G.; Oleksijew, A.; Vaidya, K.S.; Boghaert, E.R.; Tucker, L.; Zhang, Q.; Han, E.K.; Palma, J.P.; Naumovski, L.; et al. ABBV-399, a c-Met Antibody-Drug Conjugate that Targets Both MET-Amplified and c-Met-Overexpressing Tumors, Irrespective of MET Pathway Dependence. Clin. Cancer Res. 2017, 23, 992–1000. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.C.; Zhang, J.; Li, M.; Huang, X.S.; Yang, X.N.; Zhong, W.Z.; Xie, L.; Zhang, L.; Zhou, M.; Gavine, P.; et al. Establishment of patient-derived non-small cell lung cancer xenograft models with genetic aberrations within EGFR, KRAS and FGFR1: Useful tools for preclinical studies of targeted therapies. J. Transl. Med. 2013, 11, 168. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Wei, S.; Li, Q.; Zeng, J.; Xiao, W.; Zhou, C.; Yoneda, K.Y.; Zeki, A.A.; Li, T. Simvastatin Overcomes Resistance to Tyrosine Kinase Inhibitors in Patient-derived, Oncogene-driven Lung Adenocarcinoma Models. Mol. Cancer Ther. 2024, 23, 700–710. [Google Scholar] [CrossRef]
- Roper, N.; El Meskini, R.; Maity, T.; Atkinson, D.; Day, A.; Pate, N.; Cultraro, C.M.; Pack, S.; Zgonc, V.; Weaver Ohler, Z.; et al. Functional Heterogeneity in MET Pathway Activation in PDX Models of Osimertinib-resistant EGFR-driven Lung Cancer. Cancer Res. Commun. 2024, 4, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Qin, B.; Jiao, X.; Yuan, L.; Liu, K.; Zang, Y. Advances in Patient Derived Tumor Xenograft (PDTX) Model from Lung Cancer. Zhongguo Fei Ai Za Zhi 2017, 20, 715–719. [Google Scholar]
- Lu, P.; Takai, K.; Weaver, V.M.; Werb, Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb. Perspect. Biol. 2011, 3, a005058. [Google Scholar] [CrossRef]
- Gao, H.; Korn, J.M.; Ferretti, S.; Monahan, J.E.; Wang, Y.; Singh, M.; Zhang, C.; Schnell, C.; Yang, G.; Zhang, Y.; et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 2015, 21, 1318–1325. [Google Scholar] [CrossRef]
- Ilie, M.; Nunes, M.; Blot, L.; Hofman, V.; Long-Mira, E.; Butori, C.; Selva, E.; Merino-Trigo, A.; Venissac, N.; Mouroux, J.; et al. Setting up a wide panel of patient-derived tumor xenografts of non-small cell lung cancer by improving the preanalytical steps. Cancer Med. 2015, 4, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Zhao, J.; Bai, H.; Duan, J.; Wang, Z.; An, T.; Wang, J. High-fidelity of non-small cell lung cancer xenograft models derived from bronchoscopy-guided biopsies. Thorac. Cancer 2016, 7, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.A.; Anderson, W.C.; Santaguida, M.T.; Dylla, S.J. Patient-derived xenografts, the cancer stem cell paradigm, and cancer pathobiology in the 21st century. Lab. Investig. 2013, 93, 970–982. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, G.J. Applications of patient-derived tumor xenograft models and tumor organoids. J. Hematol. Oncol. 2020, 13, 4. [Google Scholar] [CrossRef]
- Lin, S.; Huang, G.; Cheng, L.; Li, Z.; Xiao, Y.; Deng, Q.; Jiang, Y.; Li, B.; Lin, S.; Wang, S.; et al. Establishment of peripheral blood mononuclear cell-derived humanized lung cancer mouse models for studying efficacy of PD-L1/PD-1 targeted immunotherapy. MAbs 2018, 10, 1301–1311. [Google Scholar] [CrossRef]
- Shultz, L.D.; Brehm, M.A.; Garcia-Martinez, J.V.; Greiner, D.L. Humanized mice for immune system investigation: Progress, promise and challenges. Nat. Rev. Immunol. 2012, 12, 786–798. [Google Scholar] [CrossRef]
- Abdel-Aziz, A.K.; Saadeldin, M.K.; D’Amico, P.; Orecchioni, S.; Bertolini, F.; Curigliano, G.; Minucci, S. Preclinical models of breast cancer: Two-way shuttles for immune checkpoint inhibitors from and to patient bedside. Eur. J. Cancer 2019, 122, 22–41. [Google Scholar] [CrossRef]
- Brehm, M.A.e.a. Lack of acute xenogeneic graft- versus-host disease, but retention of T-cell function following engraftment of human peripheral blood mononuclear cells in NSG mice deficient in MHC class I and II expression. FASEB J. 2019, 33, 3137–3151. [Google Scholar] [CrossRef] [PubMed]
- Yanagawa, S.; Tahara, H.; Shirouzu, T.; Kawai, S.; Tanaka, Y.; Ide, K.; Akimoto, S.; Ohdan, H. Development of a humanized mouse model to analyze antibodies specific for human leukocyte antigen (HLA). PLoS ONE 2021, 16, e0236614. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Zheng, Z.; Cheng, T. New paradigms on hematopoietic stem cell differentiation. Protein Cell 2020, 11, 34–44. [Google Scholar] [CrossRef]
- Hess, N.J.; Lindner, P.N.; Vazquez, J.; Grindel, S.; Hudson, A.W.; Stanic, A.K.; Ikeda, A.; Hematti, P.; Gumperz, J.E. Different Human Immune Lineage Compositions Are Generated in Non-Conditioned NBSGW Mice Depending on HSPC Source. Front. Immunol. 2020, 11, 573406. [Google Scholar] [CrossRef]
- Verma, B.; Wesa, A. Establishment of Humanized Mice from Peripheral Blood Mononuclear Cells or Cord Blood CD34+ Hematopoietic Stem Cells for Immune-Oncology Studies Evaluating New Therapeutic Agents. Curr. Protoc. Pharmacol. 2020, 89, e77. [Google Scholar] [CrossRef] [PubMed]
- Pearson, T.; Greiner, D.L.; Shultz, L.D. Creation of “humanized” mice to study human immunity. Curr. Protoc. Immunol. 2008, 81, 15–21. [Google Scholar] [CrossRef]
- Hasgur, S.; Aryee, K.E.; Shultz, L.D.; Greiner, D.L.; Brehm, M.A. Generation of Immunodeficient Mice Bearing Human Immune Systems by the Engraftment of Hematopoietic Stem Cells. Methods Mol. Biol. 2016, 1438, 67–78. [Google Scholar] [PubMed]
- Brehm, M.A.; Cuthbert, A.; Yang, C.; Miller, D.M.; DiIorio, P.; Laning, J.; Burzenski, L.; Gott, B.; Foreman, O.; Kavirayani, A.; et al. Parameters for establishing humanized mouse models to study human immunity: Analysis of human hematopoietic stem cell engraftment in three immunodeficient strains of mice bearing the IL2rgamma(null) mutation. Clin. Immunol. 2010, 135, 84–98. [Google Scholar] [CrossRef]
- Lepus, C.M.; Gibson, T.F.; Gerber, S.A.; Kawikova, I.; Szczepanik, M.; Hossain, J.; Ablamunits, V.; Kirkiles-Smith, N.; Herold, K.C.; Donis, R.O.; et al. Comparison of human fetal liver, umbilical cord blood, and adult blood hematopoietic stem cell engraftment in NOD-scid/gammac−/−, Balb/c-Rag1−/−gammac−/−, and C.B-17-scid/bg immunodeficient mice. Hum. Immunol. 2009, 70, 790–802. [Google Scholar] [CrossRef]
- Mold, J.E.; Venkatasubrahmanyam, S.; Burt, T.D.; Michaelsson, J.; Rivera, J.M.; Galkina, S.A.; Weinberg, K.; Stoddart, C.A.; McCune, J.M. Fetal and adult hematopoietic stem cells give rise to distinct T cell lineages in humans. Science 2010, 330, 1695–1699. [Google Scholar] [CrossRef] [PubMed]
- Matas-Cespedes, A.; Brown, L.; Mahbubani, K.T.; Bareham, B.; Higgins, J.; Curran, M.; de Haan, L.; Lapointe, J.M.; Stebbings, R.; Saeb-Parsy, K. Use of human splenocytes in an innovative humanised mouse model for prediction of immunotherapy-induced cytokine release syndrome. Clin. Transl. Immunol. 2020, 9, e1202. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Beltran, W.F.; Claiborne, D.T.; Maldini, C.R.; Phelps, M.; Vrbanac, V.; Karpel, M.E.; Krupp, K.L.; Power, K.A.; Boutwell, C.L.; Balazs, A.B.; et al. Innate Immune Reconstitution in Humanized Bone Marrow-Liver-Thymus (HuBLT) Mice Governs Adaptive Cellular Immune Function and Responses to HIV-1 Infection. Front. Immunol. 2021, 12, 667393. [Google Scholar] [CrossRef]
- Hegde, P.S.; Chen, D.S. Top 10 Challenges in Cancer Immunotherapy. Immunity 2020, 52, 17–35. [Google Scholar] [CrossRef] [PubMed]

| Cell Line | Histology | Driver Oncogene(s) | Other Mutations | Reference |
|---|---|---|---|---|
| H1975 | Adenocarcinoma | EGFR L858R, T790M | PIK3CA, TP53 | [41] |
| H3255 | Adenocarcinoma | EGFR L858R | TP53 | [42,43] |
| HCC2935 | Adenocarcinoma | EGFR Exon19 del (E746-A750) | TP53, APC | [44] |
| HCC4006 | Adenocarcinoma | EGFR Exon 19 del (L747-E749) | TP53, PIK3CA | [45] |
| H1650 | Adenocarcinoma | EGFR Exon19 del (E746-A750), T790M | TP53 | [43] |
| HCC827 | Adenocarcinoma | EGFR Exon19 del (E746-A750) | TP53 | [43,46] |
| PC9 | Adenocarcinoma | EGFR Exon 19 del (E746-A750) | TP53 | [47] |
| H1573 | Adenocarcinoma | KRAS G12A, NRASQ61K | PTPN1, TP53 | [48] |
| H23 | Adenocarcinoma | KRAS G12C | TP53, ATM, STK11 | [49] |
| H460 | Large cell carcinoma | KRAS Q61H | STK11, PIK3CA, TP53 | [50] |
| A549 | Adenocarcinoma | KRAS G12S | STK11, TP53 | [41] |
| H2122 | Adenocarcinoma | EML4-ALK variant 3a/b | TP53 | [51] |
| H358 | Adenocarcinoma | KRAS G12C | CTNNB1, TP53 deletion | [49] |
| H1299 | Adenocarcinoma | N-RAS Q61K | TP53 deletion | [49,52] |
| H596 | Adenocarcinoma | MET exon 14 skipping | PIK3CA, RB1, TP53 | [53] |
| H522 | Adenocarcinoma | KRAS G12S | TP53 | [50] |
| H2228 | Adenocarcinoma | EML4-ALK fusion v3 ALK-PTPN3 | TP53 | [54] |
| H661 | Large cell carcinoma | ARHGAP35 K179* mutation | CDKN2A, LASP1, TP53 | [55] |
| H2126 | Adenocarcinoma | - | SMARCA4, TP53 | [56] |
| H1437 | Adenocarcinoma | - | TP53 | [56] |
| H1563 | Adenocarcinoma | - | CDKN2A | [56] |
| H661 | Large cell carcinoma | - | CDKN2A, LASP1, TP53 | [56] |
| H1770 | Carcinoma | - | TP53 | [57] |
| H2170 | Squamous cell carcinoma | - | RHOA, TP53 | [58] |
| H69PR | SCLC | - | PIK3X, TP53, RB1 | [59] |
| DMS235 | SCLC | - | [60] | |
| H2066 | SCLC | - | TP53 | [61] |
| COR-L279 | SCLC | - | EP300, TP53 | [62] |
| SHP-77 | SCLC | - | ABL1, KRAS, RAC1, TP53 | [63] |
| NCI-H727 | SCLC | - | PKD1L-TNS3 fusion, KRAS, TP53 | [64] |
| In Vitro Models | Advantages | Disadvantages | Applications |
|---|---|---|---|
| Human lung cancer cell line models | Inexpensive, scalable, and widely available. | Limited representation of parent tumor heterogeneity; no TME. | Study cancer at molecular and cellular levels. |
| Primary cell cultures | Closer to the patient genomic profile. | Low success rate in establishing patient-derived primary cell cultures. | Study cancer at molecular and cellular levels. |
| Conditionally reprogrammed cell | Remaining the original karyotypes. | No TME. | Study cancer at molecular and cellular levels. |
| Cancer spheroids and organoids | Highly mimic original histopathology of tumors, rapid and robust growth. | Expensive, lack of TME and tumor heterogeneity. | Study self-renewal, drug resistance, heterogeneity oncogenic transformation, and drug screening. |
| Co-culture system of patient-derived immune cells and patient-derived tumors | Mimic the TME. | Difficulty in reproducing results and interpreting results. | Study the interaction between immune system and tumor cell. |
| In Vivo Models | Advantages | Disadvantages | Applications | |
|---|---|---|---|---|
| Carcinogen-induced mouse models | Tumor formation time is similar to human tumor growth progress. | 1. Relatively long latency. 2. Uncontrollable experimental results. 3. Not popular anymore. | Study tumor morphology, histology, and molecular characteristics. | |
| Xenografts | CDX | Readily available, over 300 models, cost-effective, easy to manipulate genetically, and widely used for high-throughput screening of drugs. | 1. Limited representation of tumor heterogeneity and microenvironment; results may not always translate to clinical outcomes. 2. Except the subcutaneous implantation, other transplant methods are technically difficult and need special technology to monitor the tumor growth. | Best suited for initial drug screening and mechanistic studies involving genetic and molecular pathways. |
| PDX | Retain the genetic, histological, and morphological features of original patient’s tumors. | 1. Require immunocompromised mice, which is not suitable to evaluate immunotherapy. 2. Snapshot of patient tumor that cannot reflect the heterogeneous of whole tumor. 3. Orthotopic transplantation requires higher technical skill, has high cost, and needs in vivo imaging tool to monitor the tumor growth. 4. Takes 1–9 months to establish a model. 5. Mouse-derived cells gradually replaced human stromal cells. | Valuable for validating drug efficacy of chemotherapy and/or molecularly targeted therapy for individual patients and tailoring treatment plans for specific genetic profiles. | |
| GEMM | Mimic genetic mutations observed in human cancers, allowing for the study of tumor initiation, progression, and therapy resistance within an immune-competent context. | 1. Time-consuming and expensive to develop; can lack the full range of human tumor heterogeneity. 2. CRISPR/Cas9 system can evaluate off-target activity. | 1. Study the function of tumor gene mutations and mechanisms of drug resistance. 2. Evaluate the mutation effect of immunotherapy. | |
| Syngeneic models | 1. Genetically identical to the host, enabling studies for immunotherapy and drug toxicity within a fully functional immune system. 2. Cost-effective compared to humanized models. | 1. Tumor lines are typically murine in origin, which may not accurately reflect human tumor biology. 2. Adapt to mouse biology only, not sure if the outcome is suitable for the human immune system. | Useful for evaluating the interaction between tumor cells and immune cells, as well as testing immunotherapeutic agents. | |
| Humanized PDX models | 1. Hu-PBMC model is fast to grow. 2. Incorporate a functional human immune system, allowing for long-term research and low or miner GVHD rate. | 1. Complex and expensive to establish. 2. Variability in immune reconstitution can affect reproducibility (low reconstitution rates of NK and B cells). 3. Short study period due to mouse developing GVHD. | Ideal for studying ICIs, CAR-T cell therapies, ADC, and other immunomodulatory treatments. | |
| Patient-derived LCOs | 1. Offer a high success rate in establishing cultures that maintain the histopathological and genomic fidelity of primary tumors. 2. Enable rapid drug screening and correlate well with clinical outcomes. | Organoids may not fully replicate the TME, including interactions with the immune system and stroma. | Emerging as a crucial tool for personalized medicine, facilitating the testing of various treatment regimens and helping guide clinical decision-making. | |
| Database | Website | Information | Reference |
|---|---|---|---|
| Mouse Genome Database (MGD) | http://www.informatics.jax.org/ | Gene characterization, nomenclature, mapping, gene homologies among vertebrates, sequence links, phenotypes, allelic variants and mutants, and strain data | [162] |
| Gene Expression Database (GXD) | http://www.informatics.jax.org/expression.shtml | Gene expression information from the laboratory mouse | [163] |
| Mouse Models of Human Cancer database (MMHCdb) | http://tumor.informatics.jax.org/mtbwi/index.do | Spontaneous and induced tumors in mice including GEMM, PDX | [164] |
| Name | Method | Advantages | Disadvantages |
|---|---|---|---|
| Humanized PBMC (hu-PBMC) Mouse Models | Tail vein injection of hu-PBMCs, which include lymphocytes (T, B cells and NK cells), neutrophils, and monocytes |
|
|
| Humanized CD34+ (hu-HPSC) Mouse Models | Tail vein injection of hu-HPSCs, which include hematopoietic stem cells (HSC) and hematopoietic progenitor cells |
|
|
| Knock-in Humanized Mouse Models | Knock-in human gene to replace murine gene |
|
|
| Human Fetal Bone, Liver, and Thymic Tissue (BLT) Engraftment | Subcapsular injection of HLA-matched fetal thymus or other immune organs into kidney capsule; more intact TME |
|
|
| Spleen Mononuclear Cell (SPMC) Engraftment | Intraperitoneal injection of single cells from donor splenic tissue; More B cells and TM cells than PBMCs |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.-Z.; Kiss, Z.; Ma, W.; Liang, R.; Li, T. Preclinical Models for Functional Precision Lung Cancer Research. Cancers 2025, 17, 22. https://doi.org/10.3390/cancers17010022
Yu J-Z, Kiss Z, Ma W, Liang R, Li T. Preclinical Models for Functional Precision Lung Cancer Research. Cancers. 2025; 17(1):22. https://doi.org/10.3390/cancers17010022
Chicago/Turabian StyleYu, Jie-Zeng, Zsofia Kiss, Weijie Ma, Ruqiang Liang, and Tianhong Li. 2025. "Preclinical Models for Functional Precision Lung Cancer Research" Cancers 17, no. 1: 22. https://doi.org/10.3390/cancers17010022
APA StyleYu, J.-Z., Kiss, Z., Ma, W., Liang, R., & Li, T. (2025). Preclinical Models for Functional Precision Lung Cancer Research. Cancers, 17(1), 22. https://doi.org/10.3390/cancers17010022






